Summary
Differentiated cells can be reprogrammed into induced pluripotent stem cells (iPSCs) after overexpressing four transcription factors, of which Oct4 is essential. To elucidate the role of Oct4 during reprogramming, we investigated the immediate transcriptional response to inducible Oct4 overexpression in various somatic murine cell types using microarray analysis. By downregulating somatic-specific genes, Oct4 induction influenced each transcriptional program in a unique manner. A significant upregulation of pluripotent markers could not be detected. Therefore, OCT4 facilitates reprogramming by interfering with the somatic transcriptional network rather than by directly initiating a pluripotent gene-expression program. Finally, Oct4 overexpression upregulated the gene Mgarp in all the analyzed cell types. Strikingly, Mgarp expression decreases during the first steps of reprogramming due to a KLF4-dependent inhibition. At later stages, OCT4 counteracts the repressive activity of KLF4, thereby enhancing Mgarp expression. We show that this temporal expression pattern is crucial for the efficient generation of iPSCs.
Highlights
-
•
OCT4 interferes with somatic transcriptional networks in a cell-type-specific manner
-
•
OCT4 does not activate the pluripotent program at the early stages of reprogramming
-
•
OCT4 and KLF4 regulate Mgarp transcriptional activity in an antagonistic manner
-
•
A specific time pattern of Mgarp expression is crucial for inducing pluripotency
Schöler, Tapia, and colleagues show that efficient reprogramming to pluripotency requires Mgarp downregulation at the early steps of reprogramming but Mgarp upregulation at the late steps of reprogramming. Mgarp repression is promoted by KLF4 whereas Mgarp activation is enhanced by OCT4. These results demonstrate that KLF4 and OCT4 oppositely regulate Mgarp in a time-specific manner to ensure induction of pluripotency.
Introduction
Somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) after the forced expression of three or four transcription factors: Oct4, Sox2, Klf4, and optionally, Myc (Nakagawa et al., 2008; Takahashi and Yamanaka, 2006). Oct4 is indispensable for establishing pluripotency in the embryo (Nichols et al., 1998) and for maintaining pluripotency in mouse embryonic stem cells (ESCs) (Niwa et al., 2000). Under physiological conditions, the OCT4 protein needs to interact with SOX2 for activating most of its target genes and for maintaining the self-renewal of ESCs (Boyer et al., 2005; Reményi et al., 2003; Rodda et al., 2005). Nevertheless, overexpression of Oct4 can rescue ESC self-renewal in the absence of Sox2 (Masui et al., 2007), indicating that Sox2 is not essential for supporting pluripotency. In addition, Sox2 can be replaced by other Sox factors or by transforming growth factor β inhibitors in the reprogramming of somatic cells into iPSCs (Ichida et al., 2009; Nakagawa et al., 2008). Likewise, Klf4 is dispensable for maintaining ESC self-renewal (Jiang et al., 2008) and for inducing pluripotency (Nakagawa et al., 2008), as Klf2 and Klf5 can replace Klf4 in both functions. In addition, Esrrb can also substitute Klf4 in iPSCs generation (Feng et al., 2009). Therefore, Oct4 is the only transcription factor in the conventional reprogramming cocktail that is essential for pluripotency.
To date, the role of OCT4 in reprogramming has been studied only in the context of its interaction with SOX2 (Buganim et al., 2012; Hansson et al., 2012; Polo et al., 2012; Sridharan et al., 2009; Stadtfeld et al., 2008). As exogenous Sox2 is not required for inducing pluripotency (Ichida et al., 2009; Maherali and Hochedlinger, 2009), we decided to investigate the specific effect of OCT4 alone in the first steps of reprogramming. To circumvent the inevitable heterogeneity generated by viral factor delivery, we established a set of different somatic cell types from tetracycline-inducible Oct4 transgenic mice. This approach facilitates the study of rare events in cell populations that simultaneously activate Oct4. Finally, we performed global gene-expression profile analyses to identify the early transcriptional changes caused by ectopic Oct4 expression in somatic cells. Overall, our study provides insights into the specific OCT4-dependent events that promote the induction of pluripotency.
Results
Generation and Characterization of Different Oct4-Inducible Somatic Cell Types
To analyze the immediate transcriptional response triggered by Oct4 overexpression in somatic cells, we derived mouse embryonic fibroblasts (MEFs), neural stem cells (NSCs), and bone marrow cells (BMCs) from mice containing both a tetracycline transactivator and a tetracycline-inducible Oct4 transgene and termed these cells tetracycline-operon-controlled Oct4-expression cassette (TO)-MEFs, TO-NSCs (cell line number 1), and TO-BMCs, respectively. In addition, we established an NSC line from mice containing only the tetracycline transactivator (CtrlNSC) as a control to detect nonspecific tetracycline transactivator activities unrelated to OCT4. Finally, nontransgenic MEFs were also used as a control to identify effects caused by the tetracycline treatment itself (CtrlMEF) (Figure 1A).
Figure 1.
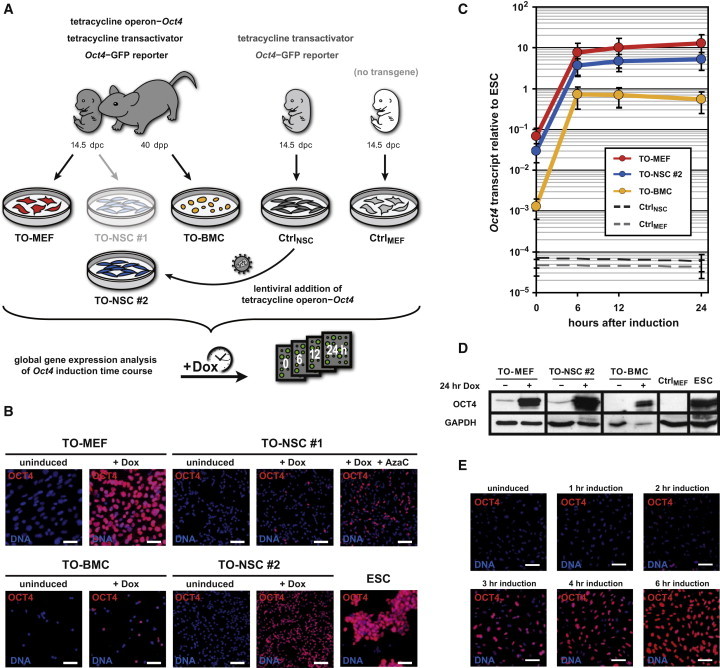
Generation and Characterization of Tetracycline-Inducible Oct4 Cells and Controls Used for Dynamic Global Gene-Expression Analysis
(A) Fibroblasts (TO-MEFs), neural stem cells (TO-NSCs number 1), and bone marrow cells (TO-BMCs) contain a tetracycline transactivator, a tetracycline-operon Oct4-expression cassette, and an Oct4-GFP reporter. NSCs with a tetracycline transactivator and an Oct4-GFP reporter but lacking a tetracycline-operon Oct4 cassette (CtrlNSC) served as a control for nonspecific transactivator activity. Wild-type C57BL/6 fibroblasts were used as a control for drug response artifacts (CtrlMEF). TO-NSCs number 2 were generated after transducing CtrlNSC with a lentiviral vector coding for a tetracycline-operon Oct4. Transcriptional effects of Oct4 expression were investigated by microarray analysis at different time points after doxycycline induction.
(B) Immunological staining for OCT4 protein (red) in the different cell types after 24 hr of doxycycline induction. Nuclei were counterstained with Hoechst (blue). The scale bars represent 50 μm.
(C) Expression of Oct4 mRNA (qRT-PCR), normalized to an embryonic stem cell (ESC) control. The error bars represent SE of calculations based on ΔCt values obtained from two different housekeeping genes: Gapdh and Actb (n = 1).
(D) OCT4 protein immunoblot of induced and uninduced samples. GAPDH was used as a loading control.
(E) OCT4 protein translation time course after doxycycline induction. OCT4 protein and nuclei are shown in red and blue, respectively. The scale bars represent 50 μm.
Abbreviations: dpc, days postcoitum; dpp, days postpartum; Dox, doxycycline; AzaC, 5-azacytidine; TO, tetracycline-inducible. See also Figure S1 as well as Tables S5 and S6.
Using immunocytochemistry, we assessed the level of OCT4 protein expression after 24 hr of doxycycline induction in each generated cell type. We counted 98% of TO-MEFs to be positive for OCT4 staining, with intensity levels comparable to those observed in ESCs (Figure 1B). In contrast, only 40% of TO-BMCs and 10% of TO-NSCs (number 1) exhibited strongly induced OCT4 expression. Interestingly, 40% of TO-NSCs (number 1) became OCT4 positive after treatment with 5-azacytidine (Figure 1B), suggesting the presence of a DNA-methylation-based mechanism for transgene silencing in this cell type. For this reason, we decided to exclude TO-NSC line number 1 from our study. Instead, we transduced CtrlNSC with a lentiviral vector coding for the same TO cassette that is present in the other cell types (Stadtfeld et al., 2008; Figure 1A). This newly generated TO-NSC line (number 2) could efficiently induce OCT4 expression in 98% of the cells after doxycycline treatment and was thus the only TO-NSC line used in the subsequent experiments (Figure 1B). Next, the induction of exogenous Oct4 expression was analyzed in a time course manner by quantitative RT-PCR (qRT-PCR). After 6 hr, all cell types exhibited ESC-like Oct4 transcript levels (Figure 1C). Furthermore, immunoblotting confirmed OCT4 expression at the protein level (Figure 1D). We also surveyed the onset of OCT4 translation in more detail by immunocytochemistry. OCT4 protein emerged as early as 2 to 3 hr after induction and reached saturation levels after approximately 6 hr (Figure 1E). Consistent with previous observations (Hochedlinger et al., 2005; Wang et al., 2011), in-vitro-cultured somatic cells showed impaired proliferation after Oct4 overexpression (Figure S1 available online).
To assess whether the Oct4 induction levels are sufficient for iPSC generation, we transduced the different cell types with retroviral vectors coding for Sox2, Klf4, and Myc. After inducing Oct4, we generated Oct4-GFP-positive, ESC-like colonies from all somatic cell types and established clonal, doxycycline-independent cell lines (Figure 2A). The reprogramming efficiency and kinetics using the inducible Oct4 transgene were similar to those achieved using viral delivery of Oct4 (data not shown). The generated iPSC lines stained positive for the pluripotency markers alkaline phosphatase (ALP; Figure 2A), NANOG, and SSEA-1 (Figure 2B). As ESCs, iPSCs exhibited unmethylated Oct4 and Nanog promoters (Figure 2C). Moreover, all iPSC lines were capable of differentiating into all three germ layers in vitro and in vivo after embryoid body formation (Figure 2D) and blastocyst injection (Figure 2E), respectively. Finally, germline contribution was demonstrated by the presence of Oct4-GFP-positive germ cells in the fetal gonads (Figure 2E). In summary, we have derived three different somatic cell types that can efficiently induce Oct4 expression to levels supporting iPSC generation.
Figure 2.
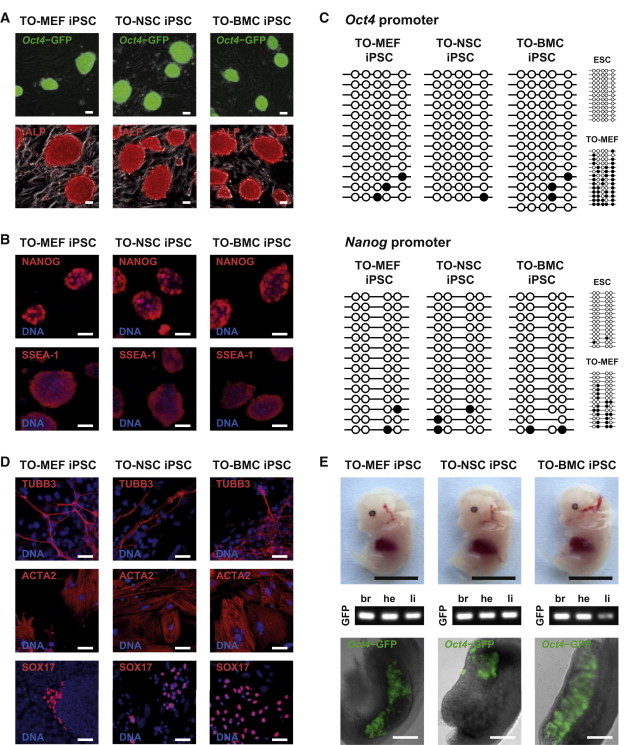
Characterization of iPSCs Generated from Tetracycline-Inducible Oct4 Cells
(A) Clonal induced pluripotent stem cell (iPSC) lines generated by tetracycline-induced Oct4 expression in combination with virally delivered Sox2, Klf4, and Myc. Phase-contrast overlay with Oct4-GFP reporter fluorescence and ALP staining. The scale bars represent 50 μm.
(B) Immunological stainings for NANOG and SSEA-1 (red), overlaid with nuclear Hoechst staining (blue). The scale bars represent 50 μm.
(C) DNA methylation of Oct4 and Nanog promoters in iPSCs and controls. Empty and full circles represent unmethylated and methylated CpG cytosine residues, respectively.
(D) Immunocytochemistry for specific marker proteins (red) showed in vitro differentiation of iPSC clones into all three germ layers. Nuclei were counterstained with Hoechst (blue). The scale bars represent 50 μm.
(E) PCR genotyping for the GFP transgene demonstrated that iPSCs could contribute to organs representative of all germ layers in 14.5 dpc chimeric embryos. Germline contribution was proven by the presence of Oct4-GFP-positive cells in the fetal gonads. The black scale bars represent 5 mm. The white scale bars represent 250 μm.
Abbreviations: TUBB3, β-III-tubulin (ectoderm; neurons); ACTA2, α-II-actin (mesoderm; smooth muscle cells); SOX17, SRY-box-containing 17 (definitive endoderm); br, brain; he, heart; li, liver. See also Tables S5 and S6.
Oct4 Induction Affects the Gene-Expression Profile in a Cell-type-Specific Manner
To identify the OCT4 targets during the first steps of reprogramming, we collected samples of the different Oct4-inducible cells and controls at 0, 6, 12, and 24 hr after Oct4 induction with doxycycline treatment. Global gene-expression profiles were then recorded by microarray analysis. Upregulation of Oct4 expression was clearly confirmed in all inducible cell types, but not in the controls (Figure 3A). The two corresponding Oct4-detecting array probes were not further considered in the subsequent analyses. Induced TO-MEF and TO-NSC cultures, which express the Oct4 transgene most uniformly, exhibited a stronger transcriptional response than TO-BMCs. CtrlMEF cells showed hardly any transcriptional response to the doxycycline treatment. However, differential expression of some genes was observed in the tetracycline-transactivator-containing CtrlNSC cells, suggesting the need for a more-refined analytic strategy that could precisely distinguish OCT4-dependent from OCT4-independent effects (Figure 3A). Therefore, we assigned a score to each probe in each cell type. This score is based primarily on the total signal change within 24 hr but also incorporates additional parameters such as expression changes in the controls, the dynamic behavior throughout the time course, and the absolute signal intensity as an indicator for noise artifacts. Probes with a score higher than 1 or lower than −1 were defined as up- or downregulated, respectively (for details, see Experimental Procedures and Figure S2). Surprisingly, the genes that were most strongly upregulated differed greatly from one cell type to another, indicating that the immediate targets of OCT4 are unique to each cell type (Figure 3B). Interestingly, fewer than 50% of the genes that were upregulated by OCT4 in the different cell types are expressed in ESCs (Figure 3B) and only a small subset (<4%) of these ESC-expressed genes is ESC-specific, i.e., not expressed in any other cell type examined (Table S1). The genes that were most strongly downregulated by OCT4 expression differed among the cell types, and many genes were specific to each cell type (Figure 3C). In fact, 21.0%, 12.4%, and 70.1% of genes downregulated in TO-MEFs, TO-NSCs, and TO-BMCs, respectively, were not expressed in any other cell type in our data set, suggesting a possible early effect of Oct4 expression in the disruption of the respective somatic regulatory program. Surprisingly, many downregulated genes are also highly expressed in ESCs (55.4%, 79.1%, and 16.6% in TO-MEFs, TO-NSCs, and TO-BMCs, respectively; Figure 3C). To further investigate whether the OCT4 target genes reflect common cellular processes, we performed gene ontology analyses (Tables S2–S4). The few statistically enriched ontology terms appeared to be remarkably unique for each cell type. Strikingly, no ontology terms for upregulated genes were found, with the exception of the unexpected term “neuron projection” in TO-MEFs. In contrast, the downregulated genes were enriched for ontologies related to the specific cell type. Moreover, TO-NSCs showed downregulation of genes related to the “mitotic cell cycle” and other similar terms. Several of these genes were also downregulated in induced TO-MEFs. This result correlates with the observed proliferation impairment caused by Oct4 overexpression in both cell types (Figure S1).
Figure 3.
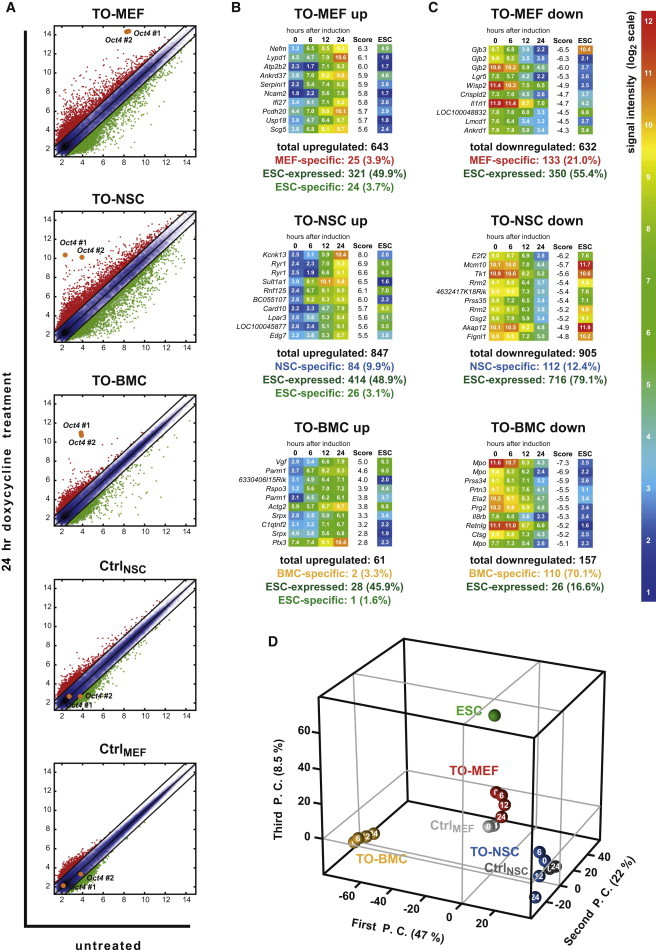
Global Gene-Expression Analysis after Induction of Oct4 Alone
(A) Scatter plots comparing expression profile before and after Oct4 overexpression for each cell type examined (log2 scale). Probes with increased and decreased intensity upon Oct4 induction are depicted in red and green, respectively. The blue area contains equally expressed probes (<2-fold difference). The two Oct4 probes are highlighted.
(B) Identification of the OCT4-induced upregulated genes. Heatmaps show the temporal changes in signal intensities for the ten genes presenting the highest score value. The expression levels in an ESC control are shown for comparison.
(C) Identification of OCT4-induced downregulated genes. Heatmaps show the temporal changes in signal intensities for the ten genes presenting the lowest score.
(D) Principal-component analysis (PCA) of global transcriptome data. The three principal components that contribute most strongly to the variability among the individual samples are shown. The numbers inside the circles denote hours after Oct4 induction. P.C., principal component.
See also Figures S2, S3, and Tables S1–S4.
An unsupervised hierarchical clustering (Figure S3A) and a principal-component analysis (PCA) (Figure 3D) showed all the time points for a specific cell type clustering together, indicating that the OCT4-induced changes in global gene expression were rather small, compared to the overall differences distinguishing one cell type from the other. However, TO-MEFs and TO-NSCs after 24 hr of Oct4 induction were more different from their uninduced counterparts than were CtrlMEF and CtrNSC cells, respectively. We also compared the numbers of differentially expressed genes between each pair of samples (Figure S3B). Importantly, none of the cell types became more similar to ESCs upon Oct4 induction, as assessed by gene expression. PCA confirmed that the transcriptional program of the different cell types shifted away from that of ESCs and revealed that the direction of gene-expression changes was unique to each cell type (Figure 3D). Overall, these results suggest that, early in reprogramming, Oct4 expression interferes with the somatic cell transcriptional network in a cell-type-specific manner, but does not directly induce a pluripotent cell fate or a gene-expression signature common to all cell types examined.
Mgarp Is One of the Few Genes that Are Commonly Upregulated in All Cell Types
To identify common targets of OCT4, we directly compared the sets of up- and downregulated genes for each individual cell type but only detected a few overlaps (Figures 4A and 4B). From a total of 1,441 upregulated probes, 102 (7.1%) were upregulated in two different cell types, and only four genes (0.3%), namely Parm1, Mgarp, Slc24a3, and Tmem53, were commonly upregulated in all three different cell types (Figure 4A). The intersections within the downregulated genes were even smaller, as 107 (6.7%) of 1,587 probes were downregulated in two different cell types. Not a single gene was commonly downregulated in TO-NSCs, TO-MEFs, and TO-BMCs (Figure 4B). The vast majority of both up- and downregulated genes thus represent unique effects of Oct4 expression that occur only in a specific cellular environment. Of note, the nonuniform induction pattern observed in TO-BMCs might contribute to this finding, as uninduced cells will dilute the measurable gene-expression changes. However, TO-MEFs and TO-NSCs, which exhibit homogeneous Oct4 induction, have in common only 6.5% of their up- and downregulated genes. We also identified 71 genes that were oppositely regulated by Oct4 expression, i.e., they were upregulated in at least one cell type while being downregulated in at least another (Figure 4C). Furthermore, 83.2% of the commonly downregulated genes are expressed in ESCs. In contrast, only 58.7% of the commonly upregulated genes are expressed in ESCs and as few as 5.8% are ESC-specific (Figure 4C). Surprisingly, these ESC-specific genes do not include any key pluripotency markers (Table S1). Finally, we selected the four commonly upregulated as well as two commonly downregulated and two oppositely regulated genes (Figure 5A) and validated their dynamic gene-expression changes using qRT-PCR (Figure 5B). Our results indicate that ectopic Oct4 expression does not activate a common transcriptional program in different somatic cell environments and show that only four genes are commonly upregulated in all the cell types examined.
Figure 4.
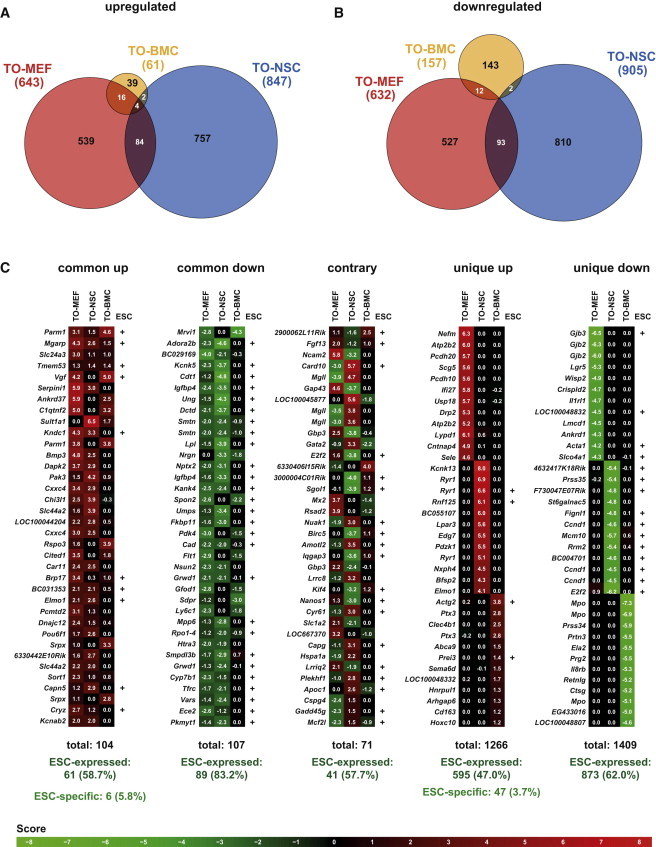
Shared and Unique Oct4 Targets
(A) Venn diagram depicting overlaps of upregulated gene sets among the different cell types after Oct4 induction.
(B) Venn diagram depicting overlaps of downregulated gene sets.
(C) Common up, genes with highest scores that are upregulated in at least two different cell types. Common down, genes with lowest scores that are downregulated in at least two different cell types. Contrary, genes that are oppositely expressed among at least two different cell types. Unique up, genes with highest scores that are upregulated in only one cell type. Unique down, genes with lowest scores that are downregulated in only one cell type.
Figure 5.
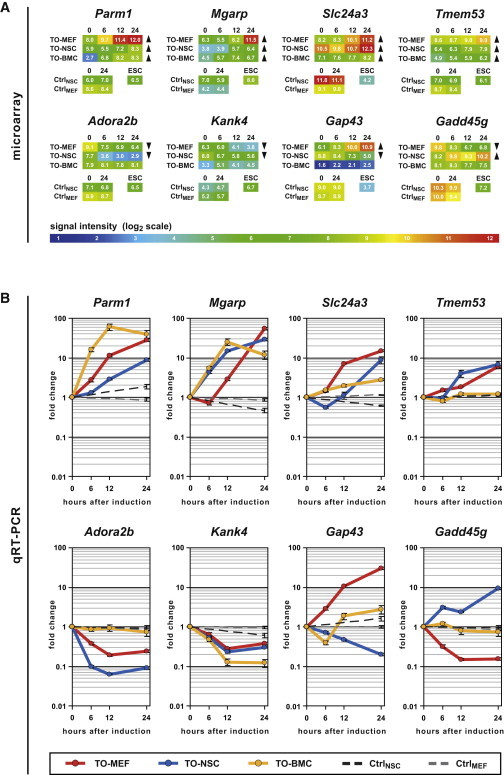
Validation of OCT4 Target Genes Identified in the Microarray Analysis
(A) The only four commonly upregulated (Parm1, Mgarp, Slc24a3, and Tmem53) and two examples of commonly downregulated (Adora2b and Kank4) as well as contrarily regulated genes (Gap43 and Gadd45 g) were chosen. Microarray signal intensities for all samples and controls at different time points after induction are shown as heatmaps. Arrows indicate in which cell types the criteria for up- or downregulation were fulfilled.
(B) Dynamic gene-expression analysis of the same genes using qRT-PCR, normalized to the respective uninduced cells. The error bars represent SE of calculations based on ΔCt values obtained from two different housekeeping genes: Gapdh and Actb (n = 1).
See also Table S5.
OCT4-Dependent Upregulation of Mgarp Is Counteracted by KLF4 to Ensure Successful Reprogramming
We then investigated Parm1 and Mgarp, the two genes most strongly upregulated by OCT4 in all the three cell types examined. Overexpression and knockdown of Parm1 had no effect on iPSC generation (data not shown), and there is no indication of an association between this gene and pluripotency or reprogramming. However, Mgarp (mitochondria-localized glutamic acid rich protein) is present in both ESCs and epiblast stem cells. Mgarp expression was not detected in early primordial germ cells but was induced to ESC-like levels in male germ cells at 13.5 days postcoitum (dpc) and maintained in spermatogonial stem cells after birth. In contrast, female germ cells did not express Mgarp (Figure S4). Furthermore, Mgarp is transcribed in several organs such as ovary, testis, the visual system, and the brain (Kinouchi et al., 2006; Li et al., 2009; Matsumoto et al., 2009; Qi et al., 2011).
To investigate a possible role of Mgarp in reprogramming, we generated a set of viral vectors to increase or decrease the Mgarp expression level in a constitutive or tetracycline-inducible manner (Figure S5). MEFs, NSCs, and BMCs bearing an Oct4-GFP reporter transgene were transduced with viruses coding for mouse Oct4, Sox2, Klf4, and Myc plus Mgarp or small hairpin RNA against Mgarp (shMgarp). Three weeks later, we assessed for ALP activity (Figure 6A) and counted the number of Oct4-GFP-positive iPSC colonies (Figure 6B). Strikingly, Mgarp constitutive overexpression caused a dramatic decline in colony numbers to fewer than 10% of the respective control (Figure 6B). This indicates that, although Mgarp is expressed in pluripotent stem cells and upregulated by the key reprogramming factor OCT4, high Mgarp levels are detrimental to the reprogramming process. Unexpectedly, constitutive knockdown of Mgarp did not have the opposite effect—but it also significantly reduced the number of iPSC colonies in all the analyzed cell types (Figure 6B). To better understand this ambivalent role of Mgarp in reprogramming, we analyzed the effect of the reprogramming factors OCT4, SOX2, KLF4, and MYC, individually and in combination, on the transcriptional activation of Mgarp. Consistent with our previous observations, OCT4 strongly and stably induced the upregulation of Mgarp expression. SOX2 and MYC did not have any effect on Mgarp expression. In contrast, KLF4 strongly inhibited the transcription of Mgarp (Figure 6C). A very interesting pattern of expression was observed after the combined overexpression of Oct4 and Klf4 alone or together with Sox2 and Myc. In fact, Mgarp expression levels first decreased until day 6 after transduction but then started to increase. This temporal expression pattern was maintained at lowered levels in the presence of two different constitutive shMgarp constructs and was completely disrupted by constitutive Mgarp overexpression (Figure 6C). Based on these results, we hypothesized that continual high levels of Mgarp (Oct4, Sox2, Klf4, and Myc [OSKM] + Mgarp) as well as constant low levels of Mgarp (OSKM + shMgarp) interfere with the initial Mgarp downregulation and the subsequent Mgarp upregulation steps induced by KLF4 and OCT4, respectively (Figure 6C). To directly test this hypothesis, we transduced MEFs with Oct4, Sox2, Klf4, and Myc plus tetracycline-inducible vectors for Mgarp overexpression or knockdown. The expression or inhibition was induced at different time points (Figure 6D). Mgarp overexpression within the first 3 days of the reprogramming process significantly reduced colony numbers, as it interfered with the initial step in which the level of Mgarp needs to decrease. Consistently, Mgarp induction at later stages did not affect the reprogramming efficiency. As expected, Mgarp knockdown, which interferes with late steps, was detrimental for the induction of pluripotency independently of the time of onset (Figure 6D). Therefore, our results demonstrate that a specific two-step time course of Mgarp expression is required for reprogramming and that disturbing either of the two steps impairs iPSC generation.
Figure 6.
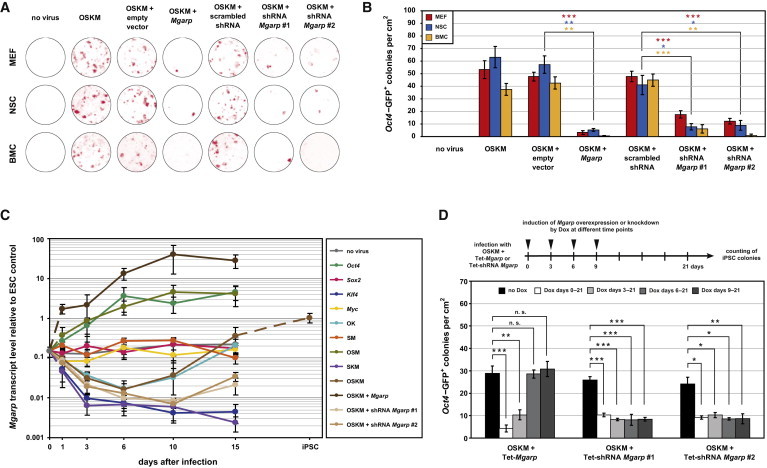
Different Mgarp Expression Levels Required at Early and Late Stages during Reprogramming of Somatic Cells into iPSCs Are Competitively Regulated by OCT4 and KLF4
(A) Reprogramming of MEFs, NSCs, and BMCs into iPSCs using different factor combinations. Representative wells were stained for ALP activity (red) on day 21 after infection.
(B) Oct4-GFP-positive iPSC colonies were counted on day 21 after infection with different factor combinations. The error bars represent SD of the mean (biological replicates, n = 5). p values refer to a two-tailed, unpaired Student’s t test: ∗p < 10−3; ∗∗p < 10−4; ∗∗∗p < 10−5.
(C) Monitoring of Mgarp expression levels (qRT-PCR) during a 15-day period after infection of MEFs with different combinations of genes, normalized to ESCs. The error bars represent SD of the mean (biological replicates, n = 4).
(D) Oct4-GFP-positive iPSC generated after infection of MEFs with four factors plus tetracycline-inducible (Tet-) vectors for Mgarp overexpression or knockdown. The expression or inhibition was induced at different time points, as indicated. The error bars represent SD of the mean (biological replicates, n = 5). p values refer to a two-tailed, unpaired Student’s t test: ∗p < 10−3; ∗∗p < 10−4; ∗∗∗p < 10−5; n.s., not significant (p > 0.1). Abbreviations: shRNA, small hairpin RNA; OSKM, Oct4, Sox2, Klf4, and Myc.
See also Figures S4–S7 and Table S5.
Next, we investigated whether OCT4 and KLF4 regulate Mgarp by directly binding to regulatory regions within its genomic locus. OCT4 protein has previously been shown to bind near the Mgarp gene in mouse ESCs using a chromatin immunoprecipitation (ChIP)-chip approach (Mathur et al., 2008). Indeed, we identified a putative OCT4 binding motif (Chen et al., 2008) in the first intron of Mgarp (Figure 7A). Furthermore, the Mgarp promoter features a sequence with a high similarity to the consensus KLF4 binding motif (Chen et al., 2008; Figure 7A). To assess whether OCT4 and KLF4 bind to the Mgarp locus, we designed a set of primers that span approximately 5 kb up- and downstream of the Mgarp transcription start site (Figure 7A) and performed ChIP-quantitative PCR (qPCR) in MEFs overexpressing Oct4 or Klf4. We detected an enrichment of both OCT4 (Figure 7B) and KLF4 (Figure 7C) in proximity of the identified candidate binding sites. Interspecies genome comparisons further supported the notion that Mgarp is a direct target gene of OCT4 (Figure 7D) and KLF4 (Figure 7E), as both binding motifs are well conserved across a variety of mammalian species.
Figure 7.
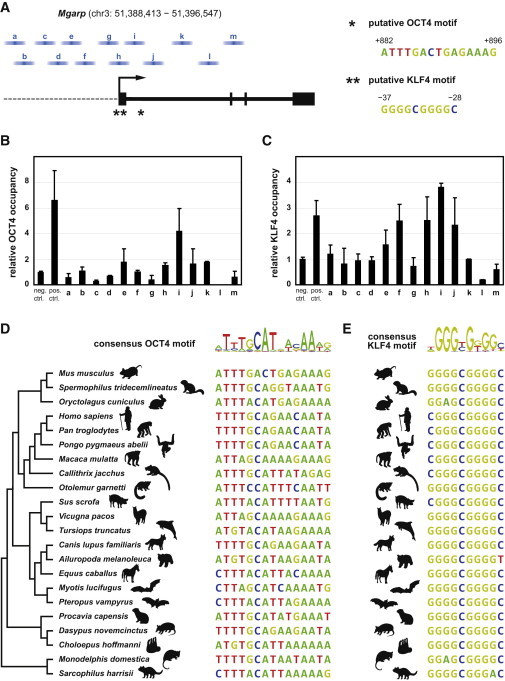
OCT4 and KLF4 Bind to the Mgarp Genomic Locus
(A) Position of 13 ChIP-qPCR amplicons (a–m) spanning approximately 5 kb up- and downstream of the Mgarp transcriptional start site. Blue bars indicate the covered regions, as estimated by the average size of crosslinked DNA fragments. The asterisk indicates position of a putative OCT4 binding motif within the first intron of Mgarp; double asterisks indicate position of a putative KLF4 binding motif within the Mgarp promoter.
(B) ChIP-qPCR analysis after α-OCT4 immunoprecipitation of crosslinked DNA isolated from Oct4-overexpressing MEFs. Relative occupancies to a negative control region (neg. ctrl., rDNA-28S). a–m, amplicons as specified in (A); pos. ctrl., OCT4 binding site positive control within regulatory region of Oct4.
(C) ChIP-qPCR analysis after α-KLF4 immunoprecipitation of crosslinked DNA isolated from Klf4-overexpressing MEFs. pos. ctrl., KLF4 binding site positive control within regulatory region of Cdh1. The error bars in (B) and (C) represent SEM (technical replicates, n = 3).
(D and E) Conservation of the identified OCT4 (D) and KLF4 (E) motif in different mammalian genomes.
The MGARP protein is localized in the mitochondria and was reported to alter mitochondrial distribution and transport (Li et al., 2009). Interestingly, a crucial step in reprogramming is the transition from a somatic-oxidative to a pluripotent-glycolytic metabolism by remodeling the mitochondrial network (Folmes et al., 2011; Kelly et al., 2013). To test whether the role of Mgarp in reprogramming is related to mitochondria, we overexpressed Mgarp in MEFs. However, we did not observe any effects on mitochondria numbers or respiratory activity (Figure S6). Finally, Mgarp, which is expressed in ESCs and required to establish pluripotency, does not play a role in ESC self-renewal, as Mgarp knockdown did not affect ESC morphology or proliferation (Figure S7).
In summary, our data suggest that the initial decrease in Mgarp expression is a consequence of direct inhibition by KLF4 and that the subsequent increase in Mgarp expression is a direct effect of activation by OCT4. Most importantly, iPSC generation is severely impaired by interference with either the initial Mgarp decrease (OSKM + Mgarp) or the subsequent Mgarp increase (OSKM + shMgarp). Therefore, the counteracting regulatory activities of OCT4 and KLF4 ensure the specific Mgarp time pattern of expression that is essential to induce pluripotency.
Discussion
In this study, we derived different Oct4-inducible somatic cell types to identify early transcriptional events induced by the exogenous OCT4 protein during cellular reprogramming. Our global gene-expression analyses revealed that OCT4 does not induce expression of pluripotency-related genes in somatic cells during the first 24 hr after its overexpression. On the contrary, many ESC-expressed genes are downregulated by OCT4 in somatic cells. In addition, our data demonstrate that OCT4 activates and inhibits different genes in a cell-type-dependent manner. The OCT4-upregulated genes are highly variable from one cell type to another and do not present any common ontological pattern. Some genes were upregulated in one cell type while being downregulated in other cell types. Thus, our results suggest that OCT4 alone can induce a general perturbation in the somatic cell transcriptional network at the early steps of reprogramming, partially through the downregulation of cell-type-specific genes. Indeed, OCT4 has been reported to not only activate pluripotent genes but also repress developmental regulators in ESCs (Boyer et al., 2005; Rodda et al., 2005). However, the sequences to which OCT4 is bound in ESCs do not maintain the same accessibility in different types of somatic cells (Koche et al., 2011; Soufi et al., 2012). Therefore, different cell-type-dependent chromatin signatures could explain the differences in gene expression between MEFs, NSCs, and BMCs after Oct4 induction.
Fibroblasts can be directly reprogrammed into neurons (Vierbuchen et al., 2010), cardiomyocytes (Ieda et al., 2010), and NSCs (Han et al., 2012) using tissue-specific combinations of transcription factors. However, the pluripotent reprogramming cocktail of Oct4, Sox2, Klf4, and Myc can also convert fibroblasts into cardiomyocytes (Efe et al., 2011) and NSCs (Kim et al., 2011; Thier et al., 2012) under suitable culture conditions. In addition, the overexpression of only Oct4 has been reported to reprogram fibroblasts toward the hematopoietic lineage (Szabo et al., 2010). In these studies, the overexpression of Oct4 alone or in combination with other transcription factors is thought to force the cells into an unstable and plastic intermediate (Orkin and Hochedlinger, 2011) that can be pushed toward the desired final cell type by specific environmental cues. We speculate that the initial stochastic phase of the reprogramming process (Buganim et al., 2012; Hanna et al., 2009) involves a transient transcriptionally unstable state that may be a prerequisite to the hierarchical events that take place during the late steps of reprogramming (Buganim et al., 2012). In addition, the reprogramming transgenes are usually expressed at higher levels during reprogramming than their corresponding endogenous counterparts in ESCs. These nonphysiological conditions may promote nonspecific binding to low-affinity binding sites and contribute to transcriptional chaos and instability. Our data suggest that Oct4 overexpression at early stages of reprogramming plays a role in the initiation of such an unstable intermediate through the inhibition of the somatic cell-type-specific program rather than through the activation of the pluripotent transcriptional network. Our results are consistent with previous four factor studies showing that fibroblast-specific genes are downregulated at early stages and that pluripotency-related genes are not upregulated until the late stages of reprogramming (Brambrink et al., 2008; Stadtfeld et al., 2008). Although MYC was suggested to be the reprogramming factor primarily responsible for suppressing the cell-type-specific program (Sridharan et al., 2009), our data show that OCT4 alone can also inhibit somatic transcriptional networks. Finally, current available data suggest that OCT4, SOX2, and KLF4 act synergistically, as OCT4 heterodimerizes with SOX2 in order to maintain ESC self-renewal (Boyer et al., 2005) and OCT4, SOX2, and KLF4 co-occupy the promoters of many reprogramming-related genes (Soufi et al., 2012; Sridharan et al., 2009). In contrast, we show that OCT4 alone is able to initiate certain steps in reprogramming, such as the activation of Mgarp, and that OCT4 and KLF4 play an antagonistic role on Mgarp transcriptional regulation.
Whereas each cell type exhibited a unique pattern of up- and downregulated genes, four genes were upregulated by OCT4 in all examined somatic cell types, including Mgarp. Our results show that the process of inducing de novo pluripotency does not simply consist of the activation of ESC-specific genes to the levels present in ESCs. Instead, some genes such as Mgarp need to be first downregulated and subsequently upregulated during the reprogramming process. We have elucidated the mechanism underlying this counterintuitive temporal expression pattern. In fact, KLF4 alone completely abolishes Mgarp expression whereas OCT4 alone induces high levels of Mgarp. This competitive interplay ensures that appropriate expression levels of Mgarp are maintained at different time points, which appear to be crucial for a successful reprogramming process, as either the permanent inhibition or the premature activation of Mgarp prevents the efficient generation of iPSCs. To our knowledge, Mgarp is the first gene described to date upon which KLF4 and OCT4 exhibit antagonistic effects. The mechanism that regulates the switch from KLF4- to OCT4-regulated Mgarp expression remains to be identified.
In summary, we have dissected the specific role that OCT4 plays during the first steps of reprogramming independently of the other reprogramming factors. We have shown that OCT4 does not require SOX2 to interfere with cell-type-specific gene-expression profiles or to initiate an unstable transcriptional state that facilitates reprogramming. Finally, we have discovered a mechanism by which OCT4 and KLF4 compete to differentially regulate Mgarp expression, resulting in distinct expression levels at different stages of reprogramming. Therefore, our study sheds new light on the function of OCT4 in the reprogramming process.
Experimental Procedures
Cell Derivation and Culture
Different somatic cell types were derived from transgenic mice containing a tetracycline-inducible transactivator (rtTA-M2), a TO (Hochedlinger et al., 2005), and a GOF18 Oct4-GFP transgene in which the GFP is driven by 18 kb of the Oct4 regulatory region (Yoshimizu et al., 1999). In addition, a control cell line was generated from transgenic mice, comprising a tetracycline-inducible transactivator (irtTA–VP16–GBD; Anastassiadis et al., 2002) plus an OG2 Oct4-GFP reporter transgene (Yoshimizu et al., 1999) but lacking a TO cassette. MEFs and NSCs were derived from 14.5 dpc embryos and cultured as previously described (Conti et al., 2005). BMCs were isolated from 6-week-old mice after flushing the femora and tibiae and plated onto dishes with a combined coating of gelatin (PAA Laboratories), laminin (Santa Cruz Biotechnology), and poly-L-lysine (Sigma), and cultivated in Dulbecco’s modified Eagle’s medium (DMEM) Low Glucose (Gibco) with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM/l glutamine (all PAA). The transgenic Oct4 was induced by either 6 μg/ml doxycycline (Sigma) or 2 μg/ml doxycycline plus 10−7 M dexamethasone (Sigma), depending on the respectively contained tetracycline transactivator. 5-azacytidine (10 nM; Sigma) was applied as indicated. Generation, culture, and differentiation of ESCs and iPSCs were performed as previously described (Tiemann et al., 2011). Animal handling was in accordance with the Max Planck Institute animal protection guidelines and the German animal protection laws.
Viral Vectors
Production and transduction of viral vectors were performed as reported (Han et al., 2010). Retroviral pMXs vectors encoding for Oct4, Sox2, Klf4, and Myc (Takahashi and Yamanaka, 2006) as well as the lentiviral vectors TetOP-CMV-Oct4 and M2-rtTA tetracycline-transactivator (Stadtfeld et al., 2008) were purchased from Addgene (plasmids numbers 13366, 13367, 13370, 13375, 19766, and 20342). Mgarp cDNA was amplified and cloned into the pMXs and TetOP-CMV backbones. shRNA constructs targeting Mgarp were cloned into the lentiviral vectors pLVTHM (Wiznerowicz and Trono, 2003; Addgene number 12247) and Tet-pLKO-puro (Wee et al., 2008; Addgene number 21915). A Klf4-2a-tomato overexpression cassette was also cloned into the pLVTHM backbone (Han et al., 2011).
iPSC Characterization
Immunocytochemistry, immunoblotting, ALP staining, methylation analysis, and blastocyst injection were performed as previously described (Han et al., 2011, 2013; Tiemann et al., 2011). Relative transcript expression levels were calculated based on the ΔΔCt method after normalization to Gapdh and Actb housekeeping controls (Kim et al., 2008). Primers and antibodies are listed in Tables S5 and S6, respectively.
Measurement of Mitochondrial Activity and Copy Number
Mitochondria were stained for 30 min with 30 nM tetramethylrhodamine methyl ester (Invitrogen) and quantified on a BD FACSAria II flow cytometer (Becton Dickinson). Copy numbers of mitochondrial DNA were assessed by quantitative PCR (Kelly et al., 2013).
ChIP
MEFs overexpressing Oct4 (TO-MEFs after doxycycline treatment) or Klf4 (MEFs sorted for tomato expression after transduction with LVTHM-Klf4-2a-tomato) were used for ChIP. Cell cultures were crosslinked with 1% formaldehyde for 10 min at room temperature and sonicated using a Bioruptor device (Diagenode). Chromatin was immunoprecipitated with the corresponding antibodies, and the precipitated DNA was analyzed by qRT-PCR. Primers and antibodies are listed in Tables S5 and S6, respectively.
Microarray Gene-Expression Analysis
Sample preparation and analysis was performed as previously described (Tapia et al., 2012). Score values representing up- and downregulated genes were calculated from the processed raw data as follows (Figure S2). Signal changes for each probe and each cell type were calculated by subtraction of the logarithmic values between 0 hr and 24 hr (Δtotal). To reduce the impact of high relative changes in noisy background expression, the signal change was set to zero if the signal intensity never reached a threshold of six. Next, two correction parameters were introduced. The first parameter compensates for signal changes that do not show a gradual tendency throughout the time course. To this end, subtraction values between 6 and 0 hr, 12 and 6 hr, and 24 and 12 hr were calculated only for those signal intensities greater than six. If any of these intermediate steps presents a sign (positive or negative) that differs from that of the overall trend, this value is subtracted from the Δtotal value. The second correction parameter compensates for signal changes that are observed not only in the sample cell type but also in one or both controls. In this case, the subtraction value between 24 and 0 hr of the control cell line that presents the most similar behavior to the studied sample was subtracted from Δtotal. Finally, the sign of the score after subtracting the two correction parameters cannot become different from the original sign of Δtotal. Probes with a score higher than 1 or lower than −1 were considered up- or downregulated, respectively, which corresponds to more than a 2-fold total change in expression.
Gene ontology analyses were performed using the Functional Annotation tool of the DAVID bioinformatics database (Huang da et al., 2009).
Author Contributions
The indicated authors contributed as follows: U.T., conception and design, collection and assembly of data, data analysis and interpretation, and manuscript writing; A.G.M., provision of study material and collection and assembly of data; K.A., collection and assembly of data; G.W., collection and assembly of data; G.U.L.F., provision of study material; M.J.A.-B., data analysis and interpretation; H.R.S., conception and design, data analysis and interpretation, supervision of the study, manuscript writing, and financial support; and N.T., conception and design, data analysis and interpretation, supervision of the study, and manuscript writing.
Acknowledgments
We thank Tobias Cantz, Frank Edenhofer, and Boris Greber for their helpful comments; Martin Stehling for technical assistance; and Areti Malapetsas for editing the manuscript. This research was supported by the Max-Planck-Gesellschaft and a grant of the Deutsche Forschungsgemeinschaft (DFG SI 1695/1-2 SPP 1356: Pluripotency and Cellular Reprogramming). U.T. and A.G.M. are supported by a fellowship of the International Max Planck Research School – Molecular Biomedicine.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Hans Robert Schöler, Email: office@mpi-muenster.mpg.de.
Natalia Tapia, Email: ntapia@mpi-muenster.mpg.de.
Accession Numbers
The Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/gds) accession number for the microarray data reported in this paper is GSE47061.
Supplemental Information
References
- Anastassiadis K., Kim J., Daigle N., Sprengel R., Schöler H.R., Stewart A.F. A predictable ligand regulated expression strategy for stably integrated transgenes in mammalian cells in culture. Gene. 2002;298:159–172. doi: 10.1016/s0378-1119(02)00979-4. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T., Foreman R., Welstead G.G., Lengner C.J., Wernig M., Suh H., Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Faddah D.A., Cheng A.W., Itskovich E., Markoulaki S., Ganz K., Klemm S.L., van Oudenaarden A., Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Conti L., Pollard S.M., Gorba T., Reitano E., Toselli M., Biella G., Sun Y., Sanzone S., Ying Q.L., Cattaneo E., Smith A. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe J.A., Hilcove S., Kim J., Zhou H., Ouyang K., Wang G., Chen J., Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- Feng B., Jiang J., Kraus P., Ng J.H., Heng J.C., Chan Y.S., Yaw L.P., Zhang W., Loh Y.H., Han J. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat. Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- Folmes C.D., Nelson T.J., Martinez-Fernandez A., Arrell D.K., Lindor J.Z., Dzeja P.P., Ikeda Y., Perez-Terzic C., Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.W., Tapia N., Joo J.Y., Greber B., Araúzo-Bravo M.J., Bernemann C., Ko K., Wu G., Stehling M., Do J.T., Schöler H.R. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell. 2010;143:617–627. doi: 10.1016/j.cell.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Han D.W., Greber B., Wu G., Tapia N., Araúzo-Bravo M.J., Ko K., Bernemann C., Stehling M., Schöler H.R. Direct reprogramming of fibroblasts into epiblast stem cells. Nat. Cell Biol. 2011;13:66–71. doi: 10.1038/ncb2136. [DOI] [PubMed] [Google Scholar]
- Han D.W., Tapia N., Hermann A., Hemmer K., Höing S., Araúzo-Bravo M.J., Zaehres H., Wu G., Frank S., Moritz S. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- Han D.W., Tapia N., Araúzo-Bravo M.J., Lim K.T., Kim K.P., Ko K., Lee H.T., Schöler H.R. Sox2 Level Is a Determinant of Cellular Reprogramming Potential. PLoS ONE. 2013;8:e67594. doi: 10.1371/journal.pone.0067594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Saha K., Pando B., van Zon J., Lengner C.J., Creyghton M.P., van Oudenaarden A., Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J., Rafiee M.R., Reiland S., Polo J.M., Gehring J., Okawa S., Huber W., Hochedlinger K., Krijgsveld J. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2012;2:1579–1592. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K., Yamada Y., Beard C., Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ichida J.K., Blanchard J., Lam K., Son E.Y., Chung J.E., Egli D., Loh K.M., Carter A.C., Di Giorgio F.P., Koszka K. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Chan Y.S., Loh Y.H., Cai J., Tong G.Q., Lim C.A., Robson P., Zhong S., Ng H.H. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- Kelly R.D., Sumer H., McKenzie M., Facucho-Oliveira J., Trounce I.A., Verma P.J., St John J.C. The effects of nuclear reprogramming on mitochondrial DNA replication. Stem Cell Rev. 2013;9:1–15. doi: 10.1007/s12015-011-9318-7. [DOI] [PubMed] [Google Scholar]
- Kim J.B., Zaehres H., Wu G., Gentile L., Ko K., Sebastiano V., Araúzo-Bravo M.J., Ruau D., Han D.W., Zenke M., Schöler H.R. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Kim J., Efe J.A., Zhu S., Talantova M., Yuan X., Wang S., Lipton S.A., Zhang K., Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. USA. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinouchi R., Kinouchi T., Hamamoto T., Saito T., Tavares A., Tsuru T., Yamagami S. Distribution of CESP-1 protein in the corneal endothelium and other tissues. Invest. Ophthalmol. Vis. Sci. 2006;47:1397–1403. doi: 10.1167/iovs.05-0602. [DOI] [PubMed] [Google Scholar]
- Koche R.P., Smith Z.D., Adli M., Gu H., Ku M., Gnirke A., Bernstein B.E., Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lim S., Hoffman D., Aspenstrom P., Federoff H.J., Rempe D.A. HUMMR, a hypoxia- and HIF-1alpha-inducible protein, alters mitochondrial distribution and transport. J. Cell Biol. 2009;185:1065–1081. doi: 10.1083/jcb.200811033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N., Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A.A. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Mathur D., Danford T.W., Boyer L.A., Young R.A., Gifford D.K., Jaenisch R. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome Biol. 2008;9:R126. doi: 10.1186/gb-2008-9-8-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T., Minegishi K., Ishimoto H., Tanaka M., Hennebold J.D., Teranishi T., Hattori Y., Furuya M., Higuchi T., Asai S. Expression of ovary-specific acidic protein in steroidogenic tissues: a possible role in steroidogenesis. Endocrinology. 2009;150:3353–3359. doi: 10.1210/en.2008-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Orkin S.H., Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S., Wang Y., Zhou M., Ge Y., Yan Y., Wang J., Zhang S.S., Zhang S. A mitochondria-localized glutamic acid-rich protein (MGARP/OSAP) is highly expressed in retina that exhibits a large area of intrinsic disorder. Mol. Biol. Rep. 2011;38:2869–2877. doi: 10.1007/s11033-010-9948-x. [DOI] [PubMed] [Google Scholar]
- Reményi A., Lins K., Nissen L.J., Reinbold R., Schöler H.R., Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda D.J., Chew J.L., Lim L.H., Loh Y.H., Wang B., Ng H.H., Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- Soufi A., Donahue G., Zaret K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., Tchieu J., Mason M.J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Maherali N., Breault D.T., Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo E., Rampalli S., Risueño R.M., Schnerch A., Mitchell R., Fiebig-Comyn A., Levadoux-Martin M., Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tapia N., Reinhardt P., Duemmler A., Wu G., Arauzo-Bravo M.J., Esch D., Greber B., Cojocaru V., Rascon C.A., Tazaki A. Reprogramming to pluripotency is an ancient trait of vertebrate Oct4 and Pou2 proteins. Nat. Commun. 2012;3:1279. doi: 10.1038/ncomms2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thier M., Wörsdörfer P., Lakes Y.B., Gorris R., Herms S., Opitz T., Seiferling D., Quandel T., Hoffmann P., Nöthen M.M. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10:473–479. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Tiemann U., Sgodda M., Warlich E., Ballmaier M., Scholer H.R., Schambach A., Cantz T. Optimal reprogramming factor stoichiometry increases colony numbers and affects molecular characteristics of murine induced pluripotent stem cells. Cytometry A. 2011;79:426–435. doi: 10.1002/cyto.a.21072. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Südhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Chen K., Zeng X., Yang J., Wu Y., Shi X., Qin B., Zeng L., Esteban M.A., Pan G., Pei D. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Wee S., Wiederschain D., Maira S.M., Loo A., Miller C., deBeaumont R., Stegmeier F., Yao Y.M., Lengauer C. PTEN-deficient cancers depend on PIK3CB. Proc. Natl. Acad. Sci. USA. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M., Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimizu T., Sugiyama N., De Felice M., Yeom Y.I., Ohbo K., Masuko K., Obinata M., Abe K., Schöler H.R., Matsui Y. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Differ. 1999;41:675–684. doi: 10.1046/j.1440-169x.1999.00474.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


