Abstract
The replication and persistence of extra chromosomal Epstein-Barr virus (EBV) episome in latently infected cells are primarily dependent on the binding of EBV-encoded nuclear antigen 1 (EBNA1) to the cognate EBV oriP element. In continuation of the previous study, herein we characterized EBNA1 small molecule inhibitors (H20, H31) and their underlying inhibitory mechanisms. In silico docking analyses predicted that H20 fits into a pocket in the EBNA1 DNA binding domain (DBD). However, H20 did not significantly affect EBNA1 binding to its cognate sequence. A limited structure-relationship study of H20 identified a hydrophobic compound H31, as an EBNA1 inhibitor. An in vitro EBNA1 EMSA and in vivo EGFP-EBNA1 confocal microscopy analysis showed that H31 inhibited EBNA1-dependent oriP sequence-specific DNA binding activity, but not sequence-nonspecific chromosomal association. Consistent with this, H31 repressed the EBNA1-dependent transcription, replication, and persistence of an EBV oriP plasmid. Furthermore, H31 induced progressive loss of EBV episome. In addition, H31 selectively retarded the growth of EBV-infected LCL or Burkitt’s lymphoma cells. These data indicate that H31 inhibition of EBNA1-dependent DNA binding decreases transcription from and persistence of EBV episome in EBV-infected cells. These new compounds might be useful probes for dissecting EBNA1 functions in vitro and in vivo.
Keywords: EBNA1, small molecule, EBV, inhibitor, oriP, replication, persistence, DNA binding
Introduction
Epstein-Barr virus (EBV) usually first infects in human oropharyngeal epithelial cells (Rickinson and Kieff, 2007; Sixbey et al., 1983; Young et al., 1986). Subsequent infection of matured resting B lymphocytes can result in acute infectious mononucleosis and diverse B cell malignancies(Faulkner et al., 1999). In B cells with type III latency, EBV causes immortal proliferation of infected B lymphocytes through expression of EBV-encoded nuclear antigens (EBNA) (EBNA-LP, 1, 2, 3A, 3B, and 3C), two integral latent membrane proteins (LMP) (LMP1 and 2A), two small RNAs (EBER1 and 2), and Bam A rightward transcripts (BARTs), which encode multiple miRNAs (for review see (Kieff and Rickinson, 2007)). In the absence of effective T cell responses, infected B cell proliferation with latency III can become acute polyclonal lymphoproliferative diseases (LPD)(Rickinson and Kieff, 2007; Rivailler et al., 2004; Weiss and Movahed, 1989). EBV infected LPD, Burkitt’s lymphomas (BL), other lymphomas, and Hodgkin’s disease (HD) can emerge in people who have active T-cell immune function deficiencies due to HIV infection, transplantation or medications (Bashir et al., 1993; Bossolasco et al., 2001; MacMahon et al., 1991; Schang, 2005; Young et al., 1989). In immune competent people, Latency I or II EBV infection can cause fractions of lymphomas, Hodgkin’s disease, nasopharyngeal carcinoma (NPC) and gastric carcinoma. NPC is the most common EBV malignancy worldwide (Geser et al., 1982; Henle et al., 1978).
Upon primary infection of oropharyngeal epithelial cells with Epstein-Barr virus (EBV), the viral genomes become circularized through fusion of terminal repeats (TR), and they replicate and persist in episome by virtue of EBV-encoded nuclear antigen 1 (EBNA1) binding to a cognate sequence, oriP, which serves as a replication origin and cis-acting element for transactivation of EBV latent antigens (Rickinson and Kieff, 2007; Sixbey et al., 1983; Young et al., 1989). In latent infections, EBV usually persists as an extra chromosomal episome in multiple copies (Adams and Lindahl, 1975; Lindahl et al., 1974; Nonoyama and Pagano, 1972; Raynaud et al., 2005). EBV genome integration is unusual (Henderson et al., 1983; Hurley et al., 1990; Lawrence et al., 1989; Lawrence et al., 1988; Matsuo et al., 1984). It has been well documented that EBNA1 is essential for EBV episome persistence in dividing cells and that it enhances episome transcription (Rawlins et al., 1985; Reisman and Sugden, 1986; Sugden et al., 1985; Yates et al., 1985). The core EBNA1 DBD (a. a. 459–607) at its carboxyl terminus recognizes 20 copies of familial repeats (FR) and 4 copies of dyad symmetry (DS) in the EBV episome oriP element, while EBNA1 arginine-rich elements in residues 61–83 and 325–376 tether EBV episome to chromosomes for partitioning to progeny cell nuclei (Bochkarev et al., 1996; Hung et al., 2001; Rawlins et al., 1985; Reisman and Sugden, 1986; Sears et al., 2004; Wu et al., 2002; Yates et al., 1984; Yates et al., 1985). At least 3 EBNA1 domains are required for oriP interactions and are necessary for episome replication, enhanced episome transcription, or long-term episome persistence: arginine-glycine-rich domain (RG) 1, RG2, and the almost inseparable DD and oriP DBD (a. a. 459–607) (Ambinder et al., 1991; Bochkarev et al., 1996; Bochkarev et al., 1995; Bochkarev et al., 1998; Frappier et al., 1994; Goldsmith et al., 1993; Shah et al., 1992; Summers et al., 1996). EBNA1 RG1 and RG2 are critical for EBNA1’s effects on oriP-mediated transcriptional enhancement and episome persistence. Either RG-rich domain can mediate EBNA1 chromosomal association (Hung et al., 2001; Marechal et al., 1999; Sears et al., 2003), but both are necessary for wild-type EBNA1 chromosomal association (Hung et al., 2001; Sears et al., 2003). RG1 (also called linking region (LR1)) comprises AT-rich binding AT-Hook 1 (ATH1) (a. a. 40–65) and unique region 1 (UR1) (a. a. 66–89). UR1 mediates DD-independent dimerization and contributes to EBNA1-dependent transactivation (Aras et al., 2009).
Independent experiments demonstrated that as little as 70% specific inhibition of EBNA1 with dominant-negative EBNA1, EBNA1 antisense oligonucleotides, or EBNA1 RNAi is sufficient to eradicate EBV viral genomes or induce apoptosis of EBV-positive cells in as little as 6 days, validating EBNA1 as a reasonable target protein for the treatment of EBV-associated diseases (Hong et al., 2006; Imai et al., 2005; Nasimuzzaman et al., 2005; Roth et al., 1994; Yin and Flemington, 2006). The experimental evidence confirms that EBNA1 is essential for EBV episome persistence in dividing cells. Thus, a selective EBNA1 inhibitor could terminate latent EBV infection and abort the EBV-dependent growth of nonmalignant and malignant cells.
Several small molecules that indirectly or directly inhibit EBV have been developed. The growth of EBV-infected cells or EBV-induced primary B cell transformation in vitro was suppressed by these compounds, which include an Hsp90 inhibitor, BRACO-19, DS-binding pyrrole-imidazole polyamides, and Roscovitine (Li et al., 2010; Norseen et al., 2009; Sun et al., 2010; Thompson et al., 2010; Yasuda et al., 2011). The HSP90 inhibitor targeted the EBNA1 glycine-alanine repeats domain, decreased EBNA1 expression and translation, suppressed EBV-infected cell growth, and prevented EBV-induced B cell transformation (Sun et al., 2010). The G-quadruplex-interacting compound BRACO-19 inhibited EBNA1-dependent viral DNA replication and preferentially blocked the proliferation of EBV-positive cells relative to the proliferation of EBV-negative cell lines. BRACO-19 likely disrupts the ability of EBNA1 to tether to metaphase chromosomes (Norseen et al., 2009). EBNA1 inhibitors that directly bind to EBNA1 DBD were recently identified by high-throughput screen (Kang et al., 2008; Thompson et al., 2010). These inhibitors can fit into the EBNA1 DBD and inhibit EBNA1 DNA binding activity in vitro. Additionally, it was shown that a PARP-1 inhibitor enhances oriP-dependent plasmid maintenance (Tempera et al., 2010).
In the previous cell-based EBNA1 high-throughput screen in search of EBNA1-selective small molecule inhibitors, we identified compounds including Roscovitine, H20, and Shikonin. The inhibitory mechanism of Roscovitine was previously characterized, but the others remained uncharacterized (Kang et al., 2011). Roscovitine, a selective inhibitor of cyclin-dependent kinases, inhibits the CDK-directed phosphorylation of serine 393 in EBNA in vitro, resulting in the reduction of EBV episome and a selective growth retardation of EBV-positive cells (Kang et al., 2011). In another set of studies, we described a small molecule, EiK1, and short EBNA1 peptide 85 (P85), which can efficiently block EBNA1 dimerization (Kim et al., 2012). EiK1 is a small molecule compound that was also identified from the previous screen, and P85 is a short EBNA1 peptide that contains a. a. 560–566 of the β-sheet 3 strand of the EBNA1 dimeric interface. In the current study, we describe the functional characterization of H20 and its distant derivative H31 with respect to the underlying EBNA1 inhibitory mechanism. This study shows that (1) compound H20 binds to an EBNA1 core DBD (cDBD) (a. a. 459–607) (2) compound H31 strongly inhibits EBNA1 DNA binding activity, replication and persistence of EBV surrogate oriP replicons and EBV genomes, (3) H31 induces selective cell death of EBV-infected cells.
Materials and Methods
Plasmids, chemicals, and antibodies
The EBNA1-dependent reporters, oriPCp-FL (firefly luciferase)-SV40pPuro (OF), oriPCp-SEAP-SV40pPuro (OS), and CMVpEGFP-oriPmpFirefly luciferase-SV40pPuro (GOLP), and the EBNA1-independent reporters, pS0p-hRL (RL) (Renilla luciferase) (Promega, Madison, WI), were previously described (Kang et al., 2011). Plasmids for expressing EBNA1 DBD a. a. 379–641, 387–641, 459–641 and 459–607 in E. coli, all 6×His-tagged at the amino terminus were also previously described (Kang et al., 2011; Kim et al., 2012). Compounds H20 (Hit2lead ID 5889410), and H31 (Hit2lead ID 5551021) were purchased from vendors as described (Hit2lead) above, and others used in the SAR study were synthesized.
Cell lines and reporter assays
EBV-negative BJAB, BJAB-expressing FLAG-EBNA1 (BJ-FE1), BJAB-FE1 with stably integrated SV40p-RL (BJRL-FE1), BJRL-FE1 harboring episomal oriPCpFL-SV40p-Puro (OF) (BJRL-FE1-OF), BJ-FE1 harboring episomal oriPCpSEAP-SV40p-Puro (OS) (BJ-FE1-OS), EBV-infected AKATA (AKATAEBV(+)) cells, EBV-removed/negative AKATA cells (AKATAEBV(−)), EBV-transformed lymphoblastoid cell lines (LCL) LCL1022, LCL114, LCL SNU-265 and IB4 LCL were previously described (Hung et al., 2001; Kang et al., 2001; Kang et al., 2011; Zhao et al., 2006) (Hurley et al., 1991; Lee et al., 1998; Takada et al., 1991). For cell-based assays, equal numbers of BJRL-FE1-OF and BJ-FE1-OS cells were mixed. The BJRL-FE1-OF cells have an episomal oriPCpFL-SV40pPuro (OF) and integrated SV40pRL (RL). Cells were treated with a compound or DMSO control at various concentrations for indicated days. Reporter activities of FL and RL in cells used in HTS were sequentially measured, and secreted alkaline phosphatase (SEAP) activity was measured by a SEAP Reporter Assay Kit (Roche Applied Science, Indianapolis, IN). To rule out the possibility that compounds were nonspecific enzymatic FL or SEAP inhibitors, cell-free SEAP enzyme provided in the kit was reacted with the compound according to the protocol and as described. For the cell-free FL enzyme assay, Cell-Titer Glo luciferase solution containing the firefly luciferase enzyme and the luciferin substrate but lacking ATP was premixed with the 20 μM compound, incubated at RT for 1 hr, added to 106 BJAB cell extract as the ATP source, and assessed by luminometry. Data are the mean of triplicate experiments ± standard error. Reporter (%) numbers on the Y-axis are values relative to DMSO-treated (H20-untreated) cells. For the viability assay, Cell-Titer Glo luciferase solution was used (Promega, Madison, WI).
Biacore surface plasmon resonance (SPR)
6×His-EBNA1 a. a. 387–641 and a. a. 459–607, purified from E. coli BL21 (DE3) Rosetta pLysS cells as previously described (Kang et al., 2011), were used for the SPR study (Kim et al., 2012). Each Biacore CM5 sensor chip had 2 flow cells. EBNA1 a. a. 387–641 or a. a. 459–607 was immobilized on the sensor chip using 1 of 2 methods: through primary amine groups by activating the sensor surface with EDC/NHS chemistry or through free thiol groups by activating the sensor surface with maleimide groups. To test whether a compound bound to EBNA1 and whether it competed with FR for EBNA1 binding, H20 compound was flowed at a rate of 30 μL/min for 180 seconds for association and then for 180 sec for dissociation over immobilized protein in TBS-P/2% DMSO running buffer (25 mM Tris-HCl, 137 mM NaCl, 3 mM KCl, 0.005% P20 surfactant, 2% DMSO, pH 7.4), with or without 1× FR in the running buffer. To determine if H20 can compete with FR for EBNA1 binding, a competition binding experiment was set up to investigate the effect of H20 on EBNA1’s binding to the FR DNA sequence. A 50 nM FR solution was prepared in running buffer and aliquoted into 2 parts. H20 was added to one aliquot of 1× FR to a concentration of 2.5 μM. The H20/FR mixture was prepared by2-fold dilution of the 2.5 μM sample batch five times with running buffer containing FR, producing a dilution series of H20 containing 50 nM FR. These samples were tested for binding to EBNA1 a. a. 387–641. In addition, several 50 nM FR samples with no H20 were tested, and a sample with running buffer only was tested. In the second set of samples, a dilution series of H20 without FR was tested for binding. The first set of graphs was created by subtracting the binding curve with no FR or H20 from all curves of the FR/H20 mixture and the curve of FR alone. The graphs were then created by again subtracting FR-only binding curves from the FR/H20 binding curves. The parameters for the assay described above were as follows: association for 180 seconds, dissociation for 180 seconds, flow rate of 30 mL/min, regeneration for 30 seconds with 4 M NaCl pH 9.5 when needed, and compound dilution by 2-fold in duplicate experiments. Data were analyzed using Scrubber and Clamp XP software, available from the University of Utah. Many of the graphs in this report have been “normalized.” Normalization of the data involves transforming the Y-axis so that the theoretical maximum amount of binding for a 1:1 interaction with the protein surface corresponds to a sensor response of 100 RU. The actual calculation is as follows: Normalized Response = Raw Response/RUprotein, bound × Mwprotein/Mwsmall molecule × 100.
In silico docking of compounds to 3D EBNA1 structure
To determine whether compounds in this study can fit into EBNA1, atomic structures of the EBNA1 DNA binding domain resolved to 2.5 Å and 2.2 Å, respectively, with or without cognate DNA (PDB 1VHI, 1B3T), were used for the virtual in silico docking study (Bochkarev et al., 1995; Bochkarev et al., 1998; Li et al., 2010). Because GLIDE had the least number of inaccurate poses and 85% of GLIDE’s binding models had an RMS deviation of 1.4 Å or less from the native co-crystallized structure, the GLIDE program was employed as the primary docking protocol (Kontoyianni et al., 2004). Both native dimer (1VHI) and dissociated monomer from the dimer (1B3T) were challenged for this virtual docking.
Structure activity relationship (SAR) study
A study of structure and activity relationships for H20 was performed with a set of 33 structurally related compounds (see Table 1) by employing EBNA1-dependent transactivation and DNA-binding EMSA assays.
Table 1.
The structure and activity relationship study of H20 identifies H31 as an EBNA1 inhibitory compound for EBNA1 biochemical assays
| ID | R1 | R3 | R4 | Inhibitory Conc.(μM) |
||
|---|---|---|---|---|---|---|
| EMSA1 | Reporter2 | SPR3 | ||||
| H20 | CH3 | OH | OH | 100 | 0.63 | 0.25 |
| H22 | H | H | OH | >20 | >20 | ND |
| H23 | CH2CH3 | OH | H | >20 | >20 | ND |
| H24 | CH3 | OH | OCH2CH3 | 100 | >20 | ND |
| H25 | CH3 | H | OCH2CH3 | 10 | >20 | <30 |
| H26 | CH2CH3 | OCH3 | OH | >50 | >20 | ND |
| H27 | CYCLOHEXYL | OH | OH | 50 | >20 | >20 |
| H28 | CH3 | OH | OCH2CH3 | >50 | >20 | ND |
| H29 | H | OH | OCH2CH3 | >50 | >20 | ND |
| H30 | CYCLOHEXYL | OH | OCH2CH3 | 10 | >20 | ND |
| H31 | CH2CH2CH2CONH_C10H6(ADM) | H | OCH3 | 10 | 10 | >20 |
| H33 | CYCLOHEXYL | OCH2CH3 | OH | 10 | >20 | >20 |
| H34 | CH2CH2CH2COOH | H | OCH3 | >100 | ND | ND |
| H36 | CH2CH2CH2CONH2 | H | H | >100 | ND | ND |
| H37 | CH2CH2CH2COOH | OH | H | >100 | ND | ND |
| H43 | CH2CH2CH2CONHC_C10H6(ADM)-OH | OCH3 | H | >50 | >10 | ND |
| H45 | CH2CH2CH2COOH | OCH3 | H | >50 | >10 | ND |
| H46 | CH2CH3COOCH2CH2NCH3CH3 | H | CH3 | >100 | >20 | ND |
| H47 | CH2CH3COOH | H | CH2CH3 | >50 | >20 | ND |
EMSA, in vitro Electro Mobility Supershift Assay EBNA1 DNA binding assay
Reporter, in vivo EBNA1-dependent OriPCp-FL assay
SPR, in vitro Surface Plasmon Resonance Biacore assay
ND, Not done
Replication and persistence assays of oriP replicons
To determine whether H31 affects EBNA1-dependent oriP plasmid replication or replication-mediated persistence, BJAB or BJAB-FE1 cells were transiently transfected with 25 μg pCMVpEGFP-oriPmpFirefly luciferase-SV40pPuro (GOLP), in which replication is augmented by EBNA1 binding to the oriP element. The transfected cells were divided into 3 flasks and treated with compounds at indicated concentrations (0, 1, 5 μM). At 3 or 6 days post-transfection, episomal plasmid DNA was isolated by the Hirt plasmid method (Hirt, 1967), linearized with BglII, and digested with DpnI to eliminate unreplicated and non-persistent oriP plasmids. The digested DNAs were separated on an agarose gel, blotted onto a nylon membrane, and probed with 32P-labeled GOLP DNA by Southern hybridization. The relative efficiency of replication or persistence was determined using a Phosphoimager (Molecular Dynamics, Sunnyvale, CA). To further substantiate the inhibitory effect of the compound on EBV genome persistence, RAJI Burkitt’s lymphoma cells were treated with the compound at 10 μM for 14 days or 30 days. EBV genome persistence was determined by EBER in situ hybridization (Gulley et al., 2002) and real-time qPCR as previously described (Lestou et al., 1996) using primer sets of 5′-AGTCCTTCTT GGCTACTCTG-3′ and 5′-GAGGATCCGC GCCAAAG-3′ for the EBV polymerase EBV genome and 5′-GGCAACCCTA AGGTGAAGGC-3′ and 5′-TAGTGATGGC CTGGCTCACC-3′ for human GAPDH.
EBNA1 electro mobility supershift assay (EBNA1-EMSA)
Aliquots of 20 ng purified EBNA1 a. a. 387–641 or a. a. 459–607 (not shown) were mixed with compound H31 in 1× gel shift buffer (20 mM Tris, pH 7.5, 10% glycerol, 50 mM KCl, 10 mM MgCl2, 1 mM EDTA, 0.5 μg Poly(dI-dC). In the lanes specified, an excess of unlabeled cold wild-type (WT), mutant (MT) FR probe (Kang et al., 2005), or PBS (−) was added as a competitor. The monoclonal antibody to EBNA1 (α-E1) (Advanced Biotechnologies Inc., Columbia, MD) or isotype control antibody (IgG) was used to super-shift the EBNA1-probe complex. Control compounds (DMSO, puromycin (PU)) or H31 was added at 100, 50, 25, 10, and 5 μM, and the reactions were incubated at room temperature (RT) for 15 min. Subsequently, γ-32P-labeled probe was added to the reaction, which was incubated for another 15 min (Kang et al., 2005). An EMSA for GST-tagged RBP-Jκ was employed for control experiment. The reactions were stopped with 5× DNA gel loading buffer and separated on a 5% acrylamide gel. The gel was dried and exposed to a Phosphoimaging plate (Molecular Dynamics, Sunnyvale, CA).
EBNA1 association assay with metaphase chromosomes
To determine whether EBNA1 association with metaphase chromosomes is affected by compound H31, the effect of H31 on the subcellular localization of EGFP-EBNA1 fusion protein was investigated as described (Hung et al., 2001). Twenty millions of BJAB cells were transiently transfected with plasmids encoding either 25 mg EGFP-EBNA1 (GE1) or EGFP-C1 vector control (Clontech), and treated with DMSO or H31 at indicated concentration for 2 days. Treated cells were harvested, washed with PBS, swollen in a hypotonic buffer (1% trisodium citrate/0.5 mM CaCl2/0.5 mM MgCl2), quickly air-dried onto a microscopic slides, fixed with 4% formaldehyde in PBS for 30 min, permeabilized in 1% Triton X-100 in PBS, stained with 1 μg/ml propidium iodide (PI), 0.1 mg/ml RNase A in PBS for 30 min, and mounted with an antifade. Confocal microscopy used a Zeiss Axioskop microscope fitted with Nikon PCM-2000 confocal imaging hardware, Ar and HeNe laser sources, and C-IMAGING software (Compix, Cranberry Township, PA).
In situ hybridization
In situ hybridization for EBV-encoded small nuclear RNA probe (EBER) (PB0589) (Leica Biosystems, Newcastle, UK) were performed under manufacture’s protocols.
Results
H20 binds to core DBD of EBNA1 a. a. 459–607
In the previous cell-based EBNA1 high-throughput screen in search of compounds that selectively inhibit transactivation of EBNA1-dependent, oriP-enhanced firefly luciferase (FL), but not EBNA1-independent, SV40p-driven Renilla luciferase (RL) we described Roscovitine, Shikonin and H20 for an EBNA1 inhibitor. While the inhibitory mechanism of Roscovitine was previously characterized, H20 inhibitory mechanisms remained uncharacterized (Kang et al., 2011). In the present study, we further characterized the inhibitory activity of H20 (Fig. 1A) on EBNA1-dependent oriP-mediated transcription. H20 inhibited EBNA1-dependent, oriP-mediated firefly luciferase (FL) and SEAP reporter transcription by over 80% on average, but not EBNA1-independent, SV40 promoter-driven FL in cells (Fig. S1 A, left; in vivo reporter assay). The possibility of the compound being a nonspecific enzymatic FL or SEAP inhibitors was ruled out in the subsequent cell-free SEAP and FL enzyme assays, in which cell-free FL or SEAP activity was not affected by H20 treatment (Fig. S1 A, right), demonstrating that H20 itself was not a nonspecific inhibitor of SEAP and FL.
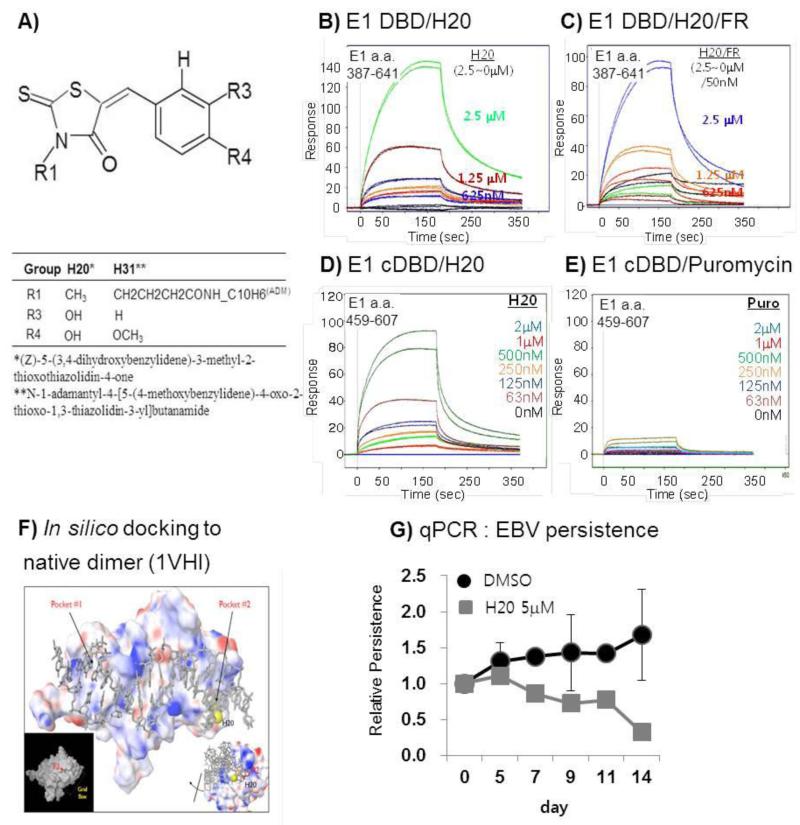
Fig. 1.
H20 bound to EBNA1 core DBD. A) Structure and residue information of H20 and derivative H31. (See also supplementary information Table 1). B) Association and dissociation curves of various H20 concentrations diluted 2-fold from 2 μM to 62.5 nM over immobilized EBNA1. H20 binds to EBNA1 DBD a. a. 387–641 in Biacore surface plasmon resonance assays. C) H20 does not affect FR binding to EBNA1 DBD. D) H20 binds to EBNA1 core DBD (cDBD) a. a. 459-607. E) Lack of puromycin binding to EBNA1 cDBD. F) in silico docking of H20 to a pocket of EBNA1 DBD a. a. 469-607 native monomer (PDB 1VH1) and dissociated monomer crystallized in complex with cognate DNA (PDB 1B3T) (see also Fig. S1 C). G) Decrease in EBV copy number in Raji in response to treatment with 5 μM H31 for 14 days was determined by real-time quantitative PCR for oriP normalized to control GAPDH. The data are the mean with standard errors from triplicate independent experiments. . In SPR, the results are representative of 5 independent SPR experiments. The Y-axis of all graphs has been normalized such that 100 response units is the theoretical maximum response for a 1:1 interaction.
To determine whether the selected H20 compound binds directly to the EBNA1 DBD, an essential domain for EBNA1 functions including transactivation, replication, and persistence of oriP-linked genomes in EBV-latent infection, a surface plasmon resonance Biacore assay was conducted. Compound H20 exhibited binding to EBNA1 in the hundred nanomolar range. H20 associated with EBNA1 DBD (a. a. 387-641) and core DBD (a. a. 459–607) in a dose-dependent manner, reaching equilibrium at concentrations less than 2.5 μM and quickly dissociating in the washing dissociation period (Fig. 1 B, C). In control SPR using puromycin that was previously known not to affect EBNA1-dependent transcription and DNA binding activity (Kang et al., 2011; Kim et al., 2012) and structurally unrelated to H20 (see the structure in Fig. S1 B), the puromycin compound did not bind to EBNA1 cDBD (Fig. 1 E). Despite the incomplete dissociations between sample injections produced less than ideal curves, the KD of H20 to EBNA1 was approximately 450 nM, although the analysis of the kinetic association and dissociation curves did not suggest the typical 1:1 Langmuir binding model. In addition, a competition binding experiment was set up to see if H20 competed with FR for EBNA1 binding in SPR (Fig. 1 D). The H20 binding curve in the presence of FR was very similar to the H20-only binding curves (Fig. 1 B, D). The data indicate that H20 and FR may bind to 2 different sites and that H20 binding does not significantly affect FR binding at the concentrations tested.
In silico docking of compounds to 3D EBNA1 structure
To determine whether and where H20 compounds fit into the EBNA1 minimal DBD, we attempted to virtually dock the H20 compound to the native EBNA1 dimer (PDB 1VH1) (Fig. 1 F) using the GLIDE program (Kontoyianni et al., 2004) and also a dissociated EBNA1 monomers from the original EBNA1 dimer (PDB 1B3T) (Bochkarev et al., 1995; Bochkarev et al., 1998), from which a monomer and cognate FR DNA were virtually removed to derive the monomeric structure (Fig. S1 C). The GLIDE program found that EBNA1 DBD a. a. 459–607, and more narrowly a. a. 469–607, has at least 2 pockets, and that pocket 2 in the native and monomeric EBNA1 are capable of accepting compound H20 (Fig. 1 F, Fig. S1 C). Pocket 2 was formed by residues of 3 α-helices: I481 of α-helix 1, N519 of α-helix 2, and L582 of α-helix 3. H20 appeared to dock in pocket 2. This H20-docking pocket 2 was distant from the hydrophilic oriP FR DNA binding pocket formed by R469, G472, K514, Y518, R521, P535, and L536, accounting for H20’s incapability of inhibiting EBNA1 DNA binding activity despite its strong association with the EBNA1 DBD. These in silico docking results suggest that H20 is likely a ligand of EBNA1. However, H20 did not affect EBNA1 DNA binding activity in vitro (data not shown) which was consistent with the lack of inhibitory effect on EBNA1 binding to FR (see below). Nevertheless, because H20 repressed EBNA1-mediated transcription in the previous reporter (Kang et al., 2011) and bound directly to EBNA1 (Fig. 1 B, C), we next investigated whether H20 affects EBNA1-mediated persistence of oriP plasmid and EBV whole genome. The BJAB-EBNA1 cells used in HTS that stably maintains oriP plasmid (Kang et al., 2011) were treated with DMSO and H20 at 0.5 and 1.5 uM for 14 days, during which time the media and compound were replaced at every 3 days. The persisted oriP plasmid DNA were isolated by a modified Hirt method for rapid purification of episomal plasmid DNA from mammalian cells (Arad, 1998) and probed to P32-labeled oriP plasmid by Southern hybridization. Relative cell viability of H20 to DMSO control was calculated from the viable number of cells at the day of harvest. While cell viability was unaffected by H20 at 1.5 μM, oriP plasmid persistence decreased by approximately 20 % at concentrations of equal to or lower than 1.5 uM in EBV-negative BJAB cells stably reconstituted with EBNA1 and EBV surrogate oriP replicon (Fig. S1 D). Contrary to this, the persistence of whole viral genome was significantly affected by more than 80% as evidenced in EBV-infected RAJI cells treated with 5 μM (Fig. 1G). These data indicate that H20 binds to EBNA1, interferes with EBNA1 mediated functions of transcription and genome persistence but does not affect DNA binding activity in vitro.
Limited SAR study of H20
A structure-activity relationship (SAR) study for H20 was undertaken to analyze H20 and further derive compounds that can inhibit EBNA1-dependent DNA binding activity or bind to EBNA1 (see Table 1). Limited SAR assays using H20 and 18 other congeners collectively revealed that increased lipophilicity at R1 and oxygen, provided either by OH, methoxy, or ethoxy at R4 (Table 1), are correlated with increased inhibition of EBNA1 dependent sequence-specific DNA binding activity in vitro to cognate DNA. Among them, H31 showed strong inhibition of both EBNA1-dependent transcription and EBNA1-specific DNA binding activity (see below in Fig. 2 A, B) (Table 1), but not RBP-Jκ DNA binding activity to Jκ site in vitro (Fig. S2 A). Although we were unable to detect H31 binding directly to the EBNA DBD in the SPR assay and in silico docking prediction (data not shown), H31 compound was selected for further analyses (Fig. 1 A) (Table 1), because of its strong inhibition of EBNA1-mediated sequence-specific DNA binding activity in vitro to EBV viral sequence, the principle function of EBNA1 required for all EBNA1 known activities including supports for transcription, replication and persistence of viral genome.
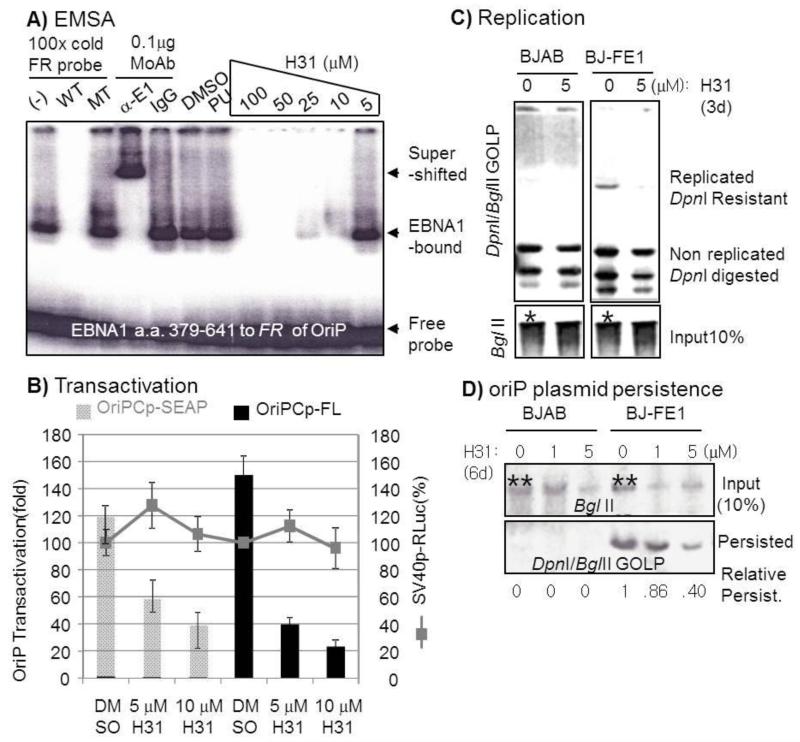
Fig. 2.
Inhibitory mechanisms of H31 on EBNA1 function. A) Dose-dependent H31 inhibition of EBNA1 DBD a. a. 379–641 (denoted 42B) in vitro DNA binding activity. EBNA1 was mixed with 100–5 μM compound H31, DMSO, or puromycin (PU) in gel shifts before the addition of γ-P32ATP-labeled double-stranded FR probes. In the lanes specified, an excess (×100) of unlabeled wild type (WT) or mutant (MT) FR probe, PBS(−), a monoclonal antibody to EBNA1 (α-E1), or isotype control antibody (IgG) was added as a competitor or to super-shift the EBNA1-probe complex. B) H31 affected EBNA1-dependent oriPCp-FL and oriPCp-SEAP in a dose-dependent manner but not EBNA1-independent SV40p-RL, reflective of H31′s having a specific inhibitory activity without non-specific toxicity to cells. Lack of toxicity was also evident from the comparable cell number and SV40p-RLuc activity in response to higher concentration of 30 μM compared to DMSO-treated control (data not shown). C)~D) H31 repressed EBNA1-dependent oriP plasmid (GOLP) replication measured on the day 3 post transfection (C) and persistence on the day 6 (D) in BJAB-FE1 cells from a duplicate experiments. Episomal GOLP plasmid DNA isolated by the Hirt plasmid method (Hirt, 1967) was linearized with BglII and digested with DpnI to eliminate non-replicated or non-persisted oriP plasmids and subjected to Southern hybridization to GOLP probe DNA. Significant decrease of persisted oriP plasmid in response to H31 was calculated by the persisted oriP plasmids normalized by the residual input amounts per lane at day 6 post transfection. Note BJAB or BJ-FE cells were transiently transfected with 25 μg of oriP plasmid and the same batch of transfected cells were divided into2~ 3 flasks into which compounds were treated at the indicated concentration for 3 or 6 days (Fig 2 C or D). Comparable transfection efficiency into BJAB or BJ-FE cells was therefore judged by similar levels of residual inputs after 3 or 6 days post transfection (denoted as * or ** in Fig. 2 C or D). Compared to untreated control (compare 0 μM), decrease in persisted plasmid in H31-treated cells (1 μM, 5 uM) indicates functional inhibition of EBNA1-mediated oriP plasmid persistence. Also, decreases of input levels in H31- treated lanes additionally reflect H31′s inhibitory activity in oriP plasmid persistence.
H31 suppresses EBNA1-mediated sequence-specific DNA binding to oriP in vitro and in vivo, but not EBNA1-mediated sequence-nonspecific chromosome association in cells
In the in vitro EBNA1 antibody-induced EBNA1 EMSA, in which DMSO or a control chemical (puromycin, PU) did not have an effect, H31 inhibited EBNA1 a. a. 379–641-mediated in vitro DNA binding to the cognate FR element in a dose-dependent manner (Fig. 2 A). The protein-probe complexes were EBNA1-bound FR because the protein-bound FR probe was super shifted by EBNA1 monoclonal antibody, but not by isotype control antibody (IgG) or BSA (data not shown), and the binding was completely competed out by an excess amount of unlabeled cold wild-type (WT) probe, but not by mutant (MT) FR probe or PBS (−). This result clearly indicates that H31 can significantly repress EBNA1-dependent sequence specific DNA binding activity, which likely correlated with functional interference of transactivation or replication in cells (see below).
We next questioned whether H31 affects EBNA1’s 2 RG domain-mediated sequence-nonspecific chromosome associations, which are required for EBNA1-dependent transactivation and persistence of the oriP plasmid (Fig. S2 B). We transfected pEGFP-EBNA1 (GE1) or pEGFP-C1 vector control into EBNA-negative BJAB cells and treated with DMSO or H31 at 10 μM for 2 days, then fixed and stained the cells with propidium iodide (PI). Fluorescence images of EGFP-EBNA1 tethered to mitotic chromosomes in metaphase cell spreads were integrated by confocal microscopy analyses. In GE1-transfected, DMSO- treated control cells, the EGFP-EBNA1 was visualized on mitotic chromosomes (denoted as “on), in which EGFP-fused EBNA1 (green) and PI (red) merged into diffuse and bright yellow fluorescence on chromosomes as shown previously(Hung et al., 2001). In H31-treated cells, EGFP-EBNA1 was found to locate on the chromosomes by displaying the same pattern of merged fluorescence on chromosomes. In the pEGFP-C1 vector –transfected, H31-treated control cells, EGFP signal was not displayed on the chromosomes (denoted as “off”) whereas PI stained on the chromosome as expected (Fig. 2 B). The lack of difference in GE1 association with host chromosomes between DMSO and H31-treated cells indicate that H31 did not affect EBNA1 RG domain-mediated association with chromosomes (Fig. S2 B). Taken together with the in vitro EMSA and in vivo confocal microscopy, these results demonstrate that H31 selectively inhibits EBNA1-mediated sequence-specific DNA binding to oriP in vitro but not EBNA1-mediated sequence-nonspecific chromosome association in cells.
H31 inhibits transcription, replication and persistence of oriP plasmids and EBV genomes
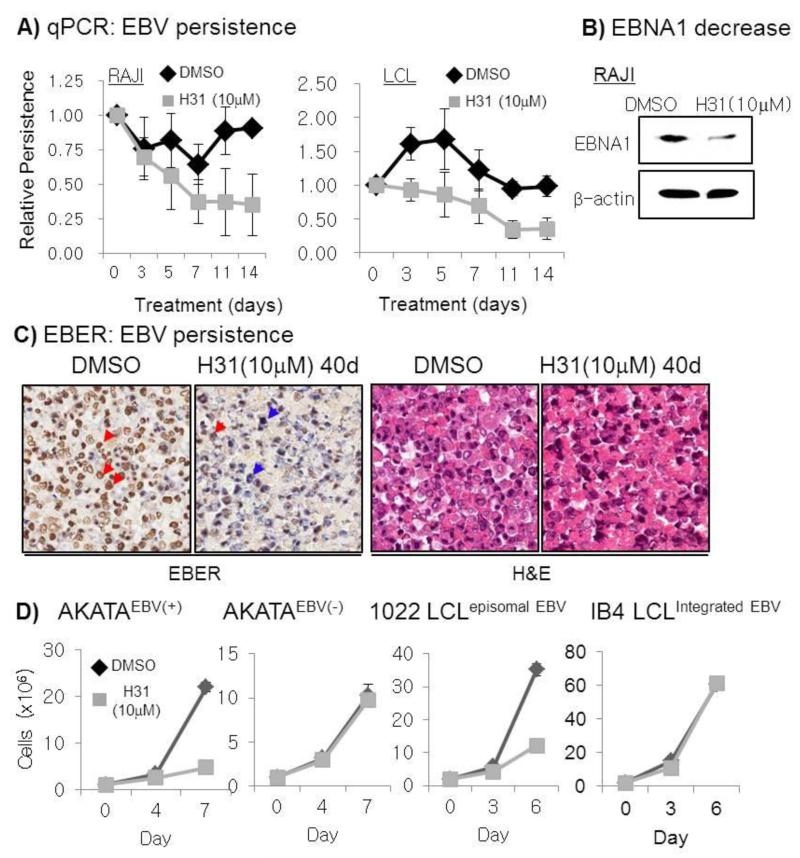
We next investigated whether H31 inhibition of EBNA1-dependent sequence-specific DNA binding represses EBNA1-dependent, oriP-mediated transactivation. Co-cultures of BJRL-FE1-OF and BJ-FE1-OS, both of which harbor EBNA1-dependent, oriP-mediated reporters, were treated with compounds for 2–3 days, subjected to firefly luciferase, Renilla luciferase, and SEAP assays. As expected, H31 inhibited EBNA1-dependent oriPCp-FL and oriPCp-SEAP in a dose-dependent manner, but not EBNA1-independent SV40p-RL and viability as measured by cell counting (Fig. 2 B). We further investigated whether H31 repressed EBNA1-dependent oriP plasmid replication and persistence. BJAB or BJAB-FE1 cells were transiently transfected with GOLP plasmid, an EBV oriP surrogate replicon, and assessed by replication and semi-quantitative persistence assays (Fig. 2 C, D). The oriP plasmids were transfected with comparable efficiency (denoted * in Fig. 2 C). H31 repressed the replication of the EBNA1-dependent oriP-harboring GOLP plasmid in BJAB-FE1 cells (Fig. 2 C). Whereas BJAB cells did not support replication of transiently transfected GOLP (see BJAB; 0, 5), BJ-FE1-expressing FLAG-EBNA1 cells did support replication (see BJ-FE1; 0), which was inhibited by 5 μM H31 treatment (see BJ-FE1; 5). Consistent with the replication assay results, further semi-quantitative persistence assays for the oriP plasmid showed that plasmid persistence decreased upon H31 treatment by ~60%, while transfection efficiencies were comparable, as judged by equal intensities from input DNA (Fig. 2 D, compare inputs for BJAB 0 uM and BJ-FE 0 uM, respectively, denoted as **). The persistence efficiencies were calculated by the persisted oriP plasmids normalized by the residual input amounts at day 6 post transfection. The inhibitory effect on oriP plasmid persistence was further substantiated by the significant decrease (>65%) of EBV whole genome copy number upon H31 treatment in the following 3 independent experiments. EBV-infected Raji Burkitt’s lymphoma cells and LCL SNU-265 cells (Lee et al., 1998) were treated with 10 μM H31 for 14 days, during which time the media and compound were replaced, and subjected to real-time quantitative PCR for EBV oriP and host GAPDH. The relative persistence was determined from EBV oriP amounts normalized by/GAPDH as previously (Kang et al., 2011). Upon H31 treatment for 14 days, the persistence of EBV genome decreased by 70~85 % in both Raji and LCL (Fig. 3 A). Accordingly to genome decrease, EBNA1 expression also decreased in Raji cells (Fig. 3 B). The Raji cells were then treated with 10 μM H31 for 40 days and subjected to EBER in situ hybridization. While the surviving cells treated with DMSO and H31 retained cellular integrity, showing a clear cytoplasm and dark nucleus in H&E staining, EBER transcription was much lower in H31-treated cells than in DMSO-treated control cells (Fig. 3 C), further substantiating the inhibitory effect of the compound on genome persistence
Fig. 3.
Negative effects of H31 on the EBV genome persistence and cell proliferation. A) Decrease in EBV copy number in Raji in response to treatment with 10 μM H31 for 14 days. The relative persistence (relative amount of EBV to GAPDH) was determined from EBV oriP amounts normalized by/GAPDH each day as previously (Kang et al., 2011). The data are the mean with standard errors from triplicate independent experiments. B) Decrease in the steady state level of EBNA1 protein in response to 10 μM H31 for 14 days in Raji from the left panel in A), reflective of EBV genome decrease. C) Significant reduction of EBER expression in RAJI cells in response to treatment with 10 μM H31 at for 40 days by EBER in situ hybridization. In contrast to the high fraction (>90%) of EBER+/hi cells in non-treated RAJI cells, the survived RAJI cells upon H31 treatment showed much less fraction (<30%) of EBER+ cells. Note that red arrow heads denote EBV(+), hence EBER+/hi cells, blue arrow head denotes EBV(−), hence EBER −/low cells. D) H31 suppressed EBV episome-dependent cell growth in EBV-infected cells. H31 selectively suppressed growth of AKATAEBV(+), 1022 LCL, but not AKATAEBV(−) or IB4 LCL. Shown are data from triplicate experiments with standard errors (note small standard error is not visible in some instances). Similar results were obtained in other independent experiments for additional cell lines (Exp 1 and Exp 2 where indicated) (see Fig. S3)
H31 suppressed EBV episome-dependent cell growth in EBV-infected cells
Because H31 interfered with EBNA1-mediated oriP transcription, replication and persistence, H31 was expected to suppress the growth of LCL and AKATA, EBV-infected Burkitt’s lymphoma cells of which proliferation is largely dependent on the EBV genomes. We determined the effect of H31 on the proliferation of 6 cell lines; In the first triplicate experiment, EBV-infected AKATA (AKATAEBV(+)), three LCLs carrying multiple copies of episomal EBV genomes, EBV-eliminated AKATA (AKTAEBV(−)), and IB4 LCL carrying 4 copies of integrated EBV genomes. As expected, H31 selectively suppressed growth of AKATAEBV(+), 1022 LCL, but not AKATAEBV(−), IB4 LCL (Fig. 3 D) or EBV-negative BJAB cells (as shown in Fig. 2 B). In second triplicate experiment, similar results were obtained in which growth of EBV-infected LCL (265 LCL, 114 LCL) and EBV-positive AKTATA cell (AKATAEBV(+), but not AKATAEBV(−), were selectively suppressed (Fig. S3). These data indicate that H31 inhibition blocks EBNA1-dependent transcription and EBV episome persistence, leading to blockade or retardation of EBV-infected cells.
Discussion
Latent Epstein-Barr virus (EBV) infection is the causative infectious agent of certain human B cell lymphomas and carcinomas. EBV genomes mostly replicate and persist as extra chromosomal episome in latently infected cells. EBV episome persistence, replication, and gene expression are dependent on at least 3 EBNA1 domains: RG1 (a. a. 33–89), RG2 (a. a. 328–386), and the almost inseparable DD and oriP DBD (a. a. 459–607) (Ambinder et al., 1991; Bochkarev et al., 1996; Bochkarev et al., 1995; Bochkarev et al., 1998; Frappier et al., 1994; Goldsmith et al., 1993; Shah et al., 1992; Summers et al., 1996). These 3 domains can be further grouped into 2 functional regions: the EBNA1 chromosome association domains RG1 and RG2 that mediate sequence-nonspecific association with chromosomes and a sequence-specific DBD that binds to multiple cognate EBV sequences in oriP.
Several small molecules that indirectly or directly inhibit EBV have been developed. The growth of EBV-infected cells or EBV-induced primary B cell transformation in vitro was suppressed by these compounds, which include an Hsp90 inhibitor, BRACO-19, DS-binding pyrrole-imidazole polyamides, and Roscovitine (Li et al., 2010; Norseen et al., 2009; Sun et al., 2010; Thompson et al., 2010; Yasuda et al., 2011). The HSP90 inhibitor targeted the EBNA1 glycine-alanine repeats domain, decreased EBNA1 expression and translation, suppressed EBV-infected cell growth, and prevented EBV-induced B cell transformation (Sun et al., 2010). The G-quadruplex-interacting compound BRACO-19 inhibited EBNA1-dependent viral DNA replication and preferentially blocked the proliferation of EBV-positive cells relative to the proliferation of EBV-negative cell lines. BRACO-19 likely disrupts the ability of EBNA1 to tether to metaphase chromosomes (Norseen et al., 2009). EBNA1 inhibitors that directly bind to EBNA1 DBD were recently identified by an in silico high-throughput screen (Kang et al., 2008; Thompson et al., 2010). These inhibitors can fit into the EBNA1 DBD and inhibit EBNA1 DNA binding activity in vitro. Additionally, it was shown that a PARP-1 inhibitor enhances oriP-dependent plasmid maintenance (Tempera et al., 2010). Our previous HTS identified Roscovitine, Shikonin, and the compound (5-(3, 4-dihydroxybenzylidene)-3-methyl-2-thioxothiazolidin-4-one), referred to as H20. In another set of studies, we described a small molecule, EiK1, and short EBNA1 peptide 85 (P85), which can efficiently block EBNA1 dimerization (Kim et al., 2012). EiK1 is a small molecule compound that was also identified from the previous screen, and P85 is a short EBNA1 peptide that contains a. a. 560–566 of the β-sheet 3 strand of the EBNA1 dimeric interface. In this study, we further characterized the small molecule H20 and its derivative H31, which selectively inhibits EBNA1-mediated, oriP sequence-specific DNA binding, leading to the selective death of EBV-infected cells in vitro and in vivo. The inhibition of DNA binding appeared to be selective for EBNA1; it did not affect RBP-Jκ binding in vitro.
EBNA1 is well suited to the virtual docking approach because the dimer structures with or without cognate DNA (1VHI, 1B3T) have been resolved to 2.5 Å and 2.2 Å, respectively (Bochkarev et al., 1995; Bochkarev et al., 1998). The GLIDE-assisted in silico docking (Kontoyianni et al., 2004) in this study suggested that H20 binding to EBNA1 occurs by docking the benzyl ring into pocket 2 with an anionic interaction (or hydrogen bond) between the R4 oxygen of H20 and a nearby residue in the surrounding the pocket of EBNA1. In addition, the SAR study suggested that the oxygen at R4 and hydrophobicity at R1 in the H20 series are important for inhibition of EBNA1, whereas R3 oxygen is dispensable (Table 1).
EBNA1 a. a. 459–607 DD/DBD forms a strong dimer in high salt conditions, and the dimerization interface formed by the β-sheet 3 strands (a. a. 560–566) is buried in the hydrophobic inner space of the globular EBNA1 structure (Bochkarev et al., 1996; Bochkarev et al., 1995; Bochkarev et al., 1998). Consequently, it should be difficult for a compound or peptide to dissociate the dimer or to target the dimeric interface. However, the EBNA1 DD was successfully targeted by using an EBNA1-specific sequence peptide or small molecule (Kim et al., 2012). Another group also found structurally unrelated compounds that targeted 2 different pockets, indicating that targeting the dimerization domain of EBNA1 with a small molecule or short peptide is experimentally achievable (Thompson et al., 2010). Therefore, it could be more practical for a small molecule to target DBD by binding to structural pockets, i.e., pocket 2, as shown in this study, or the DNA binding pocket, as shown in a previous report (Li et al., 2010). Based on the in silico docking, H20 can virtually dock to a space in pocket 2 formed by I481, N519, and L582, which are located distantly from the DNA binding pocket composed by the surrounding amino acids R469, G472, K514, Y518, R521, P535, and L536 (Kang et al., 2008). However, the purely hypothetical interaction of H20 with the pocket 2 remains to be experimentally confirmed because the proposed docking site was not proven to be the same site where H20 binds in SPR. Yet, the docking results in this study are consistent with the biochemical gel shift, in which H20 did not affect EBNA1 DNA binding below the 100 μM level. Also consistent the data described above, the SPR assay indicated that increasing H20 concentrations did not significantly disrupt the association between FR and the immobilized EBNA1 (compare Fig. 1 B with 1 D). The results demonstrate that H20 did not significantly affect FR’s binding to EBNA1; H20 does not bind the DNA binding pocket where FR binds, but probably to a different pocket (Fig. 1 B, C, D). Although H31 inhibited EBNA1 DNA binding, as shown in the EMSA (Fig. 2 A), we were unable to detect H31 binding directly to the EBNA DBD in the SPR assay (data not shown). Given that EBNA1 dimerization is required for EBNA1-mediated DNA binding and transactivation and that H31 inhibited DNA binding and transactivation, it is possible that hydrophobic H31 targets the EBNA1 dimeric interface in the pre-existing native dimer and thereby interferes with de novo dimer formation or with the dynamic interchange to dimer from monomer in a dynamically regulated equilibrium.
Alternatively, the bulky structure of the adamantine (ADM) ring in H31 may change the global structure of EBNA1 in vitro or in vivo, resulting in loss of DNA binding activity. It is equally possible that H31 can bind to functional domains other than DBD/DD, thereby inhibiting DNA binding. In that respect, we do not know yet whether H31 affects the structural integrity of the RG domains because 3D structures of the EBNA1 RG domains at the atomic level are unavailable. EBNA1 RG1 and RG2 are critical for EBNA1’s effect on oriP-mediated transcriptional enhancement and episome persistence. Although either RG-rich domain can mediate EBNA1 chromosomal association (Hung et al., 2001; Marechal et al., 1999; Sears et al., 2003), both domains are necessary for transcription and persistence mediated by the association of EBNA1 with chromosomes (Hung et al., 2001; Sears et al., 2003). No effect of H31 on EBNA1 RG1- or RG2-mediated chromosomal association was observed in this study, suggesting that H31 may not affect RG functions in vitro. Studies on the co-crystal structures of either EBNA1 full length or RG domains with H20 and H31, and the activity test of these compounds in appropriate mouse model should be attempted to better understand the inhibitory mechanism in atomic level and in vivo achieve, which will be the subject of future study.
H31 treatment for relatively short periods preferentially blocked the growth of EBV-infected cells over non-infected cells. H31 toxicity was not attributed to this difference, as H31 did not exhibit comparable toxicity measured from BJAB cell growth (see Fig. 2 B, D). Given that as little as 70% specific inhibition of EBNA1 with dominant-negative EBNA1, EBNA1 antisense oligonucleotides, or EBNA1 RNAi was sufficient to eradicate EBV viral genomes or induce apoptosis of EBV-positive cells in as early as 6 days(Hong et al., 2006; Imai et al., 2005; Miyake et al., 2008; Nasimuzzaman et al., 2005; Roth et al., 1994; Yin and Flemington, 2006), the retardation or preferential blockade of EBV-infected cell growth upon H31 would not be surprising. It is noteworthy that biochemical outcomes upon H31 treatment were similar to those upon treatment of structurally unrelated compounds from different groups; inhibition of transcription from or persistence of oriP episome and preferential suppression of EBV-infected cells growth (Kang et al., 2011; Li et al., 2010; Norseen et al., 2009; Sun et al., 2010; Yasuda et al., 2011; Zhu et al., 2009). Given that AKATA cells spontaneously loses EBV episome, resulting in EBV negative AKATA cells, which still grows but slower than the parental EBV-positive AKATA cells as also seen in our manuscript and EBV-reinfected AKATA recovered the growth potential, the presence of EBV in AKATA supports, but not essential, for growth(Komano et al., 1998; Shimizu et al., 1994). The results in this study were in fact consistent with a previous paper in which AKTATA growth was retarded by gradual loss of EBV episome ((Zhu et al., 2009). Further elaboration of H31 and other small probes to derive better compounds is required to enable control EBV –associated disorders.
In summary, we described the functional characterization of H20 and its derivative, H31, with respect to the underlying EBNA1 inhibitory mechanism. This study showed that (1) compound H20 binds to the EBNA1 minimal DNA binding domain and inhibits EBNA1-dependent functions, (2) compound H31 inhibits EBNA1 DNA binding activity, (3) H31 inhibits replication and persistence of oriP replicons and EBV genomes, and suppresses EBV-infected cells growth. These inhibitors will likely add our understandings on the outcomes such as transcription, cell death and tumorigenesis in EBV-infected Burkitt’s lymphoma, HL and NPC, when EBNA1 function was experimentally suppressed
Supplementary Material
Highlights.
Characterized mechanisms of small molecules for inhibiting Epstein-Barr virus nuclear antigen 1 (EBNA1)
H31 inhibited EBNA1 functions of DNA binding, transactivation, replication and support of viral genome persistence
It selectively suppressed the proliferation of EBV-infected cells
Acknowledgments
E.K and M.-S.K were supported by NIH 5R01CA131354-02. This study was also supported by a grant from the National R&D Program for Cancer Control, Ministry for Health and Welfare, Republic of Korea (1120010) and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0012393). We thank Mr. Jay Duffner, and Drs. Piotr Sliz and Timothy A Lewis for help in SPR, in silico docking and compound syntheses.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: M-SK designed, initiated, conducted, and supervised most of the research, collected and analyzed all data, made figures, and wrote and submitted the paper. EK collaborated on most of the research, analyzed and discussed data, and revised part of the paper. EKL, SYK, KWN, EHJ, and BZ performed, repeated most parts of the research and analyzed subsets of data.
Conflict-of-interest disclosure. The authors declare no competing financial interest.
References
- Adams A, Lindahl T. Epstein-Barr virus genomes with properties of circular DNA molecules in carrier cells. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:1477–1481. doi: 10.1073/pnas.72.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambinder RF, Mullen MA, Chang YN, Hayward GS, Hayward SD. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arad U. Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. Biotechniques. 1998;24:760–762. doi: 10.2144/98245bm14. [DOI] [PubMed] [Google Scholar]
- Aras S, Singh G, Johnston K, Foster T, Aiyar A. Zinc coordination is required for and regulates transcription activation by Epstein-Barr nuclear antigen 1. PLoS pathogens. 2009;5:e1000469. doi: 10.1371/journal.ppat.1000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir R, Luka J, Cheloha K, Chamberlain M, Hochberg F. Expression of Epstein-Barr virus proteins in primary CNS lymphoma in AIDS patients. Neurology. 1993;43:2358–2362. doi: 10.1212/wnl.43.11.2358. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Barwell JA, Pfuetzner RA, Bochkareva E, Frappier L, Edwards AM. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell. 1996;84:791–800. doi: 10.1016/s0092-8674(00)81056-9. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Barwell JA, Pfuetzner RA, Furey W, Jr., Edwards AM, Frappier L. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1. Cell. 1995;83:39–46. doi: 10.1016/0092-8674(95)90232-5. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Bochkareva E, Frappier L, Edwards AM. The 2.2 A structure of a permanganate-sensitive DNA site bound by the Epstein-Barr virus origin binding protein, EBNA1. J Mol Biol. 1998;284:1273–1278. doi: 10.1006/jmbi.1998.2247. [DOI] [PubMed] [Google Scholar]
- Bossolasco S, Nilsson A, de Milito A, Lazzarin A, Linde A, Cinque P, Chiodi F. Soluble CD23 in cerebrospinal fluid: a marker of AIDS-related non-Hodgkin’s lymphoma in the brain. AIDS. 2001;15:1109–1113. doi: 10.1097/00002030-200106150-00005. [DOI] [PubMed] [Google Scholar]
- Faulkner GC, Burrows SR, Khanna R, Moss DJ, Bird AG, Crawford DH. X-Linked agammaglobulinemia patients are not infected with Epstein-Barr virus: implications for the biology of the virus. J Virol. 1999;73:1555–1564. doi: 10.1128/jvi.73.2.1555-1564.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier L, Goldsmith K, Bendell L. Stabilization of the EBNA1 protein on the Epstein-Barr virus latent origin of DNA replication by a DNA looping mechanism. J Biol Chem. 1994;269:1057–1062. [PubMed] [Google Scholar]
- Geser A, de The G, Lenoir G, Day NE, Williams EH. Final case reporting from the Ugandan prospective study of the relationship between EBV and Burkitt’s lymphoma. Int J Cancer. 1982;29:397–400. doi: 10.1002/ijc.2910290406. [DOI] [PubMed] [Google Scholar]
- Goldsmith K, Bendell L, Frappier L. Identification of EBNA1 amino acid sequences required for the interaction of the functional elements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1993;67:3418–3426. doi: 10.1128/jvi.67.6.3418-3426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulley ML, Glaser SL, Craig FE, Borowitz M, Mann RB, Shema SJ, Ambinder RF. Guidelines for interpreting EBER in situ hybridization and LMP1 immunohistochemical tests for detecting Epstein-Barr virus in Hodgkin lymphoma. American journal of clinical pathology. 2002;117:259–267. doi: 10.1309/MMAU-0QYH-7BHA-W8C2. [DOI] [PubMed] [Google Scholar]
- Henderson A, Ripley S, Heller M, Kieff E. Chromosome site for Epstein-Barr virus DNA in a Burkitt tumor cell line and in lymphocytes growth-transformed in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:1987–1991. doi: 10.1073/pnas.80.7.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle W, Henle G, Ho JH. Epstein-Barr virus-related serology in nasopharyngeal carcinoma and controls. IARC Sci Publ. 1978;20:427–437. [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hong M, Murai Y, Kutsuna T, Takahashi H, Nomoto K, Cheng CM, Ishizawa S, Zhao QL, Ogawa R, Harmon BV, Tsuneyama K, Takano Y. Suppression of Epstein-Barr nuclear antigen 1 (EBNA1) by RNA interference inhibits proliferation of EBV-positive Burkitt’s lymphoma cells. J Cancer Res Clin Oncol. 2006;132:1–8. doi: 10.1007/s00432-005-0036-x. [DOI] [PubMed] [Google Scholar]
- Hung SC, Kang MS, Kieff E. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1865–1870. doi: 10.1073/pnas.031584698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley EA, Klaman LD, Agger S, Lawrence JB, Thorley-Lawson DA. The prototypical Epstein-Barr virus-transformed lymphoblastoid cell line IB4 is an unusual variant containing integrated but no episomal viral DNA. J Virol. 1991;65:3958–3963. doi: 10.1128/jvi.65.7.3958-3963.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley EA, McNeil JA, Lawrence JB, Thorley-Lawson DA. Genomic integration as a novel mechanism of EBV persistence. Curr Top Microbiol Immunol. 1990;166:367–374. doi: 10.1007/978-3-642-75889-8_45. [DOI] [PubMed] [Google Scholar]
- Imai S, Kuroda M, Yamashita R, Ishiura Y. Therapeutic inhibition of Epstein-Barr virus-associated tumor cell growth by dominant-negative EBNA1. Uirusu. 2005;55:239–249. doi: 10.2222/jsv.55.239. [DOI] [PubMed] [Google Scholar]
- Kang MS, Hung SC, Kieff E. Epstein-Barr virus nuclear antigen 1 activates transcription from episomal but not integrated DNA and does not alter lymphocyte growth. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:15233–15238. doi: 10.1073/pnas.211556598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MS, Lee EK, Soni V, Lewis TA, Koehler AN, Srinivasan V, Kieff E. Roscovitine inhibits EBNA1 serine 393 phosphorylation, nuclear localization, transcription, and episome maintenance. J Virol. 2011;85:2859–2868. doi: 10.1128/JVI.01628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MS, Lu H, Yasui T, Sharpe A, Warren H, Cahir-McFarland E, Bronson R, Hung SC, Kieff E. Epstein-Barr virus nuclear antigen 1 does not induce lymphoma in transgenic FVB mice. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:820–825. doi: 10.1073/pnas.0408774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MS, Soni V, Bronson R, Kieff E. Epstein-Barr virus nuclear antigen 1 does not cause lymphoma in C57BL/6J mice. J Virol. 2008;82:4180–4183. doi: 10.1128/JVI.02596-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff ED, Rickinson AB. Epstein-Barr Virus and Its Replication. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott WIlliams and WIlkins, a Wolters Kluwer Business; Philadelphia: 2007. pp. 2603–2654. [Google Scholar]
- Kim SY, Song KA, Kieff E, Kang MS. Small molecule and peptide-mediated inhibition of Epstein-Barr virus nuclear antigen 1 dimerization. Biochemical and biophysical research communications. 2012;424:251–256. doi: 10.1016/j.bbrc.2012.06.095. [DOI] [PubMed] [Google Scholar]
- Komano J, Sugiura M, Takada K. Epstein-Barr virus contributes to the malignant phenotype and to apoptosis resistance in Burkitt’s lymphoma cell line Akata. J Virol. 1998;72:9150–9156. doi: 10.1128/jvi.72.11.9150-9156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyianni M, McClellan LM, Sokol GS. Evaluation of docking performance: comparative data on docking algorithms. J Med Chem. 2004;47:558–565. doi: 10.1021/jm0302997. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Marselle LM. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989;57:493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Villnave CA, Singer RH. Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell. 1988;52:51–61. doi: 10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- Lee WK, Kim SM, Sim YS, Cho SG, Park SH, Kim CW, Park JG. B-lymphoblastoid cell lines from cancer patients. In vitro cellular & developmental biology. Animal. 1998;34:97–100. doi: 10.1007/s11626-998-0090-1. [DOI] [PubMed] [Google Scholar]
- Lestou VS, Strehl S, Lion T, Gadner H, Ambros PF. High-resolution FISH of the entire integrated Epstein-Barr virus genome on extended human DNA. Cytogenetics and cell genetics. 1996;74:211–217. doi: 10.1159/000134416. [DOI] [PubMed] [Google Scholar]
- Li N, Thompson S, Schultz DC, Zhu W, Jiang H, Luo C, Lieberman PM. Discovery of selective inhibitors against EBNA1 via high throughput in silico virtual screening. PloS one. 2010;5:e10126. doi: 10.1371/journal.pone.0010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Klein G, Reedman BM, Johansson B, Singh S. Relationship between Epstein-Barr virus (EBV) DNA and the EBV-determined nuclear antigen (EBNA) in Burkitt lymphoma biopsies and other lymphoproliferative malignancies. Int J Cancer. 1974;13:764–772. doi: 10.1002/ijc.2910130605. [DOI] [PubMed] [Google Scholar]
- MacMahon EM, Glass JD, Hayward SD, Mann RB, Becker PS, Charache P, McArthur JC, Ambinder RF. Epstein-Barr virus in AIDS-related primary central nervous system lymphoma. Lancet. 1991;338:969–973. doi: 10.1016/0140-6736(91)91837-k. [DOI] [PubMed] [Google Scholar]
- Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas JC. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J Virol. 1999;73:4385–4392. doi: 10.1128/jvi.73.5.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Heller M, Petti L, O’Shiro E, Kieff E. Persistence of the entire Epstein-Barr virus genome integrated into human lymphocyte DNA. Science. 1984;226:1322–1325. doi: 10.1126/science.6095452. [DOI] [PubMed] [Google Scholar]
- Miyake A, Dewan MZ, Ishida T, Watanabe M, Honda M, Sata T, Yamamoto N, Umezawa K, Watanabe T, Horie R. Induction of apoptosis in Epstein-Barr virus-infected B-lymphocytes by the NF-kappaB inhibitor DHMEQ. Microbes and infection / Institut Pasteur. 2008;10:748–756. doi: 10.1016/j.micinf.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Nasimuzzaman M, Kuroda M, Dohno S, Yamamoto T, Iwatsuki K, Matsuzaki S, Mohammad R, Kumita W, Mizuguchi H, Hayakawa T, Nakamura H, Taguchi T, Wakiguchi H, Imai S. Eradication of epstein-barr virus episome and associated inhibition of infected tumor cell growth by adenovirus vector-mediated transduction of dominant-negative EBNA1. Mol Ther. 2005;11:578–590. doi: 10.1016/j.ymthe.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Nonoyama M, Pagano JS. Separation of Epstein-Barr virus DNA from large chromosomal DNA in non-virus-producing cells. Nat New Biol. 1972;238:169–171. doi: 10.1038/newbio238169a0. [DOI] [PubMed] [Google Scholar]
- Norseen J, Johnson FB, Lieberman PM. Role for G-quadruplex RNA binding by Epstein-Barr virus nuclear antigen 1 in DNA replication and metaphase chromosome attachment. J Virol. 2009;83:10336–10346. doi: 10.1128/JVI.00747-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins DR, Milman G, Hayward SD, Hayward GS. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- Raynaud FI, Whittaker SR, Fischer PM, McClue S, Walton MI, Barrie SE, Garrett MD, Rogers P, Clarke SJ, Kelland LR, Valenti M, Brunton L, Eccles S, Lane DP, Workman P. In vitro and in vivo pharmacokinetic-pharmacodynamic relationships for the trisubstituted aminopurine cyclin-dependent kinase inhibitors olomoucine, bohemine and CYC202. Clin Cancer Res. 2005;11:4875–4887. doi: 10.1158/1078-0432.CCR-04-2264. [DOI] [PubMed] [Google Scholar]
- Reisman D, Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickinson AB, Kieff ED. Epstein-Barr Virus. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins, a Wolters Kluwer Business; Philadelphia: 2007. pp. 2655–2700. [Google Scholar]
- Rivailler P, Carville A, Kaur A, Rao P, Quink C, Kutok JL, Westmoreland S, Klumpp S, Simon M, Aster JC, Wang F. Experimental rhesus lymphocryptovirus infection in immunosuppressed macaques: an animal model for Epstein-Barr virus pathogenesis in the immunosuppressed host. Blood. 2004;104:1482–1489. doi: 10.1182/blood-2004-01-0342. [DOI] [PubMed] [Google Scholar]
- Roth G, Curiel T, Lacy J. Epstein-Barr viral nuclear antigen 1 antisense oligodeoxynucleotide inhibits proliferation of Epstein-Barr virus-immortalized B cells. Blood. 1994;84:582–587. [PubMed] [Google Scholar]
- Schang LM. Advances on cyclin-dependent kinases (CDKs) as novel targets for antiviral drugs. Curr Drug Targets Infect Disord. 2005;5:29–37. doi: 10.2174/1568005053174609. [DOI] [PubMed] [Google Scholar]
- Sears J, Kolman J, Wahl GM, Aiyar A. Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by Epstein-Barr nuclear antigen 1. J Virol. 2003;77:11767–11780. doi: 10.1128/JVI.77.21.11767-11780.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J Virol. 2004;78:11487–11505. doi: 10.1128/JVI.78.21.11487-11505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah WA, Ambinder RF, Hayward GS, Hayward SD. Binding of EBNA-1 to DNA creates a protease-resistant domain that encompasses the DNA recognition and dimerization functions. J Virol. 1992;66:3355–3362. doi: 10.1128/jvi.66.6.3355-3362.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Tanabe-Tochikura A, Kuroiwa Y, Takada K. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt’s lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J Virol. 1994;68:6069–6073. doi: 10.1128/jvi.68.9.6069-6073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixbey JW, Vesterinen EH, Nedrud JG, Raab-Traub N, Walton LA, Pagano JS. Replication of Epstein-Barr virus in human epithelial cells infected in vitro. Nature. 1983;306:480–483. doi: 10.1038/306480a0. [DOI] [PubMed] [Google Scholar]
- Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers H, Barwell JA, Pfuetzner RA, Edwards AM, Frappier L. Cooperative assembly of EBNA1 on the Epstein-Barr virus latent origin of replication. J Virol. 1996;70:1228–1231. doi: 10.1128/jvi.70.2.1228-1231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Barlow EA, Ma S, Hagemeier SR, Duellman SJ, Burgess RR, Tellam J, Khanna R, Kenney SC. Hsp90 inhibitors block outgrowth of EBV-infected malignant cells in vitro and in vivo through an EBNA1-dependent mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3146–3151. doi: 10.1073/pnas.0910717107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K, Horinouchi K, Ono Y, Aya T, Osato T, Takahashi M, Hayasaka S. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus genes. 1991;5:147–156. doi: 10.1007/BF00571929. [DOI] [PubMed] [Google Scholar]
- Tempera I, Deng Z, Atanasiu C, Chen CJ, D’Erme M, Lieberman PM. Regulation of Epstein-Barr virus OriP replication by poly(ADP-ribose) polymerase 1. J Virol. 2010;84:4988–4997. doi: 10.1128/JVI.02333-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S, Messick T, Schultz DC, Reichman M, Lieberman PM. Development of a high-throughput screen for inhibitors of Epstein-Barr virus EBNA1. Journal of biomolecular screening. 2010;15:1107–1115. doi: 10.1177/1087057110379154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LM, Movahed LA. In situ demonstration of Epstein-Barr viral genomes in viral-associated B cell lymphoproliferations. The American journal of pathology. 1989;134:651–659. [PMC free article] [PubMed] [Google Scholar]
- Wu H, Kapoor P, Frappier L. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein-Barr nuclear antigen 1. J Virol. 2002;76:2480–2490. doi: 10.1128/jvi.76.5.2480-2490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda A, Noguchi K, Minoshima M, Kashiwazaki G, Kanda T, Katayama K, Mitsuhashi J, Bando T, Sugiyama H, Sugimoto Y. DNA ligand designed to antagonize EBNA1 represses Epstein-Barr virus-induced immortalization. Cancer science. 2011;102:2221–2230. doi: 10.1111/j.1349-7006.2011.02098.x. [DOI] [PubMed] [Google Scholar]
- Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- Yin Q, Flemington EK. siRNAs against the Epstein Barr virus latency replication factor, EBNA1, inhibit its function and growth of EBV-dependent tumor cells. Virology. 2006;346:385–393. doi: 10.1016/j.virol.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Young L, Alfieri C, Hennessy K, Evans H, O’Hara C, Anderson KC, Ritz J, Shapiro RS, Rickinson A, Kieff E, et al. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. The New England journal of medicine. 1989;321:1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- Young LS, Clark D, Sixbey JW, Rickinson AB. Epstein-Barr virus receptors on human pharyngeal epithelia. Lancet. 1986;1:240–242. doi: 10.1016/s0140-6736(86)90776-2. [DOI] [PubMed] [Google Scholar]
- Zhao B, Maruo S, Cooper A, M RC, Johannsen E, Kieff E, Cahir-McFarland E. RNAs induced by Epstein-Barr virus nuclear antigen 2 in lymphoblastoid cell lines. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1900–1905. doi: 10.1073/pnas.0510612103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Liao G, Shan L, Zhang J, Chen MR, Hayward GS, Hayward SD, Desai P, Zhu H. Protein array identification of substrates of the Epstein-Barr virus protein kinase BGLF4. J Virol. 2009;83:5219–5231. doi: 10.1128/JVI.02378-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.