Summary
Interleukin-22 (IL-22) is highly induced in response to infections with a variety of pathogens and its main functions are considered to be tissue repair and host defense at mucosal surfaces. Here we show that IL-22 has a previously undiscovered role during infection in that its expression suppresses the intestinal microbiota and enhances the colonization of a pathogen. IL-22 induced the expression of antimicrobial proteins, including lipocalin-2 and calprotectin, which sequester metal ions from microbes. As Salmonella Typhimurium overcomes metal starvation by lipocalin-2 and calprotectin, IL-22 boosted this pathogen’s colonization of the inflamed intestine by suppressing commensal Enterobacteriaceae, which in the absence of IL-22 overgrew S. Typhimurium. Thus, IL-22 expression can tip the balance between pathogenic and commensal bacteria in favor of a pathogen. Taken together, IL-22 induction can be exploited by pathogens to suppress the growth of their closest competitors, thereby enhancing pathogen colonization of mucosal surfaces.
Introduction
The cytokine interleukin-22 (IL-22) has important functions in tissue regeneration and in the maintenance of the skin and the mucosal barrier (reviewed in Rutz et al., 2013). In the mucosa, IL-22 expression is primarily triggered in response to the interaction of microorganisms with antigen presenting cells (APCs). APCs in turn release many cytokines including IL-23, which engages the intraepithelial and lamina propria lymphocytes to produce cytokines like IL-17 and IL-22 (reviewed in Blaschitz and Raffatellu, 2010; Khader and Gopal, 2010; Liu et al., 2009). The major function of IL-17 is to orchestrate the recruitment of neutrophils to the site of infection via the induction of CXC chemokines, like CXCL1 and CXCL8, and via the enhancement of granulopoiesis, which explains its protective role during infection with a variety of pathogens (reviewed in Blaschitz and Raffatellu, 2010). Similar to IL-17, IL-22 is rapidly induced in the intestinal mucosa in response to IL-23, but also through activation of the aryl hydrocarbon receptor (reviewed in Esser et al., 2009). IL-22 shares sequence homology with IL-10 and it signals through a receptor complex, consisting of both the IL-22 receptor 1 and the IL-10 receptor 2 subunits, that is expressed almost exclusively in non-hematopoietic cells (reviewed in Sonnenberg et al., 2011). Several sources of IL-22 have been identified in the gut, including innate lymphoid cells (ILCs) and Th17 cells (reviewed in Sonnenberg et al., 2011). Akin to and often in synergy with IL-17, IL-22 has been shown to play a protective role during infection with some pathogens, including Klebsiella pneumoniae (Aujla et al., 2008), Citrobacter rodentium (Zheng et al., 2008), vancomycin-resistant Enterococcus (Kinnebrew et al., 2010), and Plasmodium chabaudi (Mastelic et al., 2012).
One of the mechanisms by which IL-22 is thought to enhance mucosal barrier function is through the induction of antimicrobial proteins in the mucosa (Aujla et al., 2008; Zheng et al., 2007; Zheng et al., 2008), a function that partly explains its role in the containment of commensals to the intestinal niche (Sonnenberg et al., 2012). Antimicrobial proteins that are upregulated by IL-22 in epithelial cells include: lipocalin-2, which binds to the siderophore enterochelin and limits iron availability in the gut (Fischbach et al., 2006; Raffatellu et al., 2009); the C-type lectins regenerating islet-derivative protein 3 beta and 3 gamma (Reg3β and Reg3γ), which control some components of the microbiota (Stelter et al., 2011); S100A8 and S100A9, two antimicrobial peptides that heterodimerize to form the antimicrobial protein calprotectin, which sequesters zinc and manganese from microbes (Corbin et al., 2008; Damo et al., 2013; Hayden et al., 2013; Liu et al., 2012). And yet, despite these epithelial antimicrobial defenses, many pathogens colonize mucosal surfaces and establish an infection; for example, IL-22 failed to reduce oral infection with Candida albicans and genital infection with Neisseria gonorrhoeae (Feinen and Russell, 2012; Kagami et al., 2010), although the basis for this was not investigated. At present, the mechanism(s) by which IL-22 provides protection against a subset of microorganisms is not well understood.
One of the most prolific examples of a successful mucosal pathogen is Salmonella enterica serovar Typhimurium (S. Typhimurium), which colonizes the human gastrointestinal tract and causes a severe inflammatory diarrhea (Hohmann, 2001). As with other pathogens, the mucosal response to S. Typhimurium is orchestrated by T cells that express IL-17 and IL-22 within a few hours of infection (Godinez et al., 2008). IL-17 is secreted by both CD4+ (termed “Th17” cells) and γδ T cells in the intestine (Godinez et al., 2009), where it promotes the recruitment of neutrophils and prevents the dissemination of S. Typhimurium to the reticuloendothelial system (Raffatellu et al., 2008). While this arm of the host response is beneficial to the host because it controls the dissemination of this pathogen, some components of the host response promote the colonization of S. Typhimurium as this pathogen thrives in the inflamed gut (Liu et al., 2012; Raffatellu et al., 2009; Winter et al., 2010). Furthermore, S. Typhimurium successfully outcompetes the microbiota, although the mechanistic basis for this is not completely understood. While the normal gut microbiota is largely constituted by obligate anaerobes of the phyla Bacteroidetes and Firmicutes, a recent study has suggested that intestinal inflammation suppresses the colonization of obligate anaerobes and enhances the growth of facultative anaerobes like Escherichia coli that can utilize host-derived nitrate for respiration (Winter et al., 2013). However, in mice infected with S. Typhimurium, we did not observe an overgrowth of commensal Enterobacteriaceae despite high levels of intestinal inflammation (Liu et al., 2012). Taken together, these studies suggest that there exists an as yet undiscovered mechanism that suppresses the growth of commensal Enterobacteriaceae in the inflamed gut while favoring the growth of S. Typhimurium.
During S. Typhimurium infection, Il22 is the most highly induced gene in the rhesus macaque ileum, where it is upregulated approximately 10,000 fold (Raffatellu et al., 2008). Moreover, Il22 is among the most highly induced genes in the cecum of S. Typhimurium-infected mice (Godinez et al., 2008). In our present study, we set out to determine the role of IL-22 during infection with S. Typhimurium and discovered a function for IL-22 as an important modulator of the balance between microbes in the inflamed gut.
Results
S. Typhimurium colonization of the inflamed gut is enhanced in mice expressing IL-22
Our previous studies indicated that IL-22 is among the highest upregulated genes in the intestinal mucosa during infection with S. Typhimurium in both rhesus macaques and streptomycin-treated mice (Godinez et al., 2008). To determine whether this cytokine plays a role during S. Typhimurium infection, we infected C57BL/6 wild-type (WT) and Il22−/− mice (Zheng et al., 2007) that were treated with streptomycin prior to infection to induce an inflammatory response in the cecum as previously described (colitis model; (Barthel et al., 2003; Raffatellu et al., 2009)). Using this model, we first investigated whether IL-22 has a protective effect during S. Typhimurium infection by reducing dissemination of S. Typhimurium to the reticuloendothelial system. To this end, we enumerated S. Typhimurium in the mesenteric lymph nodes and the spleen of WT and Il22−/− mice at 48, 72, and 96h post infection. We recovered similar numbers of S. Typhimurium in the Peyer’s patches, terminal ileum, mesenteric lymph nodes, and spleen in WT and Il22−/− mice at every time point analyzed (Fig. 1a and Fig. S1a–c). Similar results were obtained in the absence of streptomycin pre-treatment (data not shown). Therefore, in contrast to what has been shown for other bacterial infection models (Aujla et al., 2008; Zheng et al., 2008), our data suggested that IL-22 does not reduce the dissemination of S. Typhimurium to the reticuloendothelial system.
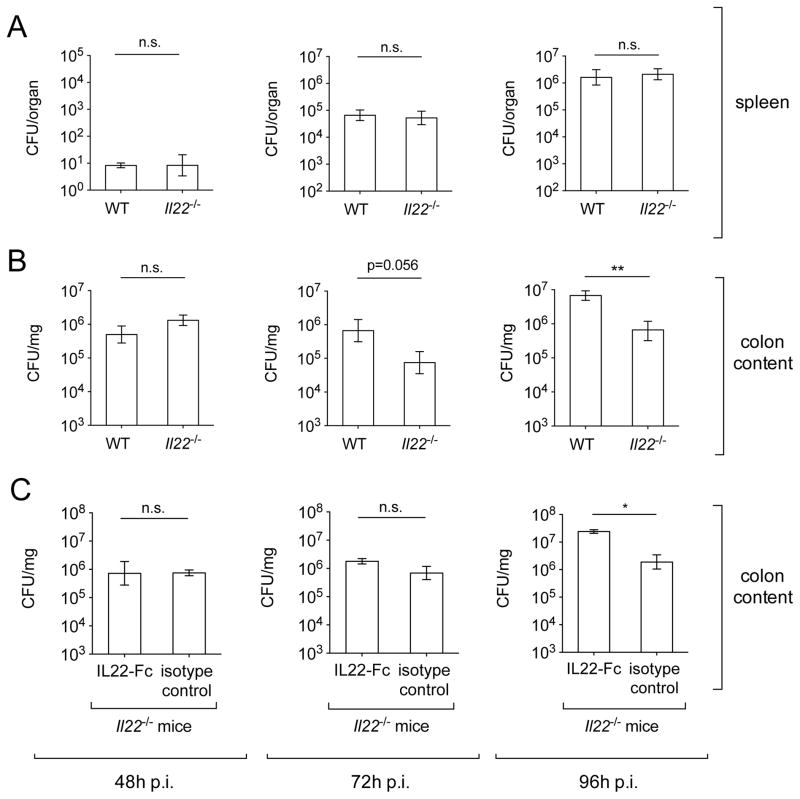
Figure 1. Colonization of S. Typhimurium in WT and Il22−/− mice.
WT and Il22−/− mice were infected with S. Typhimurium and colony forming units (CFUs) in spleen (A) and colon content (B) were determined at 48h (WT n=10, Il22−/− n=10), 72h (WT n=11, Il22−/− n=9) and 96h (WT n=11, Il22−/− n=10) after infection. (C) Il22−/− mice were administered IL22-Fc or an isotype control antibody (n=3/group) and fecal samples were collected at 48h, 72h and 96h after infection. Data shown represent CFUs per mg of fecal material for colon content and CFUs per organ for spleen. Data represent the geometric mean ± standard error. n.s.= not significant. A significant decrease over WT control is indicated by ** (P value ≤ 0.01). (See also Fig. S1 and Table S1).
Next, we examined whether IL-22 plays a role during S. Typhimurium colonization of the gut by enumerating S. Typhimurium from the colon content of infected WT and Il22−/− mice (Fig. 1b and S1d). When mice were infected with S. Typhimurium without streptomycin pre-treatment (typhoid model), S. Typhimurium numbers in the colon content were low and highly variable, and no significant differences between WT and Il22−/− mice were observed (Fig. S1d). These results are expected because S. Typhimurium does not trigger intestinal inflammation without streptomycin pre-treatment (Barthel et al., 2003), thus the role of IL-22 as well as of other inducible mucosal cytokines would not be apparent in the typhoid model of infection. We therefore measured the levels of S. Typhimurium colonization in the colon content of mice that were pre-treated with streptomycin. At 48h after infection, we found that both WT and Il22−/− mice were colonized to similar levels (Fig. 1b). However, S. Typhimurium numbers started to decline in the colon content of Il22−/− mice at 72h after infection (Fig. 1b), and we recovered approximately 90% less S. Typhimurium in the colon content of Il22−/− mice at 96h after infection (Fig. 1b). Moreover, administration of recombinant IL-22 protein (IL22-Fc) to Il22−/− mice increased the colonization of S. Typhimurium at 96h after infection to the level usually seen in WT mice (Fig. 1c). Taken together, our results suggest that S. Typhimurium colonization of the large intestine is less efficient in the absence of IL-22 in the colitis model.
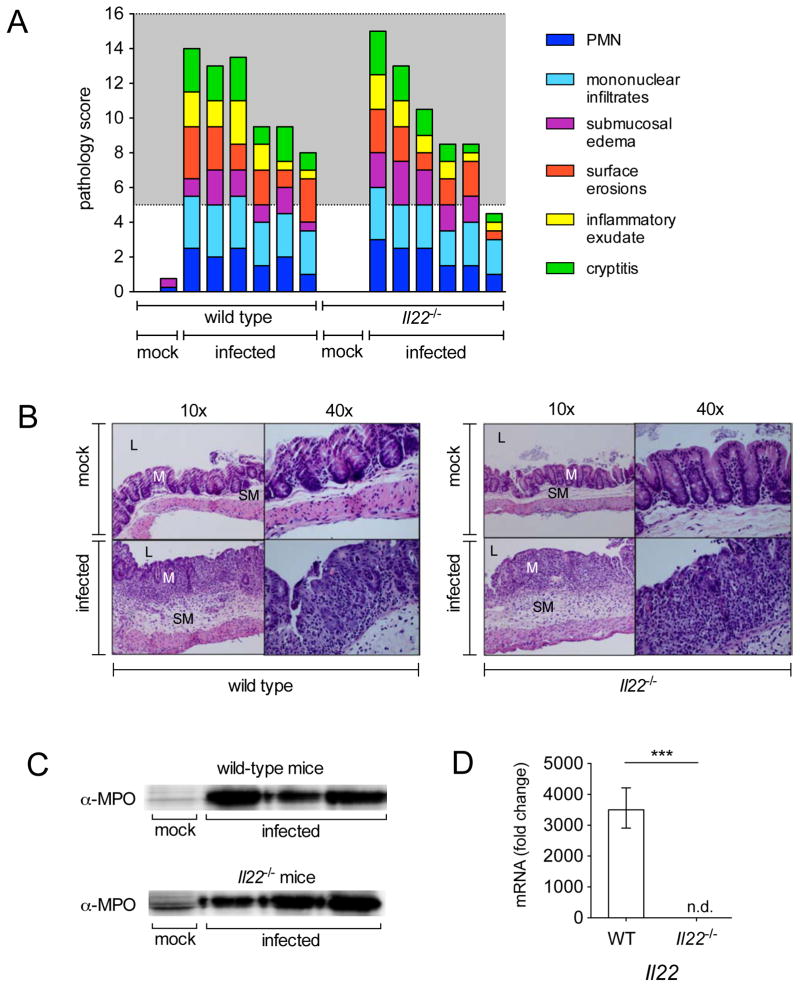
Because previous studies suggested that S. Typhimurium achieves high levels of colonization of the cecum only when this organ is inflamed, we next sought to investigate whether low levels of intestinal inflammation may explain the reduced S. Typhimurium colonization in Il22−/− mice. Histopathology indicated that both WT and Il22−/− mice developed moderate to severe inflammation when infected with S. Typhimurium, whereas mock-infected mice in both groups exhibited no abnormalities (Fig. 2). While we observed slightly less inflammation in infected Il22−/− mice in comparison to infected WT mice at 72h post infection (Fig. S2a and S2b), similar inflammation levels were detected in both groups at 96h after infection (Fig. 2a, b), i.e. at the same time we observed a significantly lower S. Typhimurium burden in Il22−/− mice (Fig. 1b). Moreover, we detected similar amounts of the neutrophil protein myeloperoxidase at 72h after infection (Fig. 2c), which was consistent with similar levels of neutrophil infiltrate that were observed by histopathology. These similarities were observed despite the fact that we detected high expression of Il22 in WT mice and could not detect Il22 in Il22−/− mice (Fig. 2d). Furthermore, we did not detect significant differences in the expression of inflammatory cytokines (gamma interferon (Ifnγ) and Il-17a (Il17a)) in WT and Il22−/− mice at both 72 and 96h after infection (Fig. S2c and S2d). The neutrophil chemoattractant Cxcl-1 (Cxcl1) was approximately 50% lower in infected Il22−/− mice at 72h post infection (Fig. S2c) in comparison to WT mice, but was detected at similar levels at 96h post infection (Fig. S2d); this is consistent with the notion that IL-22 induces the upregulation of Cxcl1 (Liu et al., 2009). Collectively, our results indicate that the lower levels of S. Typhimurium colonization observed in the cecum at 96 hours after infection in Il22−/− mice do not appear to be explained by differences in the levels of inflammation.
Figure 2. Histopathology of WT and Il22−/− mice after infection with S. Typhimurium.
(A) Blinded histopathology score indicating the score of individual mice 96h after either mock infection (treated with streptomycin but not infected) or infection with S. Typhimurium. The grey region includes scores indicative of moderate to severe inflammation. (B) H&E stained cecal sections from representative animals in each group. An image at lower magnification (10x) and one at higher magnification (40x) from the same section are shown. L=lumen; M=mucosa; SM=submucosa. Note marked edema in the submucosa and inflammation in mice infected with S. Typhimurium. (C) Myeloperoxidase (MPO) was detected 72h post infection by immunoblot in protein samples prepared from the cecum of mice that were mock infected or infected with S. Typhimurium. (D) Il22 was detected by quantitative real time PCR in the cecum of WT mice (n=6) and Il22−/− mice (n=6) 96h after infection with WT S. Typhimurium. A significant increase over mock control is indicated by *** (P value ≤ 0.001), n.d. = not detected. (See also Fig. S2 and Table S2).
IL-22 expression changes the relative abundance of Proteobacteria in the inflamed gut
Several studies have established that competition with the microbiota is essential for S. Typhimurium to colonize the inflamed gut (Barman et al., 2008; Lawley et al., 2008; Liu et al., 2012; Lupp et al., 2007; Stecher et al., 2007; Winter et al., 2010). Therefore, we hypothesized that differences in the microbiota may explain the lower S. Typhimurium burden in Il22−/− mice. To this end, we analyzed the microbiota composition in the colon content of WT and Il22−/− littermate mice before streptomycin treatment and after either mock infection or infection with S. Typhimurium (Fig. 3, Fig. S3 and table S3). The use of littermates for the analysis of the microbiota helped us to exclude any potential differences in the microbiota between WT and Il22−/− mice, as it has been shown that the baseline microbiota of Il22−/− mice may be more colitogenic and that it is transmissible to WT mice (Zenewicz et al., 2013). Illumina MiSeq analysis of DNA extracted from fecal samples confirmed previous observations that the major bacterial classes detected in animals before streptomycin treatment and in mock-infected animals at 96h post-infection were Clostridia and Bacteroidia, whereas Proteobacteria were low or undetectable (Fig. 3, Fig. S3a) (Barman et al., 2008; Lawley et al., 2008; Liu et al., 2012; Lupp et al., 2007; Stecher et al., 2007; Winter et al., 2010). A comparison of WT and Il22−/− mice showed no major differences in the composition of the microbiota, either before streptomycin treatment (baseline) or after mock infection. Five days after streptomycin treatment, the microbiota of both WT and Il22−/− mice was still constituted by Bacteroidia and Clostridia, although the relative abundance of Bacteroides was increased (Fig. 3, Fig. S3a). Groups of bacteria that are known to enhance resistance to bacterial infection were equally represented in both WT and Il22−/− mice. For instance, bacteria of the Porphyromonadaceae family have been associated with increased resistance to S. Typhimurium infection (Ferreira et al., 2011); however, in both WT and Il22−/− mice the relative abundance of the genus Parabacteroides (family Porphyromonadaceae) was similarly low before infection (0.1–0.7%) (Fig. 3, Fig. S3a). Likewise, our WT and Il22−/− mice were similarly colonized with varying levels of segmented filamentous bacteria (SFB) (Fig. S3b), Clostridia-related bacteria known to enhance resistance to Citrobacter rodentium infection through the induction of Th17 cells (Ivanov et al., 2009). Although SFB colonization levels were reduced by streptomycin treatment, no significant differences in SFB levels were observed between WT and Il22−/− mice (Fig. S3b). Altogether, our results suggest that differences in the microbiota prior to infection are unlikely to account for the higher numbers of S. Typhimurium colonization in WT mice.
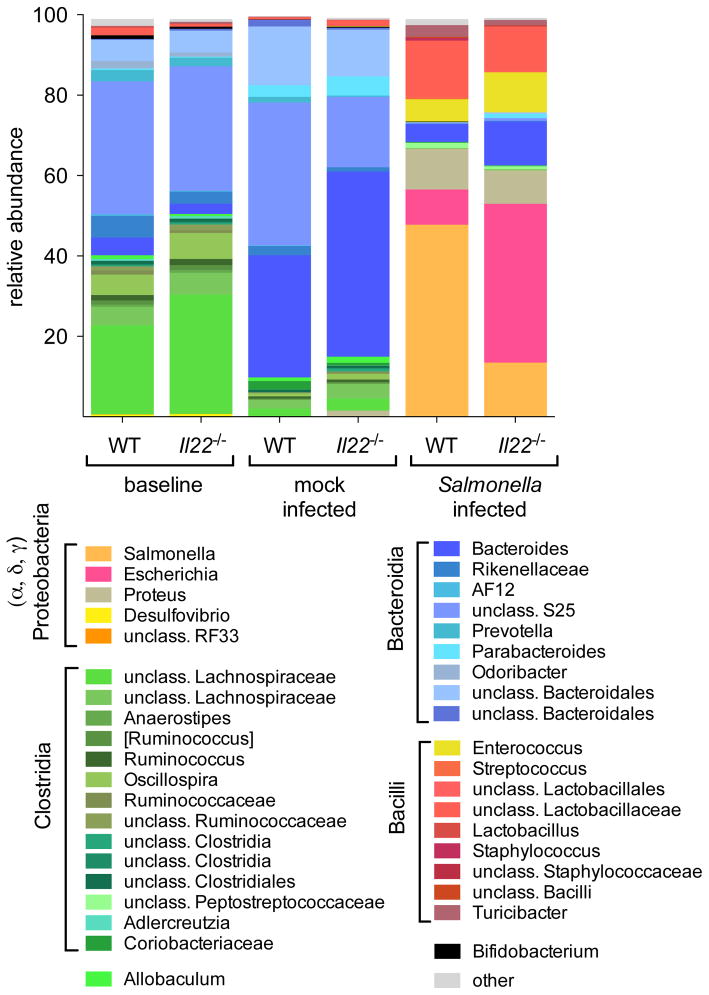
Figure 3. Analysis of colonic microbiota in WT and Il22−/− mice by sequencing.
Fecal samples were collected from mice before streptomycin treatment (n=9/group) and mock-infected animals (n=4/group) or S. Typhimurium-infected animals (n=5/group) at 96h post infection. The colonic microbiota was analyzed by sequencing using an Illumina MiSeq system. Graphed is the average relative abundance of each bacterial genus (See also Fig. S3 and Tables S3 and S4).
The most striking differences in the microbial communities were observed in mice that were infected with S. Typhimurium. As expected from prior studies (Barman et al., 2008; Lawley et al., 2008; Liu et al., 2012; Lupp et al., 2007; Stecher et al., 2007; Winter et al., 2010), Bacteroidia and Clostridia were substantially reduced in S. Typhimurium-infected mice compared to mock-infected mice (Fig. 3 and Fig. S3a). Furthermore, Proteobacteria bloomed in the inflamed gut of both WT and Il22−/− mice infected with S. Typhimurium, constituting approximately 70% of the microbiota. Indeed, the most prominent difference between WT and Il22−/− littermate mice infected with S. Typhimurium was the relative abundance of the genera Escherichia and Salmonella. While Salmonella constituted approximately 50% of the total bacteria in infected WT mice, it comprised 15% on average in infected Il22−/− mice. In contrast, Escherichia constituted the largest bacterial fraction in infected Il22−/− mice (on average, 40%) and a small fraction of bacteria in infected WT mice (on average, 9%) (Fig. 3 and Fig. S3a). These findings led us to hypothesize that, in the absence of IL-22, commensal Enterobacteriaceae can compete with S. Typhimurium in the inflamed gut.
IL-22 promotes S. Typhimurium competition with commensal Enterobacteriaceae
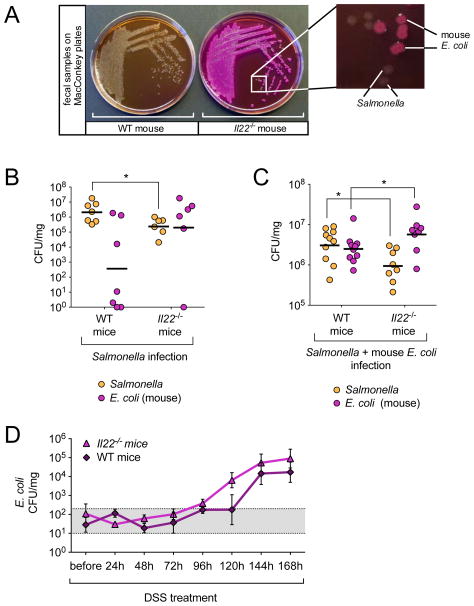
To test our hypothesis, we first sought to corroborate our microbiota sequencing data (Fig. 3) by streaking mouse fecal samples from WT and Il22−/− mice infected with S. Typhimurium on MacConkey-lactose agar, a selective and differential media commonly used in diagnostic laboratories to detect Enterobacteriaceae; enterobacterial strains that ferment lactose (e.g. Escherichia spp) form pink colonies while strains that do not ferment lactose (e.g. Salmonella spp) form colorless colonies. In agreement with our profiles, fecal samples from most WT mice infected with S. Typhimurium yielded only colorless colonies, which were determined to be S. Typhimurium (Fig. 4a). Also in line with our analysis, feces from Il22−/− mice infected with S. Typhimurium primarily yielded pink colonies, which were determined to be E. coli, although colorless S. Typhimurium colonies were also observed (Fig. 4a). Subsequent enumeration of fecal bacteria on MacConkey-lactose agar revealed that in WT mice, S. Typhimurium established high levels of colon colonization, effectively outcompeting the resident commensal E. coli in most mice by approximately 4 logs (Fig. 4b). In contrast, in Il22−/− mice S. Typhimurium was mostly outcompeted by the resident commensal E. coli (Fig. 4b), which explains the lower levels of S. Typhimurium colonization in the colon (Fig. 1b, 3, 4b). However, higher levels of E. coli colonization in Il22−/− mice were not due to differences in E. coli colonization between WT and Il22−/− mice prior to infection. Similar low numbers (approximately 102 CFU/mg) of E. coli were measured in the feces from both WT and Il22−/− mice prior to treatment with streptomycin and no E. coli were detectable after 24h (Fig. S4a and S4b). Moreover, analysis of multiple E. coli strains isolated from feces of WT and Il22−/− mice, both before and after infection with S. Typhimurium, revealed that all mice from our colony are colonized with the same E. coli strain; testing for serotype (O166, H6 or 41), colonization factors, and antibiotic susceptibility yielded the same results for all analyzed isolates (Fig. S4c and S4d). Therefore, differences in E. coli colonization between WT and Il22−/− mice infected with S. Typhimurium are not due to differences in the E. coli strains between groups.
Figure 4. Competition of S. Typhimurium with E. coli.
(A) Colon content samples collected from WT and Il22−/− mice at 96h post infection were resuspended in PBS and streaked on MacConkey agar plates for single colonies. Magnification shows the presence of lactose fermenting (pink) and lactose non-fermenting colonies in a sample from an Il22−/− mouse. (B+C) WT and Il22−/− mice were either (B) infected with S. Typhimurium (WT n=7, Il22−/− n=6) or (C) infected with S. Typhimurium and 24h later given 109 CFUs of mouse E. coli strain JB2 (WT n=10, Il22−/− n=8). Four days after infection with S. Typhimurium, colon content samples were collected and MacConkey plates were used to enumerate lactose fermenting (E. coli) and non lactose-fermenting (S. Typhimurium) colonies. Horizontal lines represent the geometric mean. (D) Fecal samples were collected from WT and Il22−/− mice before and every 24h after mice were given DSS in water. Samples were resuspended in PBS and plated on MacConkey agar plates to enumerate CFUs of E. coli (WT n=4, Il22−/− n=4). Gray area (101 to 3×102) indicates basal E. coli level in mice (See also Fig. S4).
Because our results suggested that E. coli spontaneously grew to higher amounts in Il22−/− mice than in WT mice infected with S. Typhimurium, we next tested whether the E. coli growth advantage was also seen when an equal high dose of the E. coli strain isolated in Fig. 4a (JB2) was administered to both WT and Il22−/− mice 24h post infection with S. Typhimurium. In this setting, E. coli also grew to higher numbers in Il22−/− mice than in WT mice (Fig. 4c). To determine whether the colonization advantage of E. coli in Il22−/− mice occurred independently of Salmonella infection, we induced colonic inflammation by treating WT mice and Il22−/− mice with dextran sodium sulfate (DSS). Before DSS treatment, mice were equally colonized with low numbers of E. coli (approximately 102 CFU/mg) and colonization levels did not increase up to 96h after DSS treatment (Fig. 4d). However, after the onset of intestinal inflammation, E. coli started to bloom in Il22−/− mice while remaining at baseline levels in WT mice (Fig. 4d). At later time points, E. coli also proliferated in WT mice, although at lower levels than in Il22−/− mice. Even though there was a trend towards higher E. coli colonization in Il22−/− mice, the difference did not reach statistical significance. This is probably due to differences in the magnitude and nature of inflammation caused by DSS and by S. Typhimurium. Altogether, these results demonstrate that IL-22 expression reduces the colonization of commensal Enterobacteriaceae, resulting in a colonization advantage for S. Typhimurium in the inflamed gut.
IL-22 induces the expression of antimicrobial proteins in the intestine of mice infected with S. Typhimurium
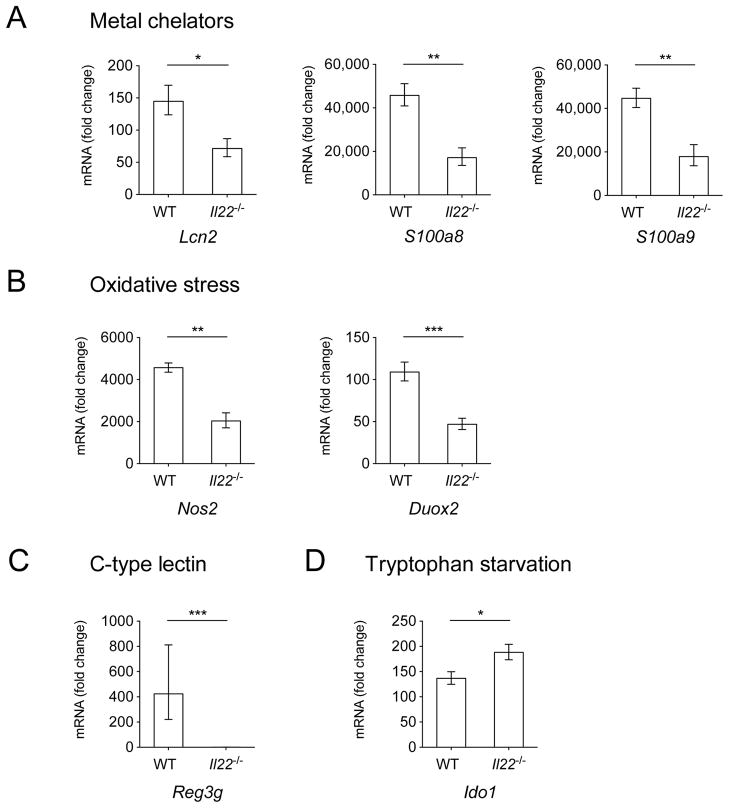
We next set out to determine the mechanism by which IL-22 provides a colonization advantage to S. Typhimurium during infection, thereby enabling it to effectively compete with commensal Enterobacteriaceae. Because IL-22 regulates antimicrobial responses, we examined whether the expression of antimicrobial genes during S. Typhimurium infection was also dependent on IL-22. To this end we analyzed the expression of: Lcn2, the gene that encodes for the antimicrobial peptide lipocalin-2, which sequesters the siderophore enterochelin and inhibits the growth of commensal Enterobacteriaceae (Flo et al., 2004); S100a8 and S100a9, which encode for the two subunits of calprotectin, an antimicrobial protein that sequesters zinc and manganese from pathogens (Corbin et al., 2008; Damo et al., 2013; Hayden et al., 2013; Liu et al., 2012); Nos2, which encodes for the inducible nitric oxide synthase (iNOS) (Mühl et al., 2011), as well as Duox2, which encodes for the dual oxidase 2 protein (Rada and Leto, 2008), each playing a role in the generation of reactive nitrogen species and reactive oxygen species, respectively; Reg3g, which encodes for regenerating islet-derivative protein 3 gamma (RegIIIγ), a C-type lectin which binds to peptidoglycan and inhibits the growth of Gram positive bacteria (Cash et al., 2006); and Ido1, which encodes for indoleamine 2,3-dioxygenase (IDO1), an antimicrobial protein that induces tryptophan starvation from microbes (Zelante et al., 2009).
Basal transcript expression of all antimicrobial genes analyzed was overall similar in mock-infected WT and Il22−/− mice, although we observed lower (approximately 1/8) expression of Reg3g and an 8–10 fold upregulation of Nos2 and Duox2 in mock-infected Il22−/− mice compared to mock-infected WT mice (Fig. S5). As shown in Figure 5, the expression of genes encoding for metal binding proteins (Lcn2, S100a8 and S100a9) (Fig. 5a), and those encoding for proteins involved in the generation of reactive oxygen and reactive nitrogen species (Nos2 and Duox2) (Fig. 5b), was significantly reduced in Il22−/− mice. Strikingly, Reg3g transcripts were upregulated approximately 400 fold in WT mice after S. Typhimurium infection, while they were nearly undetectable in Il22−/− mice (Fig. 5c). In contrast, expression of the Ido1 gene was increased in Il22−/− mice compared to WT mice (Fig. 5d). Overall, our results suggest that IL-22 contributes to the induction of the expression of antimicrobial host defense genes during infection with S. Typhimurium.
Figure 5. Expression of antimicrobial peptide genes.
(A) Lcn2, S100a8, S100a9, (B) Duox2, Nos2, (C) Reg3g, and (D) Ido1 were detected by quantitative real time PCR in the cecum of WT mice and Il22−/− mice 72h after infection with WT S. Typhimurium. Infected WT n=6, infected Il22−/− n=6, mock n=4. Data are expressed as fold increase over mock-infected WT mice. Data represent the geometric mean ± standard error. A significant increase over mock control is indicated by * (P value ≤ 0.05), ** (P value ≤ 0.01) and *** (P value ≤ 0.001). (See also Fig. S5).
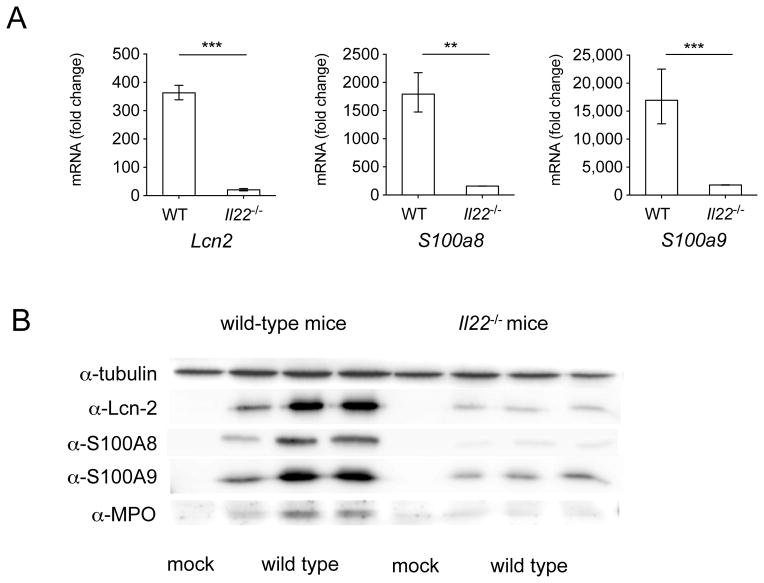
Our gene expression analysis was performed on RNA extracted from the whole cecum, however receptors for IL-22 are almost exclusively found on non-hematopoietic cells like colonocytes (Sonnenberg et al., 2011). Therefore, we set out to determine the effect of IL-22 on the induction of antimicrobial proteins expressed by colonocytes in vivo. To this end, we infected mice with either S. Typhimurium or mock and isolated crypt cells from the large intestine and cecum (Fig. 6). We found that the expression of Lcn2, S100a8 and S100a9 was significantly lower in colonocytes from Il22−/− mice compared to WT mice (Fig. 6a). Moreover, the corresponding proteins lipocalin-2, S100A8 and S100A9 were also produced significantly less in Il22−/− mice in comparison to WT mice (Fig. 6b). Similar differences in gene and protein expression were also observed at both 48 hours and 72 hours after S. Typhimurium infection (data not shown). Together, our results show that IL-22 promotes the expression of antimicrobial proteins in colonocytes during infection with S. Typhimurium.
Figure 6. Expression of antimicrobial peptide genes in colonic crypts.
(A) Lcn2, S100a8 and S100a9 were detected by quantitative real time PCR in isolated colonic crypts of WT mice and Il22−/− mice 48h after infection with WT S. Typhimurium. Infected WT n=6, infected Il22−/− n=6, mock n=2. Data are expressed as fold increase over mock-infected WT mice. Data represent the geometric mean ± standard error (for some conditions error marks are not visible due to small error). A significant increase over mock control is indicated by ** (P value ≤ 0.01) and *** (P value ≤ 0.001). (B) Lcn-2, S100A8, S100A9, myeloperoxidase (MPO), and tubulin were detected 48h post infection by immunoblot in isolated crypts of WT and Il22−/− mice that were mock-infected or infected with S. Typhimurium.
S. Typhimurium exploits IL-22-dependent host defense mechanisms
IL-22 induction of antimicrobial proteins is generally considered a mechanism of host defense against microbial infection, however this notion does not explain why S. Typhimurium colonization was increased when both IL-22 and antimicrobial proteins were expressed at high levels (Fig. 1, 3, 4, 5 and 6). One possible explanation may come from previous studies showing that S. Typhimurium is resistant to certain antimicrobial proteins (Liu et al., 2012; Raffatellu et al., 2009; Stelter et al., 2011). To test this hypothesis, we assessed whether IL-22 enhances the colonization of S. Typhimurium over isogenic mutant strains with known susceptibilities to antimicrobial proteins whose induction is dependent on IL-22. As shown in Figures 5 and 6, one of the antimicrobial proteins whose expression was reduced in Il22−/− mice is lipocalin-2, a peptide that sequesters the siderophore enterochelin and inhibits iron uptake by commensal Enterobacteriaceae including E. coli (Berger et al., 2006; Flo et al., 2004). In contrast to non-pathogenic commensals, S. Typhimurium overcomes this response by acquiring iron with the siderophore salmochelin (Crouch et al., 2008; Fischbach et al., 2006; Hantke et al., 2003). Mutants in the salmochelin receptor (iroN mutant), however, are not able to take up salmochelin and are thus susceptible to iron sequestration by lipocalin-2 in the inflamed gut (Raffatellu et al., 2009). A second antimicrobial protein whose induction was lower in Il22−/− mice is calprotectin (Fig. 5, 6). This heterodimer of the two EF-hand calcium-binding proteins S100A8 and S100A9 (Teigelkamp et al., 1991) chelates metal ions including zinc and manganese (Corbin et al., 2008), thereby exerting an antimicrobial effect against many bacteria including gut commensals (Kehl-Fie and Skaar, 2010; Sonnenberg et al., 2012). In contrast, S. Typhimurium overcomes calprotectin-mediated zinc sequestration and outgrows the microbiota by transporting zinc via the high affinity ZnuABC system (Liu et al., 2012). A mutant in this zinc transporter (znuA mutant) is susceptible to calprotectin-mediated zinc starvation and exhibits a growth defect in the inflamed gut (Liu et al., 2012). Based on this, we employed both an iroN mutant (lipocalin-2 sensitive) and a znuA mutant (calprotectin sensitive) to test whether IL-22 enhances the colonization of S. Typhimurium WT by inducing the expression of lipocalin-2 and calprotectin.
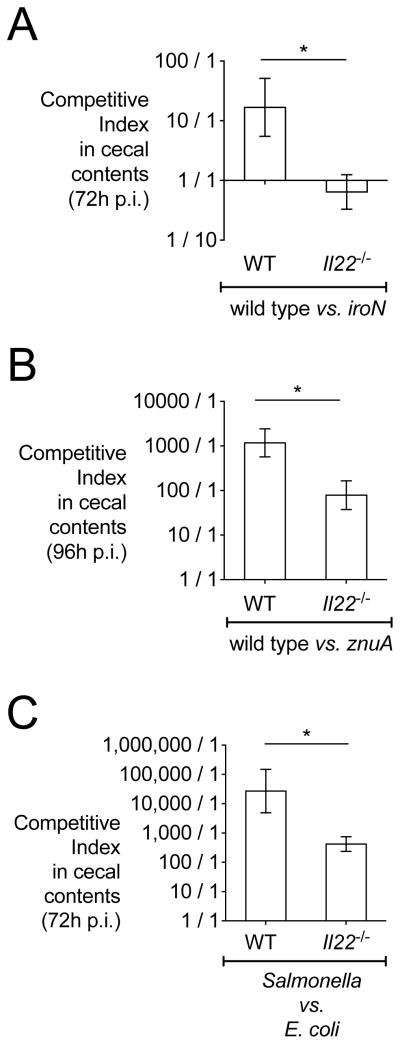
To this end, we infected both WT and Il22−/− mice with an equal mixture of S. Typhimurium WT and either the iroN or the znuA mutant (Fig. 7); this experimental setting ensured that high levels of inflammation were induced by infection with WT S. Typhimurium (data not shown). In WT mice, WT S. Typhimurium outcompeted the iroN mutant by 12 to 1 (Fig. 7a), consistent with our previously published data (Raffatellu et al., 2009). In contrast, the competitive advantage of WT S. Typhimurium was abrogated in Il22−/− mice, as both WT S. Typhimurium and the iroN mutant were recovered at similar levels (Fig. 7a). Of note, this outcome is comparable to what we had previously observed in Lcn2−/− mice (Raffatellu et al., 2009), which lack lipocalin-2, and it is consistent with lower levels of lipocalin-2 in Il22−/− mice (Fig. 5, 6). Similarly, we recovered approximately 1000 times more WT S. Typhimurium than znuA mutant in WT mice (Fig. 7b), which is comparable to our previous results (Liu et al., 2012). In contrast, the competitive advantage of WT S. Typhimurium was significantly diminished in Il22−/− mice and approximately 15-fold lower than in WT mice (Fig. 7b). To further examine the advantage that antimicrobial upregulation by IL-22 provides for S. Typhimurium, we tested if the observed lower amounts of antimicrobials in Il22−/− mice affected the competition between S. Typhimurium and a commensal E. coli strain (MG1655) with known susceptibility to lipocalin-2 (Berger et al., 2006; Flo et al., 2004). While in WT mice we recovered approximately 30,000 fold more S. Typhimurium than commensal E. coli, the growth advantage of S. Typhimurium was significantly reduced in Il22−/− mice, where we recovered only 400 fold more S. Typhimurium than E. coli (Fig. 7c). Collectively, our findings indicate that S. Typhimurium exploits IL-22-mediated host antimicrobial defenses to colonize the inflamed gut and to compete with the intestinal microbiota.
Figure 7. Competition in wild-type and Il22−/− mice of S. Typhimurium WT with strains of known sensitivity to antimicrobial proteins.
Colon content samples were collected from mice three or four days after infection with S. Typhimurium. Competitive index was calculated by dividing the output CFU ratio (WT/mutant or E. coli) by the input CFU ratio (WT/mutant or E. coli). Competitive indices of S. Typhimurium strains in the colon contents of WT and Il22−/− mice (n=5/group) infected with (A) an equal mixture of WT S. Typhimurium and the iroN mutant, (B) WT S. Typhimurium and the znuA mutant or (C) WT S. Typhimurium and E. coli. Data represent the geometric mean ± standard error. A significant decrease over the competitive index in WT mice is indicated by * (P value ≤ 0.05).
Discussion
IL-22 is an important cytokine for maintaining the skin and mucosal barrier as well as for tissue repair. Because IL-22 is highly upregulated during infection and because it orchestrates antimicrobial host defenses, it is generally thought that this cytokine has a broad protective function. However, while IL-22 expression has been shown to ameliorate and control infection with some pathogens (Aujla et al., 2008; Kinnebrew et al., 2010; Mastelic et al., 2012; Zheng et al., 2008), its induction was found to play no protective role in other infection models (Conti et al., 2009; Feinen and Russell, 2012; Graham et al., 2011; Kagami et al., 2010; Poulsen et al., 2012; Wilson et al., 2010). Therefore, these studies suggested that IL-22 induces antimicrobial responses that are not equally effective against all pathogens.
Here we investigated whether IL-22 plays a role during infection with S. Typhimurium, a highly evolved mucosal pathogen that establishes a successful intestinal infection despite high levels of IL-22 expression (Godinez et al., 2009; Raffatellu et al., 2008). Consistent with a previous report showing that S. Typhimurium colonization of the liver was comparable between WT and Il22−/− mice at 72 hours post infection (Awoniyi et al., 2012), we found that expression of IL-22 did not result in a reduced S. Typhimurium burden in mesenteric lymph nodes or spleen. Therefore, in contrast to what we observed for IL-17 with S. Typhimurium (Raffatellu et al., 2008), and similar to a subset of other pathogens, IL-22 did not appear to play a protective role during S. Typhimurium infection. Strikingly, however, we found that IL-22 not only fell short in protecting the host against S. Typhimurium dissemination, but its upregulation was actually beneficial to S. Typhimurium growth.
While the microbiota is largely comprised of anaerobes like Bacteroidetes and Firmicutes (mainly Clostridia) in the normal, non-inflamed intestine, intestinal inflammation provides a more favorable environment to facultative anaerobes like Enterobacteriaceae that can utilize nitrate respiration (Winter et al., 2013). Consistent with this, we also observed a bloom of Enterobacteriaceae (e.g. E. coli and Salmonella) in the inflamed gut of both WT and Il22−/− mice. But while in wild-type mice S. Typhimurium constituted a large fraction of the intestinal bacteria and the growth of commensal E. coli was largely suppressed, there was an overgrowth of commensal E. coli that outcompeted S. Typhimurium in the gut of mice lacking IL-22. Based on these results, we propose that IL-22 tips the balance in favor of Salmonella against commensal Enterobacteriaceae, and in particular E. coli, which are its closest competitors for a niche in the inflamed gut.
IL-22-mediated responses include the induction of several antimicrobial proteins by epithelial cells, which include the C-type lectins RegIIIγ and RegIIIβ, the psoriasin S100A7, the two subunits of calprotectin S100A8 and S100A9, β-defensins 2 and 3, and lipocalin-2 (Aujla et al., 2008; Conti et al., 2009; Kagami et al., 2010; Raffatellu et al., 2009; Zheng et al., 2008). In some cases, these antimicrobial proteins were shown to mediate the beneficial effects of IL-22 on mucosal barriers and skin by protecting the host against potentially dangerous microbes. This is, for instance, the case for vancomycin resistant Enterococcus, an opportunistic pathogen whose intestinal colonization is controlled by IL-22 through the induction of RegIIIγ (Kinnebrew et al., 2010). In agreement with this study, we also observed higher colonization of Enterococcus after infection with S. Typhimurium in the absence of IL-22 (Il22−/− mice) than in the presence of IL-22 (WT mice). Nevertheless, these antimicrobial proteins are not equally effective against all microorganisms, which may contribute to explaining why IL-22 has a protective role in only some infections. For example, RegIIIγ is bactericidal only against Gram positive organisms while it has no direct effect on Gram negatives; lipocalin-2 inhibits the growth of bacteria that rely on a specific subset of catecholate siderophores for iron acquisition, but does not inhibit pathogens that have acquired diverse additional iron transport systems; calprotectin has little effect against S. Typhimurium and likely other pathogens that have high affinity zinc transporters. While we found that IL-22 induced the expression of antimicrobial proteins during S. Typhimurium infection, we demonstrated that these responses are evaded by this pathogen with specific virulence mechanisms.
When iron and zinc availability are limited by lipocalin-2 and calprotectin, respectively, iron acquisition through salmochelin and zinc acquisition through the ZnuABC transporter greatly enhanced the competitive advantage of S. Typhimurium in the intestine of WT mice (here and (Liu et al., 2012; Raffatellu et al., 2009)). In Il22−/− mice, however, where the expression of both lipocalin-2 and calprotectin is reduced, the competitive advantage was diminished. Because both lipocalin-2 and calprotectin are components of the nutritional immune response that starves microorganisms from essential metal nutrients, our results also suggest that IL-22 is one of the key regulators of nutritional immunity. Furthermore, IL-22 may also benefit other mucosal pathogens by similar mechanisms, i.e. by inducing antimicrobial responses that suppress the growth of the microbiota, thereby enhancing their colonization.
Several studies to date have proposed that intestinal inflammation enhances the colonization of S. Typhimurium and its competition with the intestinal microbiota. Our study demonstrates that IL-22 is an important arm of the host response that enhances S. Typhimurium competition with the microbiota, and in particular with commensal Enterobacteriaceae, its closest relatives in the intestine. Because other species of the Enterobacteriaceae have adapted to colonize and thrive in the inflamed intestine (Winter et al., 2013), S. Typhimurium exploits IL-22 host defenses to control their growth.
While our findings demonstrate that IL-22 expression is beneficial to S. Typhimurium and suggest that other mucosal pathogens may compete for colonization via similar mechanisms, blockade of IL-22 during the course of infection would be detrimental to the host as it would result in poor control and dissemination of the microbiota, which is susceptible to IL-22-mediated antimicrobial responses. In light of this, specific targeting of virulence mechanisms that promote the evasion of IL-22-mediated host defenses is a more promising therapeutic strategy to reduce the intestinal colonization of mucosal pathogens resistant to the IL-22 response.
Materials and Methods
A supplementary file with supplementary materials and methods, five supplementary figures and four supplementary tables is included.
Bacterial Strains and Growth Conditions
IR715 is a fully virulent, nalidixic acid resistant derivative of S. Typhimurium wild-type isolate ATCC 14028 (Stojiljkovic et al., 1995). Mutant S. Typhimurium strains used in this study were a znuA deletion strain (Liu et al., 2012) and a strain deficient in iroN (Bäumler et al., 1998). Escherichia coli strains used in this study were E. coli MG1655 and JB2, an E. coli mouse isolate from this study (Fig. 4a). See table S1 for a list of strains used in this study. All strains were grown aerobically at 37°C in Luria-Bertani (LB) broth unless otherwise noted.
Mouse Experiments
C57BL/6 wild-type mice and Il22−/− mice were used. The construction of Il22−/− mice is described in the supplementary materials and methods of (Zheng et al., 2007). Mice were treated with streptomycin and mock-infected or infected with S. Typhimurium as previously described (Raffatellu et al., 2009). For some experiments, colitis was induced by dextran sodium sulfate as described in (Winter et al., 2013; Wirtz et al., 2007). To reconstitute IL-22 in Il22−/− mice, mice were administered recombinant IL-22 (IL22-Fc, Genentech PRO312045) every other day starting the day of streptomycin treatment. Control mice were administered an isotype control antibody to ragweed (Genentech 10D9.1E11.1F12). All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee at the University of California, Irvine.
Isolation of colon crypts
Streptomycin-treated C57BL/6 mice or Il22−/− mice were infected with S. Typhimurium or mock and sacrificed at 48 hours post infection. Crypt isolation from colon and cecum was performed as described (Whitehead et al., 1993).
Analysis of the microbiota
DNA from the colon content was extracted using the QIAamp DNA stool kit (Qiagen) according to the manufacturer’s instructions with modifications explained in supplemental methods. Bacterial DNA was amplified by a two-step PCR enrichment of the 16S rDNA (V4 region) encoding sequences from each sample using primers 515F and 806R modified by addition of barcodes for multiplexing. Libraries were sequenced using an Illumina MiSeq system. Uncalled bases, incorrect primer sequence, and runs of ≥12 identical nucleotides sequences were removed. Following quality filtering, the sequences were demultiplexed and trimmed before performing sequence alignments, identification of operational taxonomic units (OTU), clustering, and phylogenetic analysis using QIIME open source software (http://qiime.org).
Western blot
Total protein was extracted from mouse cecum using Tri-Reagent (Molecular Research Center), resolved by SDS-PAGE and transferred to a PVDF membrane. Detection of mouse tubulin was performed with primary rabbit polyclonal antibodies (Cell Signaling Technology) while detection of calprotectin was performed with polyclonal goat anti-mouse S100A8 and polyclonal goat anti-mouse S100A9 antibodies (R&D Systems). Lcn-2 was detected by polyclonal goat anti-mouse antibodies (R&D Systems) and myeloperoxidase was detected using a primary polyclonal goat anti-human and mouse antibody (R&D Systems). As secondary antibodies, a goat anti-rabbit or rabbit anti-goat conjugate to horseradish peroxidase (HRP) (Jackson) were used.
Quantitative real-time PCR
Total RNA was extracted from mouse cecal tissue using Tri-Reagent (Molecular Research Center). Reverse transcription of 1 μg of total RNA was performed using the Transcriptor First Strand cDNA Synthesis kit (Roche). Quantitative real-time PCR (qRT-PCR) for the expression of Actb, Il17a, Il22, S100a8, S100a9, Duox2, Nos2, Reg3g, Ido1, Cxcl1 and Ifng was performed with the primers described in Supplemental Methods.
Histopathology
Tissue samples were fixed in formalin, processed according to standard procedures for paraffin embedding, sectioned at 5 μm, and stained with hematoxylin and eosin. The pathology score of cecal samples was determined by blinded examinations of cecal sections from a board certified pathologist using previously published methods (Barthel et al., 2003; Raffatellu et al., 2009). Each section was evaluated for the presence of neutrophils, mononuclear infiltrate, submucosal edema, surface erosions, inflammatory exudates and cryptitis. Inflammatory changes were scored from 0 to 4 according to the following scale: 0=none; 1=low; 2=moderate; 3=high; 4=extreme. The inflammation score was calculated by adding up all of the scores obtained for each parameter and interpreted as follows: 0–2= within normal limit; 3–5= mild; 6–8=moderate; 8+=severe.
Statistical analysis
Differences between treatment groups were analyzed by ANOVA followed by Student’s t test. A P value equal to or below 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We would like to acknowledge Sean-Paul Nuccio for help with editing the manuscript and Ellena Peterson for help with strain identification. Furthermore we would like to thank Nita Salzman for providing the SFB plasmid, Matthew Rolston at the UC Davis School of Medicine Host-Microbe Systems Biology Core for processing samples for Illumina MiSeq analysis, and Janet Z. Liu for generating the artwork for our model. Work in MR lab is supported by Public Health Service Grant AI083619 and by funds from the Pacific Southwest Regional Center of Excellence for Biodefense and Emerging Infectious Disease (Award Number U54AI065359 from the National Institute of Allergy and Infectious Diseases). JB was supported by an American Heart Postdoctoral Fellowship (11POST7090006).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awoniyi M, Miller SI, Wilson CB, Hajjar AM, Smith KD. Homeostatic regulation of Salmonella-induced mucosal inflammation and injury by IL-23. PLoS One. 2012;7:e37311. doi: 10.1371/journal.pone.0037311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infection and immunity. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infection and immunity. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger T, Togawa A, Duncan GS, Elia AJ, You-Ten A, Wakeham A, Fong HE, Cheung CC, Mak TW. Lipocalin 2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2006;103:1834–1839. doi: 10.1073/pnas.0510847103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschitz C, Raffatellu M. Th17 cytokines and the gut mucosal barrier. J Clin Immunol. 2010;30:196–203. doi: 10.1007/s10875-010-9368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. 2009;206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- Crouch ML, Castor M, Karlinsey JE, Kalhorn T, Fang FC. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2008;67:971–983. doi: 10.1111/j.1365-2958.2007.06089.x. [DOI] [PubMed] [Google Scholar]
- Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, Zhang Y, Betz C, Hench L, Fritz G, et al. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci U S A. 2013;110:3841–3846. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C, Rannug A, Stockinger B. The aryl hydrocarbon receptor in immunity. Trends Immunol. 2009;30:447–454. doi: 10.1016/j.it.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Feinen B, Russell MW. Contrasting Roles of IL-22 and IL-17 in Murine Genital Tract Infection by Neisseria gonorrhoeae. Front Immunol. 2012;3:11. doi: 10.3389/fimmu.2012.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RB, Gill N, Willing BP, Antunes LC, Russell SL, Croxen MA, Finlay BB. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLoS One. 2011;6:e20338. doi: 10.1371/journal.pone.0020338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Lin H, Zhou L, Yu Y, Abergel RJ, Liu DR, Raymond KN, Wanner BL, Strong RK, Walsh CT, et al. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc Natl Acad Sci U S A. 2006;103:16502–16507. doi: 10.1073/pnas.0604636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- Godinez I, Haneda T, Raffatellu M, George MD, Paixao TA, Rolan HG, Santos RL, Dandekar S, Tsolis RM, Bäumler AJ. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infection and immunity. 2008;76:2008–2017. doi: 10.1128/IAI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinez I, Raffatellu M, Chu H, Paixao TA, Haneda T, Santos RL, Bevins CL, Tsolis RM, Bäumler AJ. Interleukin-23 orchestrates mucosal responses to Salmonella enterica serotype Typhimurium in the intestine. Infection and immunity. 2009;77:387–398. doi: 10.1128/IAI.00933-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AC, Carr KD, Sieve AN, Indramohan M, Break TJ, Berg RE. IL-22 production is regulated by IL-23 during Listeria monocytogenes infection but is not required for bacterial clearance or tissue protection. PLoS One. 2011;6:e17171. doi: 10.1371/journal.pone.0017171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K, Nicholson G, Rabsch W, Winkelmann G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci U S A. 2003;100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden JA, Brophy MB, Cunden LS, Nolan EM. High-affinity manganese coordination by human calprotectin is calcium-dependent and requires the histidine-rich site formed at the dimer interface. J Am Chem Soc. 2013;135:775–787. doi: 10.1021/ja3096416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann EL. Nontyphoidal salmonellosis. Clin Infect Dis. 2001;32:263–269. doi: 10.1086/318457. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami S, Rizzo HL, Kurtz SE, Miller LS, Blauvelt A. IL-23 and IL-17A, but not IL-12 and IL-22, are required for optimal skin host defense against Candida albicans. J Immunol. 2010;185:5453–5462. doi: 10.4049/jimmunol.1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khader SA, Gopal R. IL-17 in protective immunity to intracellular pathogens. Virulence. 2010;1:423–427. doi: 10.4161/viru.1.5.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infection and immunity. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe. 2012;11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Pezeshki M, Raffatellu M. Th17 cytokines and host-pathogen interactions at the mucosa: dichotomies of help and harm. Cytokine. 2009;48:156–160. doi: 10.1016/j.cyto.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Mastelic B, do Rosario AP, Veldhoen M, Renauld JC, Jarra W, Sponaas AM, Roetynck S, Stockinger B, Langhorne J. IL-22 Protects Against Liver Pathology and Lethality of an Experimental Blood-Stage Malaria Infection. Front Immunol. 2012;3:85. doi: 10.3389/fimmu.2012.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühl H, Bachmann M, Pfeilschifter J. Inducible NO synthase and antibacterial host defence in times of Th17/Th22/T22 immunity. Cell Microbiol. 2011;13:340–348. doi: 10.1111/j.1462-5822.2010.01559.x. [DOI] [PubMed] [Google Scholar]
- Poulsen KP, Faith NG, Steinberg H, Czuprynski CJ. Bacterial load and inflammation in fetal tissues is not dependent on IL-17a or IL-22 in 10–14 day pregnant mice infected with Listeria monocytogenes. Microb Pathog. 2012;56:47–52. doi: 10.1016/j.micpath.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada B, Leto TL. Oxidative innate immune defenses by Nox/Duox family NADPH oxidases. Contrib Microbiol. 2008;15:164–187. doi: 10.1159/000136357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, Godinez I, Sankaran S, Paixao TA, Gordon MA, et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz S, Eidenschenk C, Ouyang W. IL-22, not simply a Th17 cytokine. Immunol Rev. 2013;252:116–132. doi: 10.1111/imr.12027. [DOI] [PubMed] [Google Scholar]
- Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter C, Kappeli R, Konig C, Krah A, Hardt WD, Stecher B, Bumann D. Salmonella-Induced Mucosal Lectin RegIII beta Kills Competing Gut Microbiota. Plos One. 2011;6:e20749. doi: 10.1371/journal.pone.0020749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I, Bäumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S, Bhardwaj RS, Roth J, Meinardus-Hager G, Karas M, Sorg C. Calcium-dependent complex assembly of the myeloic differentiation proteins MRP-8 and MRP-14. J Biol Chem. 1991;266:13462–13467. [PubMed] [Google Scholar]
- Whitehead RH, VanEeden PE, Noble MD, Ataliotis P, Jat PS. Establishment of conditionally immortalized epithelial cell lines from both colon and small intestine of adult H-2Kb-tsA58 transgenic mice. Proc Natl Acad Sci U S A. 1993;90:587–591. doi: 10.1073/pnas.90.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MS, Feng CG, Barber DL, Yarovinsky F, Cheever AW, Sher A, Grigg M, Collins M, Fouser L, Wynn TA. Redundant and pathogenic roles for IL-22 in mycobacterial, protozoan, and helminth infections. J Immunol. 2010;184:4378–4390. doi: 10.4049/jimmunol.0903416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz S, Neufert C, Weigmann B, Neurath MF. Chemically induced mouse models of intestinal inflammation. Nat Protoc. 2007;2:541–546. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- Zelante T, Fallarino F, Bistoni F, Puccetti P, Romani L. Indoleamine 2,3-dioxygenase in infection: the paradox of an evasive strategy that benefits the host. Microbes Infect. 2009;11:133–141. doi: 10.1016/j.micinf.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Zenewicz LA, Yin X, Wang G, Elinav E, Hao L, Zhao L, Flavell RA. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol. 2013;190:5306–5312. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, Ouyang W. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.