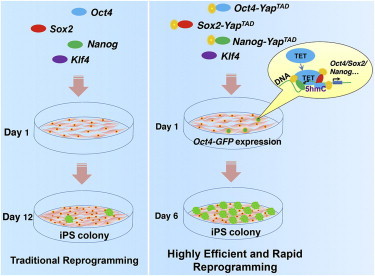
Summary
Somatic cell reprogramming toward induced pluripotent stem cells (iPSCs) holds great promise in future regenerative medicine. However, the reprogramming process mediated by the traditional defined factors (OSMK) is slow and extremely inefficient. Here, we develop a combination of modified reprogramming factors (OySyNyK) in which the transactivation domain of the Yes-associated protein is fused to defined factors and establish a highly efficient and rapid reprogramming system. We show that the efficiency of OySyNyK-induced iPSCs is up to 100-fold higher than the OSNK and the reprogramming by OySyNyK is very rapid and is initiated in 24 hr. We find that OySyNyK factors significantly increase Tet1 expression at the early stage and interact with Tet1/2 to promote reprogramming. Our studies not only establish a rapid and highly efficient iPSC reprogramming system but also uncover a mechanism by which engineered factors coordinate with TETs to regulate 5hmC-mediated epigenetic control.
Graphical Abstract
Highlights
-
•
A combination of modified reprogramming factors (OySyNyK) is developed
-
•
A highly efficient and rapid reprogramming system is established
-
•
TET1/2 proteins are involved in rapid iPSC induction by OySyNyK
-
•
OySyNyK factors coordinate with TET proteins to promote rapid reprogramming
The reprogramming process mediated by the traditional defined factors (OSMK) is slow and inefficient. Sun, Jin, Chen, and colleagues have developed a combination of modified reprogramming factors (OySyNyK) in which the transactivation domain of the Yes-associated protein is fused to defined factors and establish a highly efficient and rapid reprogramming system.
Introduction
Induced pluripotent stem cells (iPSCs) are generated by nuclear reprogramming through introducing the defined Yamanaka factors (OSMK) (Takahashi and Yamanaka, 2006; Yamanaka and Blau, 2010; Yu et al., 2009). This method not only holds great promise in future regenerative medicine, such as the generation of patient-specific cells for tissue replacement, but also provides an excellent tool to solve fundamental biological questions regarding dedifferentiation (Yamanaka and Blau, 2010). However, the molecular mechanisms underlying somatic cell reprogramming toward iPSCs have been elusive, particularly because of the technical challenges regarding the low efficiency of iPSC generation using the original method.
It has been demonstrated that three pluripotency factors, Oct4 (also known as Pou5f1), Sox2, and Nanog, play a central role in maintenance of the undifferentiated state and pluripotency of embryonic stem cells (ESCs) (Young, 2011). Although the native forms of these factors have been widely applied to iPSC generation, their relatively low transactivation activity is still a barrier for somatic cell reprogramming (Wang et al., 2011). Recent studies have shown that the modification of OCT4, SOX2, and NANOG provides a new approach to overcome the barrier of the low efficiency of iPSC generation (Hirai et al., 2011; Wang et al., 2011).
The Yes-associated protein (YAP) has been demonstrated to be a transcriptional coactivator with a potent transactivation domain (TAD) in the C-terminal region, and ectopic expression of YAP promotes cell growth and induces tumor formation (Overholtzer et al., 2006; Zhao et al., 2009). YAP also has a critical role in maintenance of stem cell pluripotency (Lian et al., 2010).
To improve the transcriptional activity of OCT4, SOX2, and NANOG, we generated a set of modified reprogramming factors by fusing the TAD of YAP with these factors. Using these modified factors (OySyNyK), we established a system that could induce highly efficient somatic cell reprogramming toward iPSCs. Importantly, we show that the reprogramming reporter Oct4-GFP could be activated by OySyNyK at the very early stage (initiated at around 24 hr versus 5 days by OSNK), suggesting that OySyNyK-induced reprogramming is very rapid. Further mechanistic studies revealed that OySyNyK factors significantly increase the expression of Tet genes, particularly TET1, at the early stage, which interact with the defined factors, SOX2 and NANOG, to promote rapid and highly efficient somatic cell reprogramming.
Results and Discussion
Establishment of a Rapid and Highly Efficient Cell Reprogramming System by Modified Factors
We first attempted to establish a highly efficient and rapid somatic cell reprogramming system. The efficiency of iPSC generation was very low (around 0.1%) when mouse embryonic fibroblasts (MEFs) were transduced with the native forms of the defined reprogramming factors (OSMK) (Takahashi and Yamanaka, 2006). Recent studies have shown that modified versions of these defined factors with improved transcriptional activity are able to increase the reprogramming efficiency of iPSC generation (Hirai et al., 2011; Wang et al., 2011). Considering that YAP functions as a potent transcriptional coactivator and plays a critical role in maintenance of stem cell pluripotency (Lian et al., 2010), we hypothesized that modified versions of the defined factors fused with the TAD of YAP would significantly promote the process of somatic cell reprogramming toward pluripotency. We therefore constructed modified forms of the reprogramming factors, including OCT4-YAPTAD (Oy), SOX2-YAPTAD (Sy), and NANOG-YAPTAD (Ny), by fusing the TAD of YAP to OCT4 (O), SOX2 (S), and NANOG (N) (Figure 1A). As shown in a luciferase reporter assay, the combination of modified factors (OySyNy) exhibited about a 22-fold greater transactivation activity than that of the combination of native OSN factors in activating the 6XCR4 reporter (Figure 1B). We next employed a retrovirus-mediated method to introduce these modified factors combined with native Klf4 (OySyNyK) into MEFs derived from Oct4-GFP mice (Lengner et al., 2007) and then measured the efficiency of GFP-positive colony formation (Figure 1C). When wild-type OSNK were introduced into MEFs carrying the Oct4-GFP transgene, we obtained 69 ± 7 GFP-positive colonies from 2.5 × 104 transduced MEFs at day 16 (Figure 1D). In contrast, when OySyNyK were transduced into Oct4-GFP MEFs, we obtained 2,371 ± 176 GFP-positive colonies from 2.5 × 104 transduced MEFs at day 7 (Figure 1D). In support of this, the flow-cytometric analysis revealed that the ratio of GFP-positive cells was significantly increased by OySyNyK with time when compared with the OSNK control (Figure 1E). Taken together, these data indicated that the combination of modified reprogramming factors significantly increased the efficiency and rate of GFP-positive colony formation.
Figure 1.
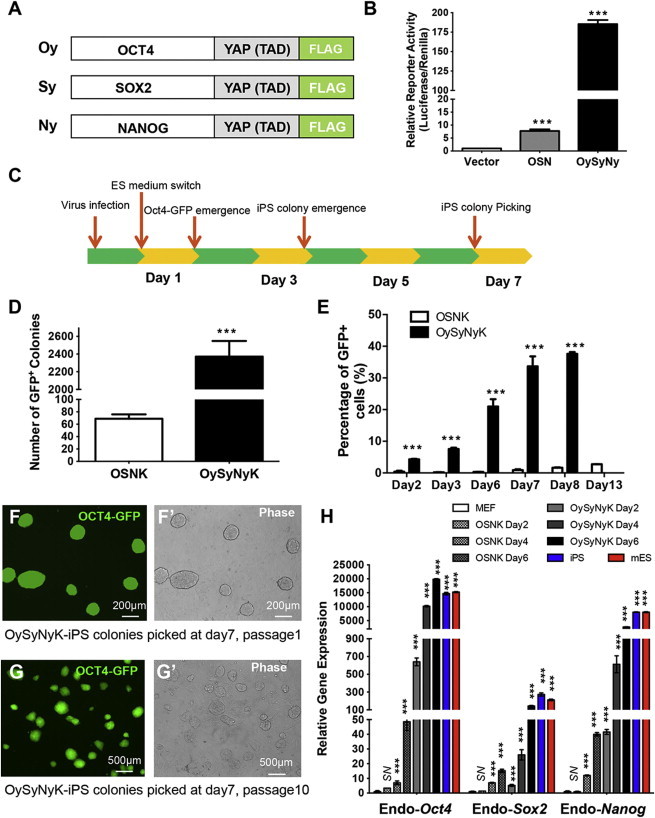
Establishment of a Rapid and Highly Efficient Cell Reprogramming System by Modified Factors
(A) Schematic diagram of construction of modified reprogramming factors, including OCT4-YAPTAD (Oy), SOX2-YAPTAD (Sy), and NANOG-YAPTAD (Ny). Coding sequences of murine Oct4, Sox2, Nanog were fused with murine Yap transcription activation domain (TAD) (amino acids 275–489) in the C terminus directly to generate Oct4-YAPTAD (Oy), Sox2-YAPTAD (Sy), and Nanog-YAPTAD (Ny) expression constructs, respectively. The modified factors were then cloned into pMXs-retroviral vector.
(B) HEK293 cells were transfected in a 24-well plate with various expression plasmids along with a pGL4.2-basic-6XCR4 vector and Renilla control vector using the calcium phosphate precipitation method. Thirty-six hours posttransfection, cells were lysed for the measurement of luciferase activity. Firefly luciferase activities were normalized based on the Renilla activity. ∗∗∗p < 0.001 versus control groups. NS, not significant.
(C) Schematic diagram describing the procedure for the generation of iPSCs induced by the modified factor combination OySyNyK. Oct4-GFP MEFs were used to generate iPSCs.
(D) Statistical summary showing the different efficiencies of GFP-positive colony formation induced by OSNK or OySyNyK. In this assay, OySyNyK- and OSNK-induced OCT4-GFP-positive colonies were counted at day 7 and day 16, respectively. ∗∗∗p < 0.001 versus control groups.
(E) Dynamics of the percentage of GFP-positive cells in OSNK- or OySyNyK-infected MEFs during somatic cell reprogramming. ∗∗∗p < 0.001 versus control groups.
(F/F′ and G/G′) Fluorescence (F and G) and bright-field (F′ and G′) images showing OySyNyK-induced iPSCs at passage 1 (F) or passage 10 (G).
(H) qRT-PCR assays showing the expression levels of endogenous pluripotency markers Oct4, Sox2, and Nanog in OSNK- and OySyNyK-transduced MEFs at different time points. Both iPSCs and ESCs were included for the comparison. OSNK- and OySyNyK-transduced MEFs groups were compared to MEF groups. ∗∗∗p < 0.001 versus control groups.
Data from (B), (D), (E), and (H) are representative of at least three independent experiments (mean and SD of triplicate assays). Groups were compared using the Student’s t test. N.S, not significant; ∗∗∗p < 0.001 versus control groups.
We noted that the OySyNyK-induced GFP-positive colonies morphologically resembled normal ESCs and typical iPSCs induced by OSNK (Figures 1F, 1F′, 1G, and 1G′). We thus established iPSC lines from the OySyNyK-induced colonies (Figure S1A available online) and further evaluated the quality of these iPSCs by performing quantitative RT-PCR (qRT-PCR) analysis to examine the expression of endogenous pluripotency genes. As shown in Figure 1H, transcript levels of endogenous Oct4, Sox2, and Nanog in OySyNyK-induced iPSCs were similar to those in ESCs, whereas the expression of transgenes was completely silenced (Figure S1B).
We next assessed the developmental potential of OySyNyK-induced iPSCs by using these iPSCs to generate teratomas and chimeric mice. As shown in Figures 2A–2E, OySyNyK-induced iPSCs not only formed teratomas when they were injected into nonobese diabetic severe combined immunodeficiency mice (Figures 2A–2C) but also efficiently produced chimeric mice when they were injected into diploid blastocysts (Figures 2D, 2E, and S2A), strongly indicating the pluripotency of OySyNyK-induced iPSCs.
Figure 2.
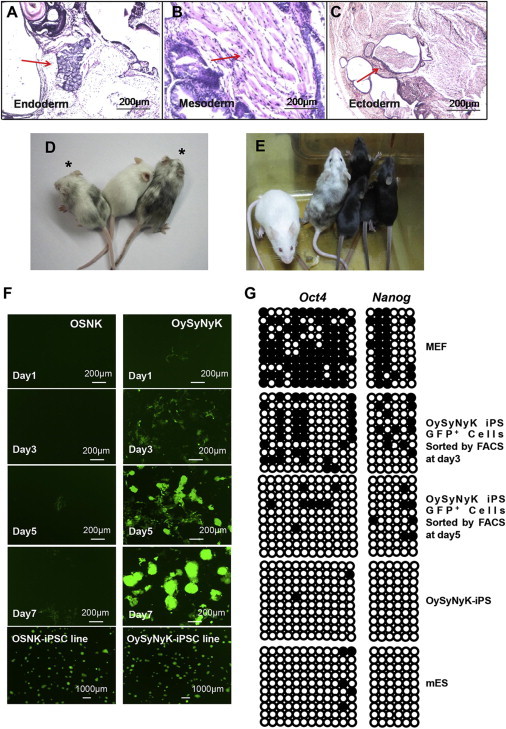
OySyNyK-Induced iPSCs Exhibit Complete Pluripotency
(A–C) Hematoxylin and eosin-stained sections of teratomas derived from OySyNyK-induced iPSCs at 8 weeks after transplantation into immunodeficient mice. Representative images show tissues from all three germ layers: endoderm (A), mesoderm (B), and ectoderm (C). Teratomas were obtained from two iPSC lines. Scale bar, 200 μm.
(D and E) Images showing 8-week-old chimeric mice as indicated by an asterisk (∗). The agouti coat color was used to indicate the iPSC contribution (D). An OySyNyK-iPSC line was microinjected into blastocysts of ICR mice. The Agouti coat color of four offspring and their chimeric mother as indicated were used to confirm the germline competence of OySyNyK-iPSCs (E).
(F) Images showing dynamic changes of the GFP expression pattern among OSNK- and OySyNyK-transduced Oct4-GFP MEFs at different time points.
(G) DNA methylation patterns at the proximal promoter of the Oct4 or Nanog gene analyzed by bisulfite sequencing. Black circles indicate methylated CpG and open circles indicate unmethylated CpG.
OySyNyK Factors Accelerate Somatic Cell Reprogramming Process at the Very Early Stage
We next employed the modified factor system to explore the molecular mechanism underlying the reprogramming process of somatic cells. We focused on investigating the early events of reprogramming by examining the dynamic expression pattern of the reprogramming reporter Oct4-GFP in MEFs transduced with OySyNyK at the early stages. Strikingly, in contrast to previous findings (Hirai et al., 2011; Wang et al., 2011), we observed a significant number of GFP-positive OySyNyK-induced cells at day 1 and day 2 after the cell cultures were switched to ESC medium (Figures 1E and 2F). Notably, the number of GFP-positive cells appeared to increase over time and formed iPSC colonies with a peak by day 7–8 (Figure 2F). In contrast, only a small number of GFP-positive cells were obtained from OSNK-induced MEFs on days 5 and 7 and reached a peak by day 16 (Figures 1D, 1E, and 2F). These findings suggested that the combination of OySyNyK not only greatly increased the efficiency of iPSC induction but also dramatically accelerated the somatic cell reprogramming process at the very early stage. Consistent with this observation, the levels of three pluripotency genes, including endogenous Oct4, Sox2, Nanog, and other reprogramming markers, such as Eras and Dax1, progressively increased in OySyNyK-transduced MEFs from day 2 to 6 as shown by qRT-PCR (Figures 1H and S2B). Similarly, immunostaining studies revealed that the SSEA1 protein, another reprogramming marker, also progressively increased at the early reprogramming stage compared to control (Figure S2C). Collectively, our data suggest that OySyNyK accelerates the somatic cell reprogramming process in the early stage compared to the OSNK native factors. Next, we tested the dynamic changes of DNA methylation at the promoters of pluripotency genes in OySyNyK-induced GFP-positive cells at day 3 and 5 and found that activation of endogenous Oct4 and Nanog was also accompanied by significant DNA demethylation of their promoters (Figure 2G).
TET1/2-Mediated 5hmC Modification Plays Important Roles in Rapid iPSC Induction by OySyNyK Factors
To determine how OySyNyK combination induces rapid early reprogramming, we examined the gene expression profiles of both OySyNyK- and OSNK-transduced MEFs from day 1 to day 5 and compared them with the profiles of iPSCs and ESCs. Cluster analyses of gene expression profiles indicated that OySyNyK-transduced MEFs at day 5 are more similar to iPSCs and ESCs (Figure S3A). Among the genes that are specifically expressed in both iPSCs and ESCs, Ten-Eleven-Translocation 1 (Tet1) was specifically elevated in OySyNyK-transduced MEFs. Ten-Eleven Translocation (TET) proteins, including TET1, TET2, and TET3, could convert 5mC to 5-hydroxymethylcytosine (5hmC), a hydroxymethylated form of 5mC (Tahiliani et al., 2009). TET protein-mediated 5hmC modification has been shown to play essential roles in the regulation of ESC pluripotency as well as myelopoiesis and zygote development (Dawlaty et al., 2011; Gu et al., 2011; Hussein et al., 2010; Ito et al., 2010; Piccolo et al., 2013). Considering that the DNA methylation level at the promoters of pluripotency genes was reduced during OySyNyK-induced reprogramming and that TET-mediated 5hmC production plays important roles in DNA methylation, we further examined the dynamic pattern of Tet expression between OySyNyK- and OSNK-transduced MEFs by qRT-PCR. As shown in Figure 3A, although the levels of Tet2 mRNA increased but no difference was observed between OySyNyK- and OSNK-transduced MEFs, Tet1 mRNA levels were rapidly and greatly increased in OySyNyK-induced MEFs in the early stages and reached higher levels at day 6 compared with those in OSNK-induced MEFs.
Figure 3.
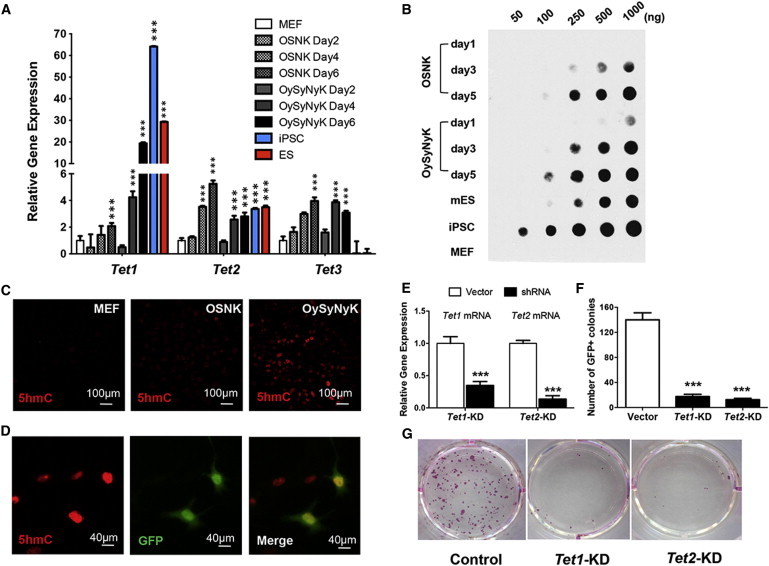
TET1/2-Mediated 5hmC Modification Plays Important Roles in Rapid iPSC Induction by OySyNyK Factors
(A) qRT-PCR assays showing the expression levels of Tet1, Tet2, and Tet3 in OSNK- and OySyNyK-transduced MEFs at different time points. Both iPSCs and ESCs were included for the comparison. OSNK- and OySyNyK-transduced MEFs groups were compared to MEF groups. ∗∗∗p < 0.001 versus control groups.
(B) Dot-blot assays showing the 5hmC level in OSNK- and OySyNyK-transduced MEFs at different time points. ESCs (CGR8), iPSCs, and MEFs cells were used as controls.
(C) Immunostaining of 5hmC in MEFs and OSNK- and OySyNyK-transduced MEFs at day 2.
(D) Immunostaining assays showing that OySyNyK-transduced GFP-positive MEFs contained high levels of 5hmC at day 2.
(E) qRT-PCR assays showing the efficiencies of Tet1 and Tet2 knockdown in OySyNyK-transduced MEFs. ∗∗∗p < 0.001 versus control groups.
(F) Statistical summary showing the number of GFP-positive colonies induced by OySyNyK under Tet1 or Tet2 knockdown. ∗∗∗p < 0.001 versus control groups.
(G) Alkaline phosphatase (AP) staining showing the efficiencies of iPSC colony induction by OySyNyK under Tet1 or Tet2 knockdown.
Data (A), (E), and (F) are from at least three independent experiments (mean and SD of triplicate assays). Groups were compared using the Student’s t test. ∗∗∗p < 0.001 versus control groups.
Given the essential roles of TET proteins in 5hmC production, we examined the dynamics of 5hmC levels during early somatic cell reprogramming induced by OySyNyK. As shown in dot-blot and immunostaining assays (Figures 3B and 3C), the levels of 5hmC were significantly increased in OySyNyK-transduced cells over time compared with those in the controls, MEFs and OSNK-transduced cells. Notably, we found that GFP-positive cells exhibited high levels of 5hmC on day 1 and 2 compared to MEF cells and OSNK-transduced cells (Figures 3D and S3B), suggesting that the increase of 5hmC levels is a significant event during the early reprogramming process. We also profiled the genome-wide 5hmC distribution in MEFs, the reprogramming intermediates at different stages induced by either the OSNK or OySyNyK method, and iPSCs using the chemical capture approach that has been established previously (Song et al., 2011). A heatmap of the top 500 ESC-specific differential 5-hydroxymethylated regions (DhMRs) showed that the reprogramming intermediates induced by the OySyNyK method were more similar to iPSCs (Figure S3C, top). Furthermore, among ESC-specific DhMRs between MEFs and iPSCs, reprogramming intermediates induced by the OySyNyK method at day 5 acquired more hydroxymethylation compared with that using the OSNK method (Figure S3C, bottom). These observations suggest that the dynamic changes of 5hmC mediated by TET proteins might play important roles in the very early stage of somatic cell reprogramming toward iPSCs.
To test this hypothesis, we further examined the timing of Oct4-GFP reporter activation and measured the final rate of GFP-positive colony (iPSC) formation in Tet1 and Tet2 knockdown MEF cells. As shown in Figures 3E–3G, knockdown of Tet1 or Tet2 significantly reduced the number of GFP-positive cells among MEFs transduced with OySyNyK in the early stage of somatic cell reprogramming and resulted in a relatively lower efficiency of iPSC colony formation than that in the control. Thus, TET proteins are important for the early reprogramming process of somatic cells and for engineered factor-induced iPSC generation. Collectively, our results suggest that the rapid reprogramming by OySyNyK may be attributed to the increased levels of Tet1/2 expression, particularly Tet1.
SOX2-YAPTAD and Its Interaction with TET1/2 Are Critical for Rapid iPSC Induction by OySyNyK Factors
Considering the important role of TET proteins in promoting somatic cell reprogramming, which is similar to that of OCT4, NANOG, and SOX2, we hypothesized that TET proteins might function in concert with these defined factors in a common biochemical pathway to regulate the reprogramming process and maintain iPSC and ESC pluripotency. We first tested whether the interaction between TET proteins and pluripotent factors (OCT4, NANOG, and SOX2) occurs in ESCs. As shown in immunoprecipitation assays, both TET1 and TET2 proteins could be detected in either NANOG or SOX2 precipitation complexes, but not in the OCT4 precipitation (Figures 4A–4C), suggesting that TET1 and TET2 proteins associate with NANOG and SOX2 in ESCs. Next, we determined whether TET proteins are associated with the modified OCT4, NANOG, or SOX2 in transfected human embryonic kidney 293 (HEK293) cells. As shown in immunoprecipitation experiments, precipitation of FLAG-tagged TET1 and TET2 from lysates of the transfected HEK293 cells resulted in appreciable coprecipitation of the modified SOX2 or NANOG (Figures S4A–S4D), whereas no or a very weak signal of OCT4 protein was detected in the precipitations of FLAG-tagged TET1 or TET2 (data not shown). Similar results were obtained in the interaction between TET1/2 protein and native forms of SOX2 and NANOG (Figures S4E–S4H). These findings suggest that endogenous TET1 and TET2 proteins physically interact with reprogramming factors and strongly associate with NANOG and SOX2.
Figure 4.
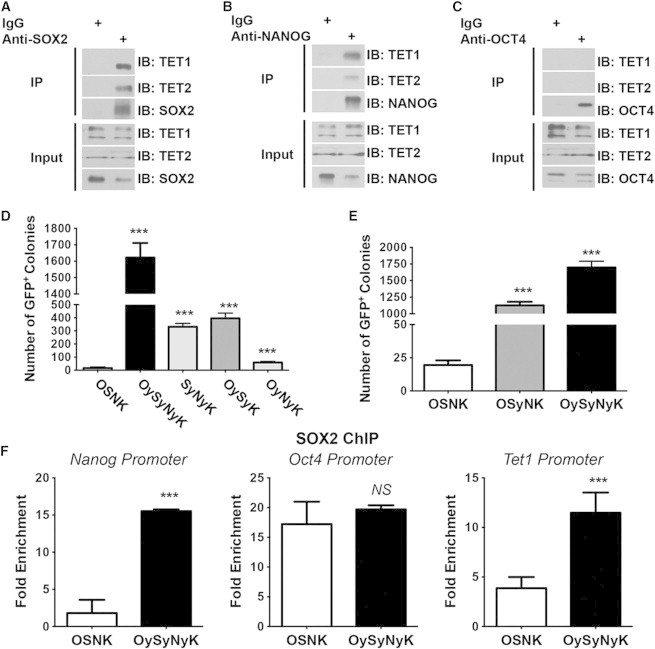
SOX2-YAPTAD and Its Interaction with TET1/2 Are Critical for Rapid iPSC Induction by OySyNyK Factors
(A–C) Anti-SOX2 (A), anti-NANOG (B), and anti-OCT4 (C) antibodies were used for immunoprecipitation from ESC lysates. Anti-TET1 and anti-TET2 antibodies were then used to perform western blotting to detect whether TET1 or TET2 was present in the SOX2 and NANOG precipitants.
(D) Statistical summary showing that absence of SOX2-YAPTAD greatly reduced the efficiency of iPSC generation. OySyNyK-induced GFP-positive colonies were counted at day 7; the other combinations were counted at day 12. ∗∗∗p < 0.001 versus control groups.
(E) Statistical summary showing that SOX2-YAPTAD plays critical role in the rapid iPSC induction by OySyNyK factors. OySyNyK-induced GFP-positive colonies were counted at day 7; the other combinations were counted at day 12. ∗∗∗p < 0.001 versus control groups.
(F) Chromatin immunoprecipitation assays showing the enrichment of native SOX2 and the modified SOX2 at the promoters of Nanog, Oct4, and Tet1 from OSNK-induced and OySyNyK-induced MEFs, respectively, at day 1. ∗∗∗p < 0.001 versus control groups.
Data from (D)–(F) are representative of at least three independent experiments (mean and SD of triplicate assays). Groups were compared using the Student’s t test. N.S, not significant; ∗∗∗p < 0.001 versus control groups.
To evaluate which modified factors are critical in rapid iPSC generation, we used different combinations of the three factors, including OySyK, OyNyK, and SyNyK, to perform the reprogramming assay. As shown in Figure 4D, OySyK and SyNyK combinations generated 396 ± 40 and 331 ± 27 GFP-positive colonies from 2.5 × 104 transduced MEFs at day 12 respectively, whereas the OyNyK combination only generated about 58 ± 9 GFP-positive colonies. Notably, the combination of OSyNK generated more than 1,127 ± 54 GFP-positive colonies (Figure 4E). These data together suggest that SOX2-YAPTAD plays critical role in the observed highly efficient generation of iPSCs.
To determine how SOX2-YAPTAD could induce rapid and efficient reprogramming, we performed a chromatin immunoprecipitation (ChIP) assay using anti-SOX2 antibody. We found that SOX2-YAPTAD was more enriched in the promoters of Nanog and Tet1, but not Oct4, in OySyNyK-induced MEFs at day 1 when compared to native SOX2 in OSNK-induced MEFs under the same conditions (Figure 4F). This result suggests that SOX2-YAPTAD can directly activate a selective subset of genes that are critical for reprogramming to promote the somatic cell reprogramming process.
Taken together, we present here a highly efficient and rapid method to generate mouse iPSCs by employing engineered reprogramming factors. Our system not only offers a tool for basic research in stem cell biology but also provides potential advantages for therapeutic applications of iPSCs in the future. Although multiple studies have reported improvements in reprogramming efficiency (Huangfu et al., 2008; Lin et al., 2009; Ma et al., 2013), the method described in this study significantly increases the reprogramming kinetics. Particularly, the reprogramming event mediated by the engineered factors was initiated at the very early stage (as early as around 24 hr). Notably, the engineered reprogramming factors also resulted in a significant increase in 5hmC levels during the early stage, suggesting that engineered reprogramming factors function in concert with TET proteins to promote 5mC demethylation during iPSC generation. Nuclear reprogramming of somatic cells toward iPSCs is under the control of a number of master transcription factors, including Klf4, Nanog, Oct4, and Sox2 (Boyer et al., 2005). These key regulators recruit other partner factors by physical interactions and then modulate the expression of downstream genes involved in reprogramming (Boyer et al., 2005; Chen et al., 2008). TET proteins, such as TET1, have recently been identified as novel interaction partners of NANOG (Costa et al., 2013; Ito et al., 2010), sharing most of the NANOG binding sites in the genome. Our study revealed that TET proteins associate not only with NANOG but also with SOX2. Importantly, we found that SOX2-YAPTAD alone could greatly increase the efficiency of iPSC generation (Figure 4E), indicating the important role of SOX2 in iPSC formation, and that hyperactivated SOX2 can overcome somatic cell reprogramming barriers.
Experimental Procedures
Plasmid Construction
pMXs-Oct4 (O), pMXs-Sox2 (S), pMXs-Nanog (N), and pMXs-Klf4 (K) retroviral vectors were obtained from Addgene. Coding sequences of mouse Oct4, Sox2, and Nanog were fused directly with the TAD domain of mouse Yap (amino acids 275–489) in the C terminus to generate Oct4-YapTAD (Oy), Sox2-YapTAD (Sy), and Nanog-YapTAD (Ny). The modified factors were then cloned into a pMXs-retroviral vector. See the Supplemental Experimental Procedures for complete details regarding plasmid construction.
Retroviral Production and Mouse iPSC Induction
HEK293 cells were transfected with pMXs retroviral vector and packaging plasmid Ecopac (1:1) by the calcium phosphate precipitation method. Detailed information regarding retroviral production and mouse iPSC induction is described in the Supplemental Experimental Procedures. All animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee of Emory University.
Additional experimental procedures are described in detail in the Supplemental Experimental Procedures.
Acknowledgments
This study was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01010306), National Basic Research Program of China (2013CB945000), and Natural Science Foundation of China (91019022, 31130036, and 31329004) and in part by the National Institutes of Health (grants NS079625 and HD073162, to P.J.; grants MH089606 and HD24064, to S.T.W.) and the Emory Genetics Discovery Fund.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Qinmiao Sun, Email: qinmiaosun@ioz.ac.cn.
Peng Jin, Email: peng.jin@emory.edu.
Dahua Chen, Email: chendh@ioz.ac.cn.
Accession Numbers
Sequence data have been deposited to the Gene Expression Omnibus with the accession number GSE46202.
Supplemental Information
References
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Costa Y., Ding J., Theunissen T.W., Faiola F., Hore T.A., Shliaha P.V., Fidalgo M., Saunders A., Lawrence M., Dietmann S. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty M.M., Ganz K., Powell B.E., Hu Y.C., Markoulaki S., Cheng A.W., Gao Q., Kim J., Choi S.W., Page D.C., Jaenisch R. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T.P., Guo F., Yang H., Wu H.P., Xu G.F., Liu W., Xie Z.G., Shi L., He X., Jin S.G. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Hirai H., Tani T., Katoku-Kikyo N., Kellner S., Karian P., Firpo M., Kikyo N. Radical acceleration of nuclear reprogramming by chromatin remodeling with the transactivation domain of MyoD. Stem Cells. 2011;29:1349–1361. doi: 10.1002/stem.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A.E., Melton D.A. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein K., Abdel-Wahab O., Lasho T.L., Van Dyke D.L., Levine R.L., Hanson C.A., Pardanani A., Tefferi A. Cytogenetic correlates of TET2 mutations in 199 patients with myeloproliferative neoplasms. Am. J. Hematol. 2010;85:81–83. doi: 10.1002/ajh.21562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., D’Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner C.J., Camargo F.D., Hochedlinger K., Welstead G.G., Zaidi S., Gokhale S., Scholer H.R., Tomilin A., Jaenisch R. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian I., Kim J., Okazawa H., Zhao J., Zhao B., Yu J., Chinnaiyan A., Israel M.A., Goldstein L.S., Abujarour R. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Ambasudhan R., Yuan X., Li W., Hilcove S., Abujarour R., Lin X., Hahm H.S., Hao E., Hayek A., Ding S. A chemical platform for improved induction of human iPSCs. Nat. Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Xie M., Laurent T., Ding S. Progress in the reprogramming of somatic cells. Circ. Res. 2013;112:562–574. doi: 10.1161/CIRCRESAHA.111.249235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M., Zhang J., Smolen G.A., Muir B., Li W., Sgroi D.C., Deng C.X., Brugge J.S., Haber D.A. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. USA. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo F.M., Bagci H., Brown K.E., Landeira D., Soza-Ried J., Feytout A., Mooijman D., Hajkova P., Leitch H.G., Tada T. Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol. Cell. 2013;49:1023–1033. doi: 10.1016/j.molcel.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.X., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X., Li Y., Chen C.H., Zhang W., Jian X. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Wang Y., Chen J., Hu J.L., Wei X.X., Qin D., Gao J., Zhang L., Jiang J., Li J.S., Liu J. Reprogramming of mouse and human somatic cells by high-performance engineered factors. EMBO Rep. 2011;12:373–378. doi: 10.1038/embor.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S., Blau H.M. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Lei Q.Y., Guan K.L. Harness the power: new insights into the inhibition of YAP/Yorkie. Dev. Cell. 2009;16:321–322. doi: 10.1016/j.devcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


