Abstract
Background/Aims
To evaluate the expression of CXC motif chemokine receptor 4 (CXCR4) in the tissues of patients with hilar cholangiocarcinoma (hilar-CCA) and to investigate the cell proliferation and frequency of neural invasion (NI) influenced by RNAi-mediated CXCR4 silencing.
Methods
An immunohistochemical technique was used to detect the expression of CXCR4 in 41 clinical tissues, including hilar-CCA, cholangitis, and normal bile duct tissues. The effects of small interference RNA (siRNA)-mediated CXCR4 silencing were detected in the hilar-CCA cell line QBC939. Cell proliferation was determined by MTT. Expression of CXCR4 was monitored by quantitative real time polymerase chain reaction and Western blot analysis. The NI ability of hilar-CCA cells was evaluated using a perineural cell and hilar-CCA cell coculture migration assay.
Results
The expression of CXCR4 was significantly induced in clinical hilar-CCA tissue. There was a positive correlation between the expression of CXCR4 and lymph node metastasis/NI in hilar-CCA patients (p<0.05). Silencing of CXCR4 in tumor cell lines by siRNA led to significantly decreased NI (p<0.05) and slightly decreased cell proliferation.
Conclusions
CXCR4 is likely correlated with clinical recurrence of hilar-CCA. CXCR4 is involved in the invasion and proliferation of human hilar-CCA cell line QBC939, indicating that CXCR4 could be a promising therapeutic target for hilar-CCA.
Keywords: Hilar cholangiocarcinoma, Neural invasion, CXCR4, RNA interference
INTRODUCTION
Extrahepatic hilar cholangiocarcinoma (hilar-CCA) is a malignant tumor originating from biliary tract epithelial cells and locates most commonly at the hepatic duct bifurcation. It is a relatively rare adenocarcinoma, with an annual incidence of one to two cases per 100,000 in the Western world.1 In China and Southeast Asia, the incidence of hilar-CCA is relatively higher.2 Although the annual incidence of this disease in the Western world is lower than that in Asia, the rate of hilar-CCA has significantly risen over the past several decades.2 Nowadays, it is becoming the most common reason for hepatic tumor-induced death.3
Due to its diagnostic difficulty and high fatality rate, hilar-CCA is one of the most extremely destructive cancers. Currently, surgery is the only therapeutic option to offer a cure. However, the post-resection recurrence rate is extremely high and the 5-year survival rate is only 5%.4 Among various factors in charge of poor survival, neural invasion (NI) represents an important prognostic factor in hilar-CCA, which is thought to be one of the main causes for the high postoperative recurrence rates of hilar-CCA.5-7 Survival rate of CCA patients without NI is significantly higher than those with NI, which indicates that NI is highly and positively correlated with the postoperative recurrence and poor prognosis of hilar-CCA.8 It is further supported by a study using 26 cases of hilar-CCA in the porta hepatis region which revealed that the incidence of NI was 100%, while the proportion of perineural invasion in resected hilar-CCA is around 85% to 88%.9
Consequently, the improvement of prognosis of hilar-CCA patients should base on the mechanistic understanding of NI during hilar-CCA pathological progression. Various molecules deemed to play critical roles in regulating the emergence and development of NI were extensively investigated.8,10 However, systematic investigations of the mode of NI induced by the interaction between chemokine released from perineural tissue and its receptor on tumor cells have not yet been carried out, although this process represents an important feature in the development of NI.
CXC motif chemokine receptor 4 (CXCR4) is a stromal cell-derived factor-1 receptor secreted by bone marrow, liver, lung, and neural cells.11 The expression of CXCR4 was detected in malignant tumor cells12-14 and was positively related with tumor cells migration and hilar-CCA cell invasion.15 However, the relationship between CXCR4 expression and NI development in vitro is still largely unknown.
In this study, the expression level of CXCR4 was measured in hilar-CCA patient samples. Then the coculture migration and MTT assay were used to analyze the influence of CXCR4 expression innervating NI in hilar-CCA cell line QBC939.
MATERIALS AND METHODS
1. Patients and preparation of tissue specimens
A total of 41 specimens were collected from the Department of General Surgery of Xiangya Hospital between June 2010 and June 2011. All specimens were diagnosed to confirm whether they were hilar-CCA by at least two independent pathologists. Of the 41 specimens, 23 were from hilar-CCA which underwent curative resection, palliative drainage or operative biopsy procedure; with mean age of 61.3 years (range, 53 to 67 years; 13 males vs 10 females). Another 18 specimens were taken from cholangitis patients and liver transplantation patients as the control group. Among which, 14 patients with intrahepatic or/and extrahepatic calculus cholangitis who underwent operative biopsy and cholangiojejunostomy (mean age, 56.8 years; range, 43 to 66 years; 6 males vs 8 females) and four normal bile duct tissues (age range, 37 to 41 years, 3 males vs 1 female). All specimens were only used for research purposes. Patients and their family members were informed about the detail procedures and research purposes of specimen collection before operations. The entire procedure of specimen collection was in conformity with the ethical principles of medicine. In accordance with the guidelines of Central South University, prior informed consent from patients for specimen collection was acquired, and the Ethics Committee of Central South University approval was acquired. The serum hematoidin level of each patient was measured by Siemens ADIVA 1650 biochemical analyzer (Siemens, Tarrytown, NY, USA). The hematoidin concentration higher than 171 µM was defined as high level of hematoidin.16 All the specimens were divided into two parts: 1) for immunohistochemical study, tissues were fixed in 4% paraform formalin and embedded in paraffin. The embedded tissue specimens were then cut at 3 µm in thickness; 2) for quantitative real-time polymerase chain reaction (PCR) study, tissues were snap-frozen in liquid nitrogen and then stored at -80℃ for further RNA extraction. Quantification of CXCR4 mRNA expression was expressed as arbitrary unit in the results.
2. Immunohistochemistry and evaluation
The expression of CXCR4 in hilar-CCA, cholangitis or normal bile duct tissue specimens was detected immunohistochemically using Histostain-SP kits (Boster, Wuhan, China) according to the manufacturer's recommendations. Briefly, specimen sections were deparaffinized, placed in 0.01 mol/L citrate buffer (pH 6.0), heated for 15 minutes to unmask the antigen, and then incubated in methanol with 3% hydrogen peroxide for 15 minutes to inactivate endogenous peroxidase activity. After washing with phosphate-buffered saline, sections were incubated with protein block serum (Boster) for 15 minutes to block nonspecific reaction and were subsequently incubated with primary anti-CXCR4 antibody (dilution 1:50; Boster) at 4℃ overnight. After that, sections were treated with biotinylated secondary antibody and peroxidase-conjugated streptavidin for 20 minutes at room temperature and then washed with distilled water. The CXCR4 immunostaining was visualized via benzidine reaction. Sections were developed with benzidine and slightly counterstained with hematoxylin. Stained sections were then scanned using light microscope.
Three independent pathologists were invited to evaluate the specimen sections single-blindly. The intensity and staining percentage of CXCR4 expression were assessed.17 The expression of CXCR4 was semiquantitatively scored by the intensity of immunostaining as negative (0+), weak (1+), medium (2+), and strong (3+). CXCR4 over-expression was identified only in samples with a score of 2+ or greater.
3. Cell lines and cell culture
Hilar-CCA cell line QBC939 was kindly provided by Professor Wang from the Third Military Medical University, Chongqing, China. Hilar-CCA cell line SNU-1196 and human perineural cell line human perineurial cells, cell line code of Sciencell USA were purchased from Korean Cell Bank and ScienCell Research Laboratories (http://www.sciencellonline.com; Catalog no. 1710, San Diego, CA, USA), respectively.
Hilar-CCA cell lines (QBC939 and SNU-1196) were maintained in RPMI 1640 (Life Technologies, Grand Island, NY, USA) medium containing 10% fetal bovine serum and penicillin-streptomycin-glutamine (Life Technologies). HPC was cultured according to the instructions of ScienCell Research Laboratories.
4. CXCR4-small interference RNA construction and transfection to Hilar-CCA cell lines
Small interference RNA (siRNA) targeting CXCR4 (GeneBank no. NM_003467) or nonspecific control sequences (Table 1) were designed and purchased from GenePharma Corporation (Shanghai, China). Transient transfection for CXCR4-siRNA to human hilar-CCA cell lines QBC939 and SNU-1196 was performed using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions.
Table 1.
The Sequences of the Designed CXC Motif Chemokine Receptor 4-Specific Small Interference RNAs
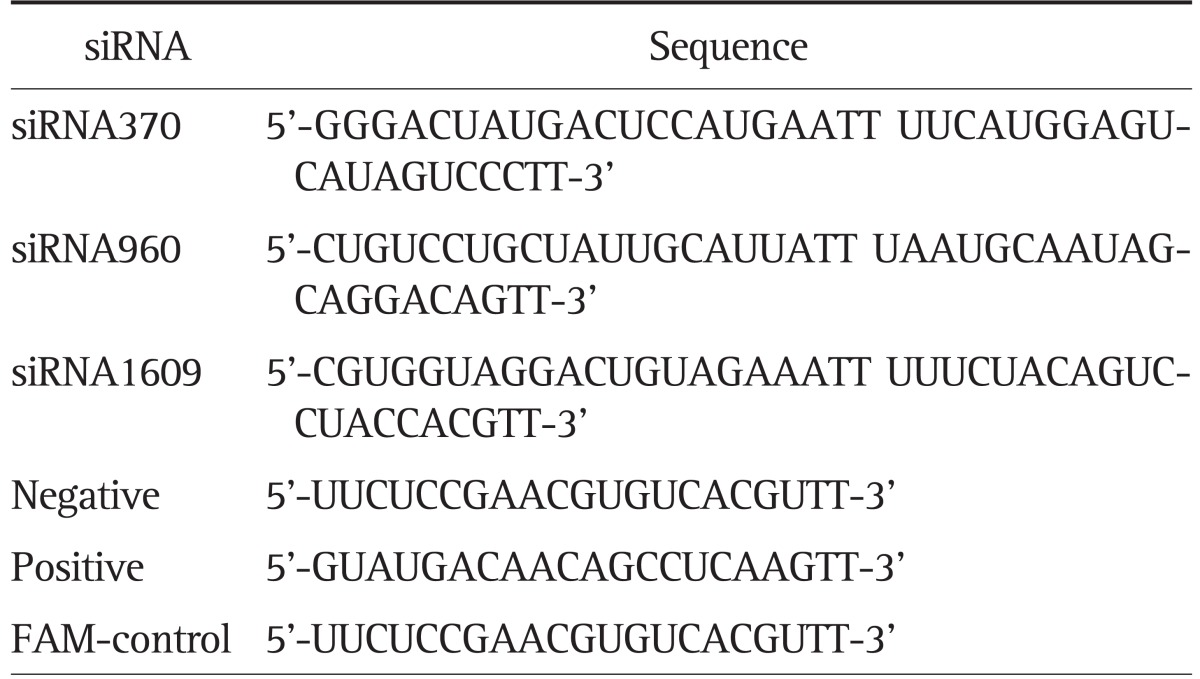
siRNA, small interference RNA; FAM, fluorescent labeled marked.
5. Measurement of CXCR4 protein and mRNA expression in hilar-CCA cell lines
CXCR4 protein expression in QBC939 was evaluated by Western blot analysis with three replicates. After washing with phosphate-buffered saline for three times, QBC939 and SNU-1196 cells were lysed in lysis buffer (mammalian protein extraction reagent; Pierce, Rockford, IL, USA) and centrifugated at 15,300 rpm for 15 minutes at 4℃. A total of 25 µg of protein was used as a sample. The electrophoresis experiment was performed in 10% sodium dodecyl sulfate-polyacrylamide gel. The CXCR4 protein was electrophoretically transferred onto a nitrocellulose membrane, and was then incubated in an ice bath with CXCR4 primary antibody (Catalog no. ab2074; Abcam HK) at voltage-stabilizer (Life Technologies). Finally, the CXCR4 protein was detected by secondary antibody conjugated to peroxidase (Zhongshan Golden Bridge Biotechnology, Beijing, China) and benzidine reaction.
The expression of CXCR4 mRNA was detected using quantitative real-time PCR assay with three replicates. Total RNA was extracted from cell lines QBC939 by Trizol reagent (Life Technologies). Quantitative PCR assay was performed by prepared CXCR4 and glyceraldehyde phosphate dehydrogenase (GAPDH) specific primers using ABI Prism 7700 sequence detection system (PE Applied Biosystems, Warrington, UK). The CXCR4 and GAPDH primers were as follows: 5'-CCGTGGCAAACTGGTACTTT-3' (CXCR4, forward); 5'-GACGCCAACATAGACCACCT-3' (CXCR4, reverse); 5'-CAATGACCCCTTCATTGACC-3' (GAPDH, forward); 5'-GACAAGCTTCCCGTTCTCAG-3' (GAPDH, reverse). PCR reaction was done with the TaqMan Universal PCR Master Mix (PE Applied Biosystems). Cycling conditions were: incubation at 50℃ for 2 minutes, 10 minutes at 95℃, and 40 cycles of 15 seconds at 95℃ and 1 minute at 60℃. CXCR4 was normalized (ΔCt) to GAPDH by subtracting the cycle threshold (Ct) value of GAPDH from the Ct value of CXCR4.
6. Perineural cell and hilar-CCA cell coculture migration assay
The NI ability of hilar-CCA was evaluated by hilar-CCA and perineural cells coculture migration assay with three replicates. Hilar-CCA cells was cocultured with perineural cells using Matrigel invasion chamber purchased from BD Biocoat Cellware (San Jose, CA, USA) as previously reported.18 Medium (0.5 µL) containing 5×105 hilar-CCA cells was added to the upper chamber, and 0.5 mL of either medium alone or medium containing 1 to 2×104 HPC cells was added to the lower chamber. Chambers were then incubated for 48 hours at 37℃ under 5% CO2. After experiment, hilar-CCA cells on the upper surface were removed, and cells that had migrated to the lower surface were stained by hematoxylin. The ratio of the migrated cells number in the siRNA interference group or of the migrated cells number in the control group was identified as migration index.
7. Apoptotic measurement using Hoechst and propidium iodine double staining
To further confirmed the apoptotic measurement by cytometry, hilar-CCA cells after CXCR4-siRNA treatments were stained with Hoechst 33342 (5 µg/mL; Sigma, St. Louis, MO, USA) and propidium iodide (PI; 5 µg/mL; Sigma) in the medium to label live cells for 15 minutes. Cells with PI positive staining were calculated against the total cell number shown by Hoechst staining.
8. Hilar-CCA proliferation assay
QBC939 cells were seeded onto 96-well plates (1×104 cells per well). The cell proliferation was assessed using 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-tetrazolium (MTT) assay (Promega, Madison, WI, USA). Plates were incubated for 24 hours at 37℃ under 5% CO2 for approximately 3 hours. After dissolve with dimethyl sulfoxide, the absorbance of cellular MTT metabolite was detected at 570 nm wavelength. Mean optical density value was documented as result, and the MTT assay was performed for three times to calculate the cell proliferation rate.
9. Statistical analysis
Statistical analysis was performed using analysis of variance, followed by the Turkey test using SigmaStat software (Systat Software Inc., San Jose, CA, USA). Results were considered statistically significant at least p<0.05.
RESULTS
1. Overexpression of CXCR4 correlates to NI and lymph node metastasis in hilar-CCA tissues
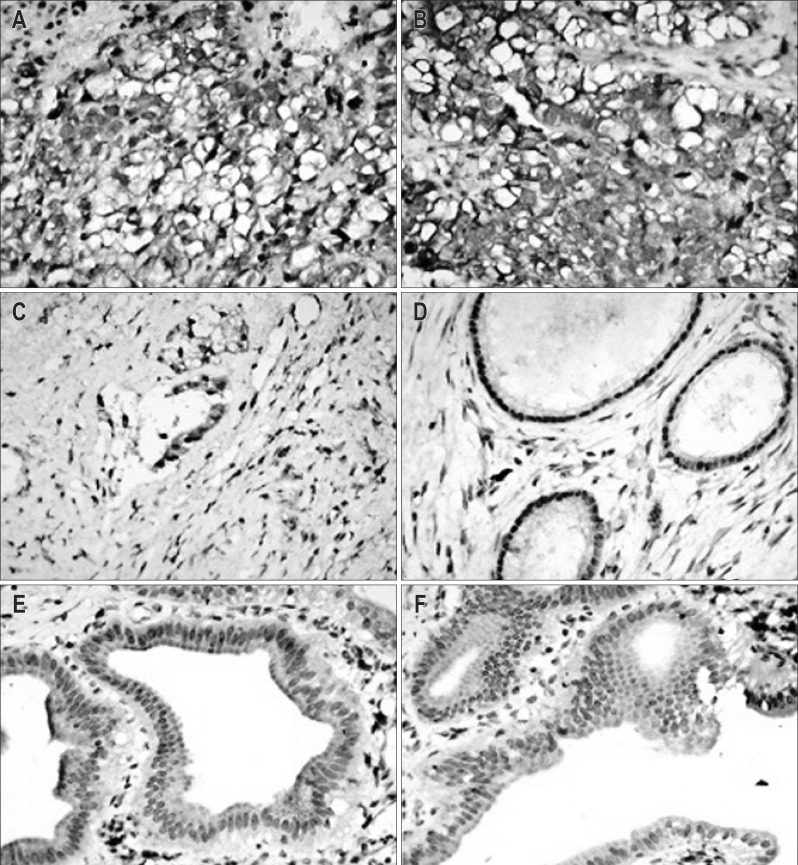
Expression pattern of CXCR4 protein in clinical samples was analyzed immunohistochemically. Results showed that CXCR4 was primarily expressed in membrane, cytoplasm, and/or biliary luminal surface. CXCR4 in 15 out of 23 (65.22%) hilar-CCA cases were detected as overexpression, five out of 14 (38.46%) cholangitis cases were detected as normal expression, whereas in all (n=15) of the normal bile duct tissues were detected as nonexpression, with a statistically significant difference (p=0.0001) (Fig. 1).
Fig. 1.
Immunohistochemical analysis of CXC motif chemokine receptor 4 (CXCR4) in different tissues. (A, B) CXCR4 overexpression in hilar-cholangiocarcinoma tissues. (C, D) CXCR4 expression in cholangitis cases. (E, F) CXCR4 expression in normal bile duct tissues (A-F, ×400).
These findings also significantly correlate with clinicopathological features such as lymph node metastasis and NI. Among the 15 CXCR4 overexpression cases (indicated by quantitative real-time PCR results), 11 cases (73.3%) showed high hematoidin concentration (above 171 µmol/L), 13 (86.7%) cases showed bigger tumor diameters (above 2 cm) than those detected in CXCR4-normal expression cases (equal or less than 2 cm) and 12 cases (80.0%) showed liver metastasis and tumor differentiation, whereas there were only three (37.5%) with liver metastasis and four (50%) with tumor differentiation in eight cases of CXCR4 nonoverexpression. In addition, 14 of 15 (93.3%) showed remarkably high association between CXCR4 overexpression with lymph node invasion and NI, with a statistically significant difference (p<0.05) (Table 2). Obviously, at the histological level, there was a significant correlation between CXCR4 overexpression and tumor development.
Table 2.
CXC Motif Chemokine Receptor 4 (CXCR4)-Overexpression-Induced Phenotypes in Human Cholangiocarcinoma Cells and the Comparison of Clinicopathological Variables Dependent on the CXCR4 mRNA Expression Level
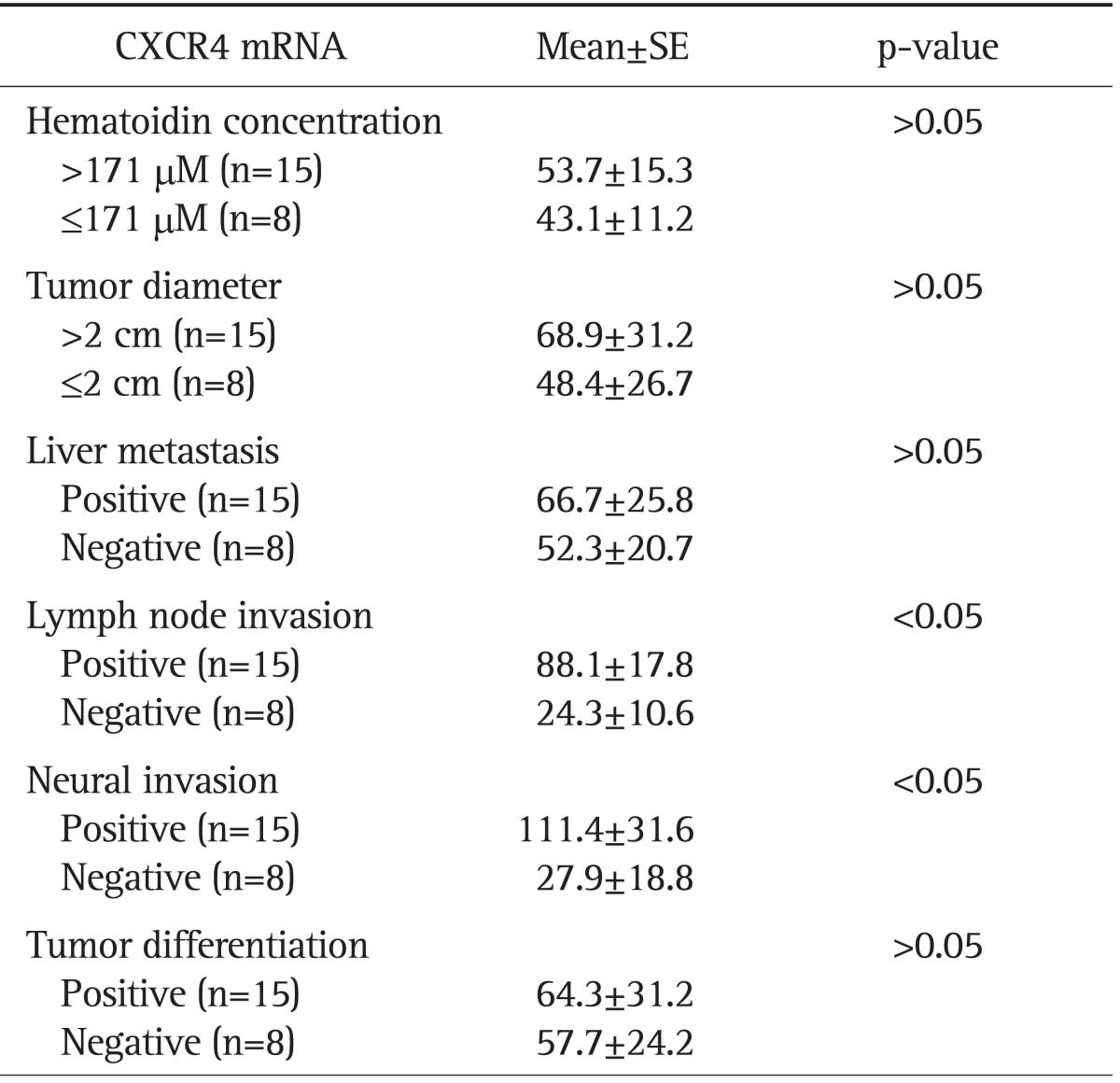
SE, standard error.
2. siRNA-CXCR4 suppresses CXCR4 expression in QBC939 cells
CXCR4 mRNA expression was examined in a panel of two hilar-CCA cell lines QBC939 and SNU-1196 by real-time PCR. Results showed that the cell line QBC939 had the highest expression level of CXCR4 mRNA (data not shown). In this regard, the cell line QBC939 was chosen for following studies.
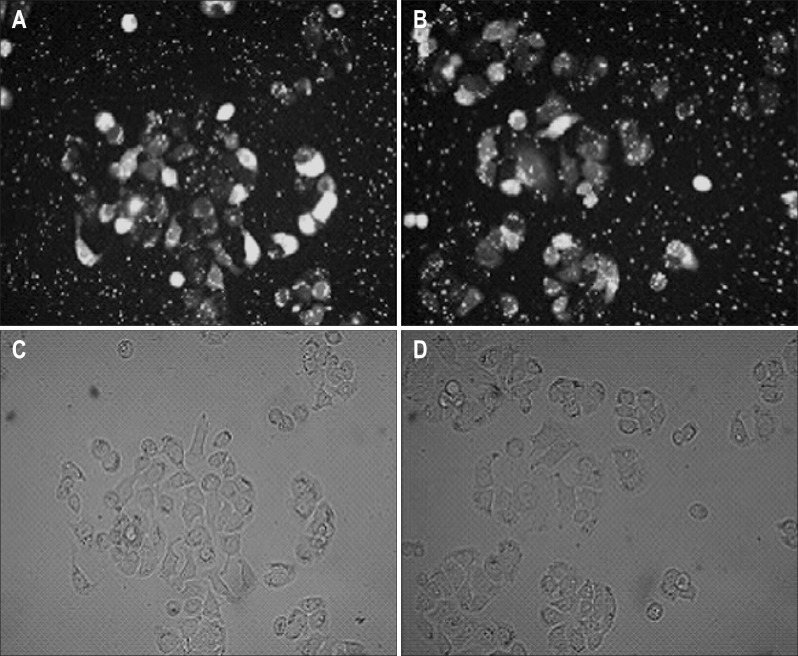
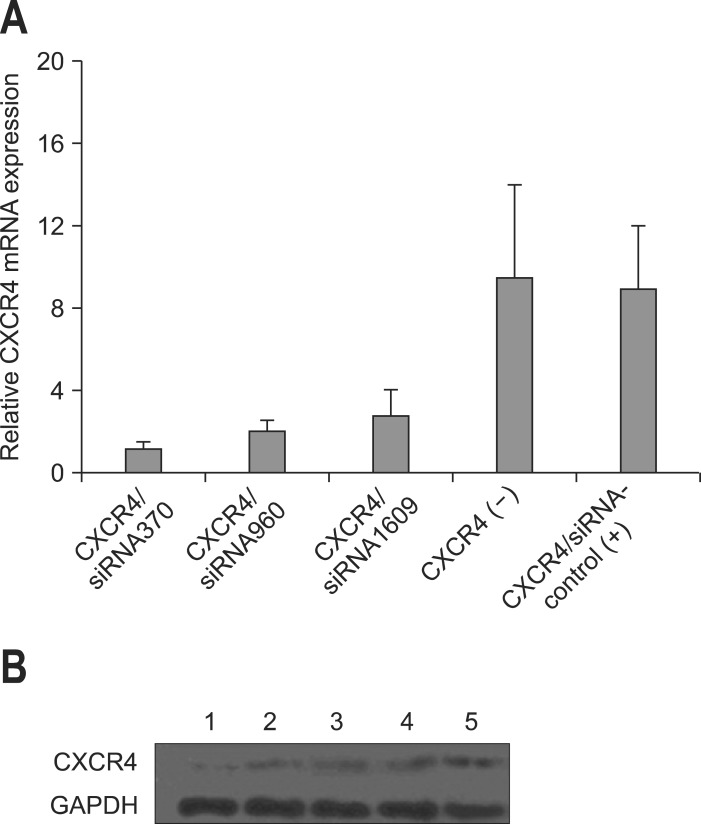
QBC939 was effectively transfected with CXCR4 siRNA oligos for 48 hours by using Lipofectamine 2000 transfection kit. Transfection efficiency was detected by fluorescent assay, which showed 80% of efficiency after 48-hour transfection (Fig. 2). The silencing effects of three different CXCR4 specific siRNAs in QBC939 cells were evaluated using real-time reverse transcription-PCR and Western blot. siRNA-CXCR4 transfectants showed a remarkably decreased expression of CXCR4. Inhibition ratio of 86.42%, 76.63%, and 69.20% for CXCR4 mRNA by using siRNA370, siRNA960, and siRNA1609 were achieved respectively when compared to untreated QBC939 cells, while siRNA-control showed no effects (Fig. 3A). Western blot analysis confirmed the down-regulation of CXCR4 protein by transfection of siRNA expressing vector (Fig. 3B).
Fig. 2.
Small interference RNA plasmid vector transfection of QBC939 cells detected by fluoroscopic examination (×400). (A, B) Representative fluorescent images of CXCR4 siRNA 370 and 960, respectively. (C, D) Representative bright field images of CXCR4 siRNA 370 and 960, respectively.
Fig. 3.
CXC motif chemokine receptor 4 (CXCR4)-small interference RNA (siRNA) results in the reduction of CXCR4 mRNA and protein levels in QBC939 cells. (A) Relative mRNA levels were analyzed by quantitative reverse transcription-polymerase chain reaction. Approximately 100 nmol/L estrogen was used to induce the control samples. (B) Western blot analysis of CXCR4 protein expression. Glyceraldehyde phosphate dehydrogenase (GAPDH) was used as a loading control. Lane 1, CXCR4/siRNA370; lane 2, CXCR4/siRNA960; lane 3, CXCR4/siRNA1609; lane 4, negative control (CXCR4[-]); lane 5, positive control (CXCR4/siRNA-control); lane 6, induced sample.
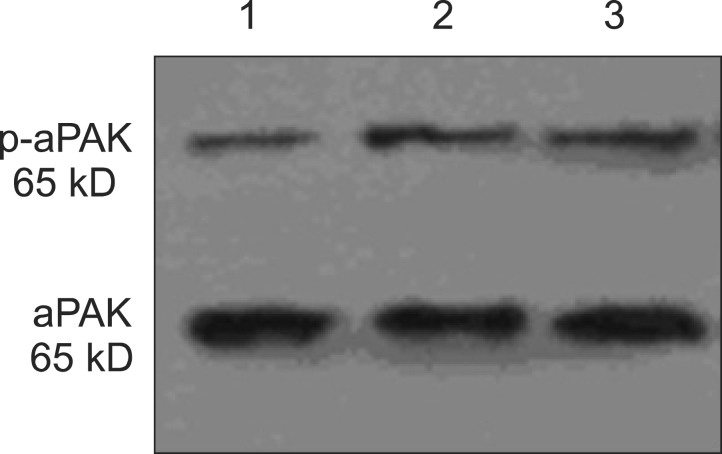
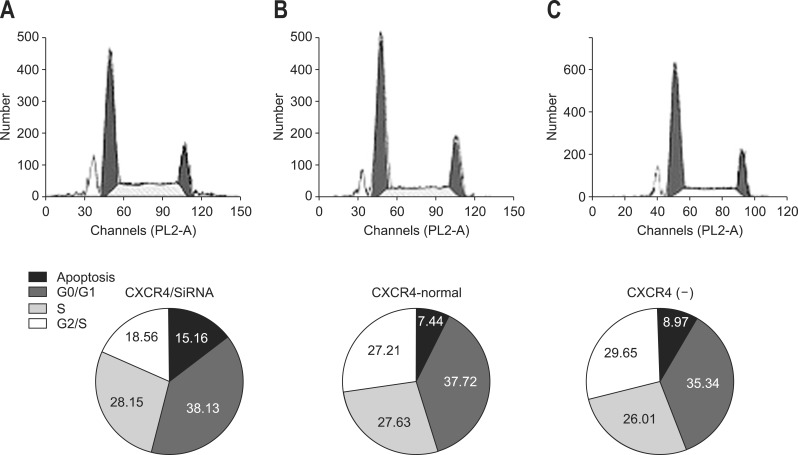
Activated p21-activated kinase (aPak) is an important tissue specific regulator of p53 in many tissues (e.g., tumor tissues). It inhibits p53 activity by interacting with ataxia-telangiectasia mutated protein kinase to prevent cell apoptosis. Western blot result showed the aPak expression level was depressed since CXCR4 was silenced by its specific siRNA (Fig. 4). In addition, the cell cycle phase observations provided direct evidence that CXCR4 inhibition increased the apoptotic rate (AR) of hilar-CCA cells, probably through the action of aPak. The proportion of cells that have undergone apoptosis increased from 2.46% of QBC939 to 15.16% of QBC939/siRNA-CXCR4 cells. The AR of QBC939 cells and QBC939/siRNA-control cells was 7.44% and 8.97%, respectively (Fig. 5). Apoptosis evaluation using Hoechst and PI further confirmed the AR results from cytometry (Table 3).
Fig. 4.
Representative Western blot results for phosphorylated activated p21-activated kinase (aPak) and total aPAK in cultured QBC939 cells. (A) CRCX4-small interference RNA (siRNA). (B) CXCR4-normal. (C) Negative control siRNA. 1, CRCX40siRNA; 2, CXCR4-normal; 3, negative control siRNA.
Fig. 5.
Cell cycle detected by flow cytometry. The upper pictures demonstrate the influence of small interference RNA (siRNA) on the cell cycle. The pie pictures demonstrate the influence of transfection siRNA on cell apoptosis. (A) CXC motif chemokine receptor 4 (CRCX4)-siRNA. (B) CXCR4-normal. (C) Negative control siRNA.
Table 3.
Apoptotic Ratio Evaluated by Hoechst 33342 and Propidium Iodine Double Staining

Cells with propidium iodine-positive staining were compared with the total cell number shown by Hoechst staining.
CXCR4, CXC motif chemokine receptor 4; siRNA, small interference RNA.
*p<0.01 compared with the CXCR4-siRNA group.
3. CXCR4 silencing reduces the NI by coculture of perineural cells and QBC939 cells in vitro
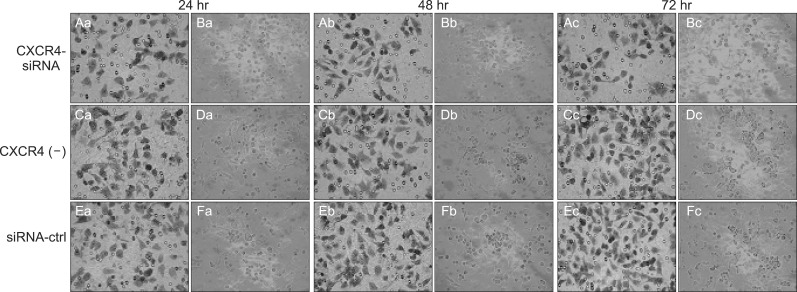
The results showed that in cocultured group during 24 to 72 hour incubation for QBC939 and QBC939/siRNA-control cells had a similar ability to pass through the Matrigel coated filter, because the numbers of invading cells were roughly equal (Fig. 6C-F). The number of QBC939/siRNA370 cells (25.60±3.28) passing through the Matrigel was significantly lower than the numbers of QBC939 (55.80±5.03) and QBC939/siRNA-control cells (p<0.01). After 72-hour coculture, a more than 300% increase of migration between QBC939/siRNA-CXCR4 cells and QBC939/siRNA-control cells was observed (Fig. 6A-D).
Fig. 6.
Coculture of human neural cells and human CCA QBC939 cells to test the effects of CXC motif chemokine receptor 4 (CXCR4)-small interference RNA on neural invasion. (A, C, E) Cellular staining of cells in the lower chamber. (B, D, F) Cellular staining of cells in the central membrane. Crystal violet staining demonstrated that the cells passed through the filter and attached to the lower chamber (×400). The data were obtained from three independent experiments.
4. CXCR4 silencing reduces the proliferation of QBC939 cells
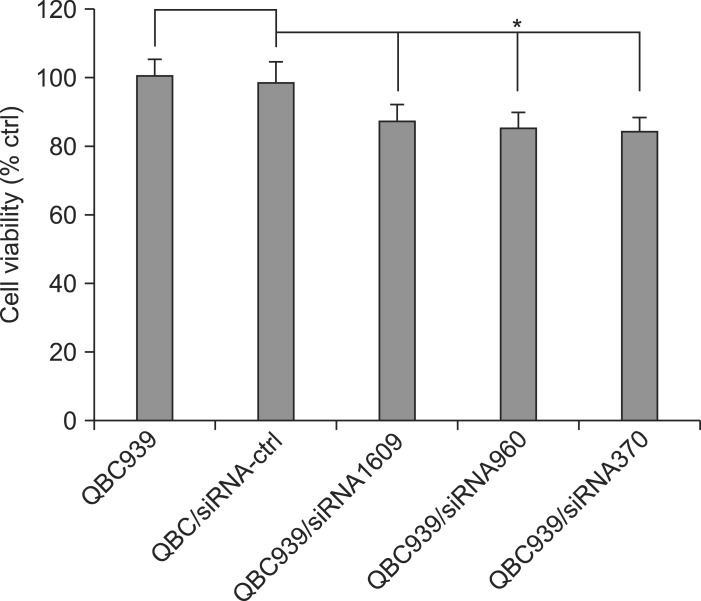
The proliferation of QBC939, QBC939/siRNA-control, and QBC939/siRNA-CXCR4 was determined respectively. As shown in Table 3, compared with QBC939, the proliferation of QBC939/siRNA-CXCR4 was significantly inhibited to 87.07% (QBC939/siRNA1609), 85.25% (QBC939/siRNA960), and 84.63% (QBC939/siRNA370) at 72 hours, respectively. There was no significant difference among those siRNA treatments (p>0.05) (Fig. 7).
Fig. 7.
Cell viability after CXC motif chemokine receptor 4 (CXCR4)-small interference RNA (siRNA) treatment. Cell viability was determined using the MTT assay.
*p<0.01 compared with QBC939/QBC939-siRNA-control and CXCR4/siRNAs (siRNA1609, siRNA960, and siRNA370).
DISCUSSION
Recent direct evidence shows that overexpression of CXCR4 promotes cancer cell progression and metastasis in several types of tumors, such as pancreatic cancer,12 breast cancer,19 colorectal cancer,20 and bile duct cancer.21 In this study, statistical analysis of immunohistochemistry data showed an obvious positive relationship between the over-expression of CXCR4 and the incidence of hilar-CCA. Down-regulation of endogenous CXCR4 by siRNA inhibition caused reduction of tumor diameter (data not shown), cancer cell apoptosis and decreased motility and invasion potentials. CXCR4 expression was effectively inhibited at both mRNA and protein levels by siRNA370, while the siRNA960 and siRNA1609 were relatively less efficient. These results indicated that siRNA targeting different sites of the same mRNA might be different in silencing efficiency. It should be noted that although we demonstrated that silencing of CXCR4 reduced the activity of aPAK, which is a key regulator of the apoptotic pathway, more investigations are warranted to study the mechanistic aspect of apoptotic regulation, including both intrinsic and extrinsic apoptotic pathways.22
It is considered that the repression of CXCR4 alone is not sufficient to explain the decreased NI and lymph node invasion. The possible involvement of matrix metalloproteinases (MMPs), glial cell line-derived neurotrophic factor, and L1 cell adhesion molecule in tumor tissue that control perineural invasion has been reported by Chinni et al.,23 and Ben et al.24 Particularly, MMPs was identified as one of triggers to introduce tumor migration by hydrolyzing extracellular matrix. Down-regulation of CXCR4 expression in hilar-CCA cells reduced the secretion of MMPs, thus inhibited the invasion ability of hilar-CCA cells through the reconstituted basement membrane in vitro.
The effect of CXCR4 silencing on the proliferation of QBC939 cells was examined. The proliferation potential of QBC939/siRNA-CXCR4 cells was slightly suppressed when compared with that of the control QBC939 cells. Activation of CXCR4 was reported to promote the proliferation of gastric cancer,25 pancreatic cancer,12 and squamous cell carcinoma in head and neck.26 Activated CXCR4 is a heterotrimeric complex with G-protein, which may activate phosphoinositide-3 kinase and phospholipase C, to further activate p21-activated kinase (PAK), leading to tumor cell polarization and adhesion for migration.27,28 However, proliferation potential depression induced by CXCR4 overexpression was found in intrahepatic and hilar-CCA cell lines.28 This suggests that CXCR4 obtained different roles for proliferation among different tumor cells. In conclusion, this study showed that the CXCR4 is probably correlated with the incidence of hilar-CCA clinically because silencing of CXCR4 by its specific RNAi inhibited NI and lymph node metastasis of human hilar-CCA cell line QBC939 in vitro. Our findings suggested that CXCR4 might be a promising target for CCA treatment against NI and lymph node metastasis.
ACKNOWLEDGEMENTS
This study was supported by the Freedom Explore Program of Central South University (2011QNZT162).
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Carriaga MT, Henson DE. Liver, gallbladder, extrahepatic bile ducts, and pancreas. Cancer. 1995;75(1 Suppl):171–190. doi: 10.1002/1097-0142(19950101)75:1+<171::aid-cncr2820751306>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 2.Shaib Y, El-Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis. 2004;24:115–125. doi: 10.1055/s-2004-828889. [DOI] [PubMed] [Google Scholar]
- 3.DeOliveira ML, Cunningham SC, Cameron JL, et al. Cholangiocarcinoma: thirty-one-year experience with 564 patients at a single institution. Ann Surg. 2007;245:755–762. doi: 10.1097/01.sla.0000251366.62632.d3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramacciato G, Nigri G, Bellagamba R, et al. Univariate and multivariate analysis of prognostic factors in the surgical treatment of hilar cholangiocarcinoma. Am Surg. 2010;76:1260–1268. [PubMed] [Google Scholar]
- 5.Zhang BY, Lu Y, Dong Q, Sun CD, Mu P. Surgical treatment and prognostic analysis of 93 cases of hilar cholangiocarcinoma. Am J Med Sci. 2010;339:221–224. doi: 10.1097/MAJ.0b013e3181c7c8b4. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen KT, Steel J, Vanounou T, et al. Initial presentation and management of hilar and peripheral cholangiocarcinoma: is a node-positive status or potential margin-positive result a contraindication to resection? Ann Surg Oncol. 2009;16:3308–3315. doi: 10.1245/s10434-009-0701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen FZ, Zhang BY, Feng YJ, et al. Current research in perineural invasion of cholangiocarcinoma. J Exp Clin Cancer Res. 2010;29:24. doi: 10.1186/1756-9966-29-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Z, Kleeff J, Kayed H, et al. Nerve growth factor and enhancement of proliferation, invasion, and tumorigenicity of pancreatic cancer cells. Mol Carcinog. 2002;35:138–147. doi: 10.1002/mc.10083. [DOI] [PubMed] [Google Scholar]
- 9.Bhuiya MR, Nimura Y, Kamiya J, et al. Clinicopathologic studies on perineural invasion of bile duct carcinoma. Ann Surg. 1992;215:344–349. doi: 10.1097/00000658-199204000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schreiber SC, Giehl K, Kastilan C, et al. Polysialylated NCAM represses E-cadherin-mediated cell-cell adhesion in pancreatic tumor cells. Gastroenterology. 2008;134:1555–1566. doi: 10.1053/j.gastro.2008.02.023. [DOI] [PubMed] [Google Scholar]
- 11.Gleichmann M, Gillen C, Czardybon M, et al. Cloning and characterization of SDF-1gamma, a novel SDF-1 chemokine transcript with developmentally regulated expression in the nervous system. Eur J Neurosci. 2000;12:1857–1866. doi: 10.1046/j.1460-9568.2000.00048.x. [DOI] [PubMed] [Google Scholar]
- 12.Katayama A, Ogino T, Bandoh N, Nonaka S, Harabuchi Y. Expression of CXCR4 and its down-regulation by IFN-gamma in head and neck squamous cell carcinoma. Clin Cancer Res. 2005;11:2937–2946. doi: 10.1158/1078-0432.CCR-04-1470. [DOI] [PubMed] [Google Scholar]
- 13.Uchida D, Begum NM, Tomizuka Y, et al. Acquisition of lymph node, but not distant metastatic potentials, by the overexpression of CXCR4 in human oral squamous cell carcinoma. Lab Invest. 2004;84:1538–1546. doi: 10.1038/labinvest.3700190. [DOI] [PubMed] [Google Scholar]
- 14.Marchesi F, Monti P, Leone BE, et al. Increased survival, proliferation, and migration in metastatic human pancreatic tumor cells expressing functional CXCR4. Cancer Res. 2004;64:8420–8427. doi: 10.1158/0008-5472.CAN-04-1343. [DOI] [PubMed] [Google Scholar]
- 15.Leelawat K, Leelawat S, Narong S, Hongeng S. Roles of the MEK1/2 and AKT pathways in CXCL12/CXCR4 induced cholangiocarcinoma cell invasion. World J Gastroenterol. 2007;13:1561–1568. doi: 10.3748/wjg.v13.i10.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, Qiu L, Ma J, Huang Q, Ren Y. Analysis of factors influencing prognosis of patients underwent hilar carcinoma without resection. J Hepatobiliary Surg. 2008;(4):281–283. [Google Scholar]
- 17.Ku JL, Yoon KA, Kim IJ, et al. Establishment and characterisation of six human biliary tract cancer cell lines. Br J Cancer. 2002;87:187–193. doi: 10.1038/sj.bjc.6600440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendoza M, Khanna C. Revisiting the seed and soil in cancer metastasis. Int J Biochem Cell Biol. 2009;41:1452–1462. doi: 10.1016/j.biocel.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holland JD, Kochetkova M, Akekawatchai C, Dottore M, Lopez A, McColl SR. Differential functional activation of chemokine receptor CXCR4 is mediated by G proteins in breast cancer cells. Cancer Res. 2006;66:4117–4124. doi: 10.1158/0008-5472.CAN-05-1631. [DOI] [PubMed] [Google Scholar]
- 20.Wang SC, Lin JK, Wang HS, Yang SH, Li AF, Chang SC. Nuclear expression of CXCR4 is associated with advanced colorectal cancer. Int J Colorectal Dis. 2010;25:1185–1191. doi: 10.1007/s00384-010-0999-1. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto K, Tajima H, Nakanuma S, et al. Angiotensin II enhances epithelial-to-mesenchymal transition through the interaction between activated hepatic stellate cells and the stromal cell-derived factor-1/CXCR4 axis in intrahepatic cholangiocarcinoma. Int J Oncol. 2012;41:573–582. doi: 10.3892/ijo.2012.1499. [DOI] [PubMed] [Google Scholar]
- 22.Ouyang L, Shi Z, Zhao S, et al. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–498. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chinni SR, Sivalogan S, Dong Z, et al. CXCL12/CXCR4 signaling activates Akt-1 and MMP-9 expression in prostate cancer cells: the role of bone microenvironment-associated CXCL12. Prostate. 2006;66:32–48. doi: 10.1002/pros.20318. [DOI] [PubMed] [Google Scholar]
- 24.Ben QW, Wang JC, Liu J, et al. Positive expression of L1-CAM is associated with perineural invasion and poor outcome in pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2010;17:2213–2221. doi: 10.1245/s10434-010-0955-x. [DOI] [PubMed] [Google Scholar]
- 25.Zhao BC, Wang ZJ, Mao WZ, et al. CXCR4/SDF-1 axis is involved in lymph node metastasis of gastric carcinoma. World J Gastroenterol. 2011;17:2389–2396. doi: 10.3748/wjg.v17.i19.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katayama A, Ogino T, Bandoh N, Nonaka S, Harabuchi Y. Expression of CXCR4 and its down-regulation by IFN-gamma in head and neck squamous cell carcinoma. Clin Cancer Res. 2005;11:2937–2946. doi: 10.1158/1078-0432.CCR-04-1470. [DOI] [PubMed] [Google Scholar]
- 27.Barbieri F, Bajetto A, Florio T. Role of chemokine network in the development and progression of ovarian cancer: a potential novel pharmacological target. J Oncol. 2010;2010:426956. doi: 10.1155/2010/426956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16:2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]