Abstract
The emergence of jawed vertebrates (gnathostomes) from jawless vertebrates was accompanied by major morphological and physiological innovations, such as hinged jaws, paired fins and immunoglobulin-based adaptive immunity. Gnathostomes subsequently diverged into two groups, the cartilaginous fishes and the bony vertebrates. Here we report the whole-genome analysis of a cartilaginous fish, the elephant shark (Callorhinchus milii). We find that the C. milii genome is the slowest evolving of all known vertebrates, including the ‘living fossil’ coelacanth, and features extensive synteny conservation with tetrapod genomes, making it a good model for comparative analyses of gnathostome genomes. Our functional studies suggest that the lack of genes encoding secreted calcium-binding phosphoproteins in cartilaginous fishes explains the absence of bone in their endoskeleton. Furthermore, the adaptive immune system of cartilaginous fishes is unusual: it lacks the canonical CD4 co-receptor and most transcription factors, cytokines and cytokine receptors related to the CD4 lineage, despite the presence of polymorphic major histocompatibility complex class II molecules. It thus presents a new model for understanding the origin of adaptive immunity.
Supplementary information
The online version of this article (doi:10.1038/nature12826) contains supplementary material, which is available to authorized users.
Subject terms: Molecular evolution, Cellular immunity, Comparative genomics, Development
Whole-genome analysis of the elephant shark, a cartilaginous fish, shows that it is the slowest evolving of all known vertebrates, lacks critical bone formation genes and has an unusual adaptive immune system.
Supplementary information
The online version of this article (doi:10.1038/nature12826) contains supplementary material, which is available to authorized users.
First genome of a cartilaginous fish
The elephant shark (Callorhinchus milii) is a cartilaginous fish native to the temperate waters off southern Australia and New Zealand, living at depths of 200 to 500 metres and migrating into shallow waters during spring for breeding. The genome sequence is published in this issue of Nature. Comparison with other vertebrate genomes shows that it is the slowest evolving genome of all known vertebrates — coelacanth included. Genome analysis points to an unusual adaptive immune system lacking the CD4 receptor and some associated cytokines, indicating that cartilaginous fishes possess a primordial gnathostome adaptive immune system. Also absent are genes encoding secreted calcium-binding phosphoproteins, in line with the absence of bone in cartilaginous fish.
Supplementary information
The online version of this article (doi:10.1038/nature12826) contains supplementary material, which is available to authorized users.
Main
The emergence of gnathostomes from jawless vertebrates marks a major event in the evolution of vertebrates. This transition was accompanied by many morphological and phenotypic innovations, such as jaws, paired appendages and an adaptive immune system based on immunoglobulins, T-cell receptors and major histocompatibility complex (MHC) molecules1,2,3,4 (Fig. 1a). How these novelties emerged and how they facilitated the divergence, adaptation and dominance of gnathostomes as the major group (99.9%) of living vertebrates are key unresolved questions. The living gnathostomes are divided into two groups, the cartilaginous fishes (Chondrichthyes) and bony vertebrates (Osteichthyes), which diverged about 450 Myr ago (Fig. 1a). A key feature distinguishing the two groups is that chondrichthyans have largely cartilaginous endoskeletons whereas osteichthyans have ossified endoskeletons. Although fossil jawless vertebrates (for example galeaspids) and jawed vertebrates (for example placoderms) possessed dermal and perichondral bone, endochondral bone is found only in osteichthyans5. Chondrichthyans include about 1,000 living species that are grouped into two lineages, the holocephalans (chimaeras) and elasmobranchs (sharks, rays and skates), which diverged about 420 Myr ago6 (Fig. 1a). A detailed whole-genome evaluation of a chondrichthyan and comparative analysis with the available genome information on osteichthyans and a jawless vertebrate7 might help us to understand features unique to chondrichthyans and provide insights into the ancestral state of gnathostome-specific morphological features and physiological systems.
Figure 1. Phylogeny of chordates.
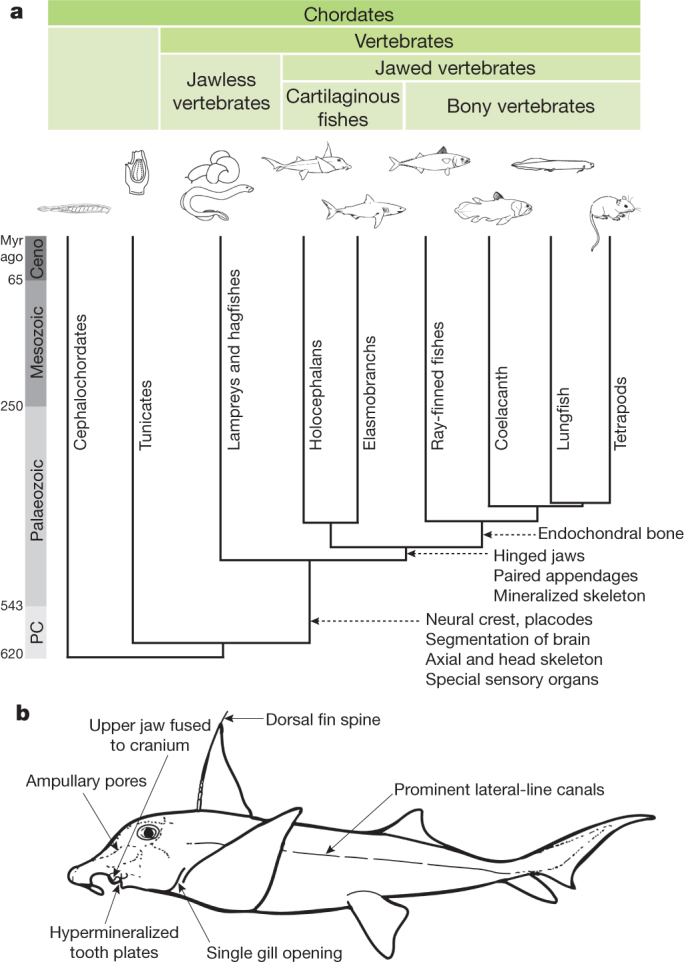
a, Major shared features of various vertebrate taxa; b, unique features of C. milii. Lampreys and hagfishes (cyclostomes) lack mineralized tissues. In contrast, cartilaginous fishes produce extensive dermal bone such as teeth, dermal denticle and fin spine. However, they lack the ability to make endochondral bone which is unique to bony vertebrates. Divergence times are from refs 6, 36. Ceno, Cenozoic era; PC, Precambrian era.
We previously identified C. milii, a holocephalan, as a chondrichthyan genome model8,9 because of its relatively small genome (∼1.0 gigabase). Compared with elasmobranchs, the unique features of holocephalans include a single gill opening, a complete hyoid arch, fusion of the upper jaw to the cranium, and non-replaceable hypermineralized tooth plates10 (Fig. 1b). Callorhinchus milii inhabits temperate waters of the continental shelves off southern Australia and New Zealand, typically at depths of 200 to 500 m (ref. 11). Here, we report the generation and analysis of a high-quality genome sequence of C. milii. Several key findings are presented here and further details on our in-depth characterization of this genome are presented in Supplementary Notes I to XI.
Genome assembly and annotation
Genomic DNA of a single male C. milii was sequenced and assembled (Supplementary Note I) to a size of 0.937 gigabases, comprising 21,208 scaffolds (N50 contig, 46.6 kilobases; N50 scaffold, 4.5 megabases; Supplementary Table I.1). The average GC content of the C. milii genome is 42.3%, and approximately 46% of the genome is organized into isochores (Supplementary Note II). Using the Ensembl annotation pipeline12 and RNA-seq transcript evidence, we predicted a total of 18,872 protein-coding genes. In addition, microRNA (miRNA) genes were identified by small-RNA sequencing and annotation of the genome assembly (Supplementary Note III). Callorhinchus milii have more miRNA gene loci (693 genes and 136 families) than do teleosts (for example, zebrafish have 344 genes and 94 families) but fewer than do humans (1,527 genes and 558 families) and other mammals (mirBase release 19). Several novel C. milii-specific miRNAs are expressed at high levels in a tissue-specific manner (Supplementary Figs III.1 and III.2). Notably, a considerable number (16%; 22/136) of C. milii miRNA families conserved in mammals have been secondarily lost in teleosts. Their explicit tissue-specific expression patterns in C. milii and mammals suggest that they have important roles in gene regulation. A total of 63,877 noncoding elements (average size, 271 base pairs) conserved between C. milii and bony vertebrates represent potential cis-regulatory elements (Supplementary Note IV). Surprisingly, only a tiny fraction (less than 1.0%) of these are found in the genomes of sea lamprey, sea squirt and amphioxus, suggesting that the emergence of gnathostomes was accompanied by major innovations in cis-regulatory elements and gene regulatory networks.
Phylogenomics and evolutionary rate
Morphological and palaeontological studies have placed Chondrichthyes as a sister group to bony vertebrates5. However, subsequent molecular phylogenetic analyses based on mitochondrial or nuclear genes have produced conflicting topologies13,14,15. Using a genome-scale data set comprising 699 one-to-one orthologues from C. milii and 12 other chordates, we provide robust support for the traditional phylogenetic tree with an unambiguous split between Chondrichthyes and bony vertebrates (Supplementary Fig. V.1). Furthermore, analysis of gains and losses of introns provided independent support for Chondrichthyes as a sister group to bony vertebrates (Supplementary Note V).
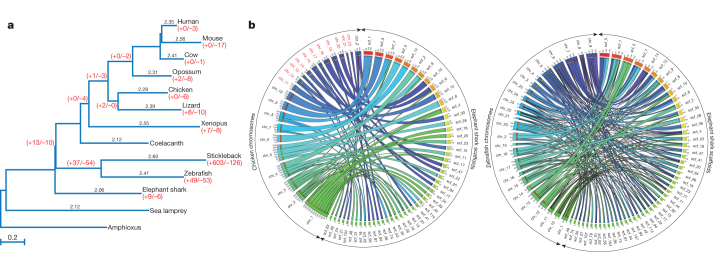
Previous studies based on a few mitochondrial and nuclear protein-coding genes indicated that the nucleotide substitution rate in elasmobranchs is an order of magnitude lower than that in mammals16,17. Using the genome-wide set of 699 orthologues, we estimated the molecular evolutionary rate of C. milii and compared it with other gnathostomes, with sea lamprey as the outgroup. Callorhinchus milii protein-coding genes have evolved significantly slower than all other vertebrates examined (P < 0.01 for all comparisons; Supplementary Tables VI.1–VI.3), including the coelacanth, which has been considered to be the slowest evolving bony vertebrate18. A neutral tree based on fourfold-degenerate sites indicated that the low evolutionary rate is a reflection of the neutral nucleotide mutation rate, and confirmed that the neutral evolutionary rate of C. milii is the lowest (Fig. 2a).
Figure 2. Callorhinchus milii possesses the slowest evolving vertebrate genome.
a, Neutral tree of 13 chordates based on fourfold degenerate sites. Pairwise distances to amphioxus are shown for each species above their respective branches. Intron gain/loss (+/−) events are shown in red below taxon labels and at ancestral nodes. b, Circos plots showing syntenic relationships between C. milii scaffolds and chicken and zebrafish chromosomes. Most C. milii scaffolds show a one-to-one relation with chicken chromosomes whereas they show a correspondence of one-to-two, or more, with zebrafish chromosomes. Chicken microchromosomes are labelled in red.
The lower rates of molecular evolution of C. milii are also evident in the fewer changes in the intron–exon organization of its genes (Supplementary Note VII). Callorhinchus milii has experienced fewer intron gains or losses than any bony vertebrate since their divergence from the gnathostome ancestor (Fig. 2a). The highest rates of change were found in two teleost fishes, the stickleback and zebrafish, with the stickleback lineage experiencing the highest number of changes (603 gains and 126 losses after it split from the zebrafish lineage) recorded in any vertebrate lineage (Fig. 2a). In addition to a lower rate of intron changes, the C. milii genome also has experienced a relatively low rate of major interchromosomal rearrangements, comparable to that of chicken, which has one of the most stable karyotypes among tetrapods19,20 (Supplementary Note VIII). An extensive conservation of synteny was observed in comparisons of C. milii scaffolds with chicken and human chromosomes, with a majority of C. milii scaffolds (93%) showing a one-to-one correspondence with chicken chromosomes (Supplementary Figs VIII.1 and VIII.2). A three-way comparison between C. milii, chicken and teleosts (medaka and zebrafish) revealed that teleosts have undergone a substantially higher number of interchromosomal rearrangements than previously demonstrated by simple tetrapod–teleost comparisons (Fig. 2b and Supplementary Tables VIII.7 and VIII.8).
Evolution of protein-coding gene families
Protein domains
Comparisons of Pfam domains (Supplementary Note IXa) identified 17 C. milii domains that are missing in bony vertebrates (Supplementary Table IX.2). Sixteen of these are present in amphioxus or other eukaryotes, and are thus ancient protein domains that are still retained in C. milii but have been lost from bony vertebrates. We note that 13 domains shared between C. milii and tetrapods are absent in teleosts (Supplementary Table IX.4), indicating that they have been secondarily lost from the teleost lineage.
Lineage-specific gene losses
Orthologues of more C. milii genes were found to be lost from the teleost lineage (271 genes; Supplementary Note IXb and Supplementary Table IX.6) relative to the tetrapod lineage (34 genes; Supplementary Table IX.7). Human orthologues of many genes lost from teleosts are associated with genetic diseases (104 genes, 38%; Supplementary Table IX.6), indicating their importance for human physiology. The loss of these genes from teleosts supports the idea that teleosts represent a more derived group than do other gnathostomes. The functional annotation of zebrafish orthologues of the 34 genes lost from tetrapods highlighted several genes that are specific to the aquatic lifestyle, such as those regulating fin and lateral-line development and those encoding receptors for water-soluble odorants (Supplementary Table IX.7).
Genetic basis of bone formation
Bone is the most widespread mineralized tissue in vertebrates, and its formation represents a major leap in vertebrate evolution. Although chondrichthyans produce dermal bone (for example teeth, dermal denticles and fin spines) and calcified cartilage5,21, unlike bony vertebrates, their cartilage is not replaced with endochondral bone. Among vertebrates, the earliest mineralized tissue was found in the feeding apparatus of extinct jawless fishes, the conodonts21. Early dermal bone was found in extinct jawless vertebrates such as heterostracans, whereas perichondral bone surrounding the cartilage was found in several fossil jawless vertebrates (osteostracans, galeaspids) and jawed vertebrates5 (placoderms, acanthodians; Fig. 3). However, the highly complex process of endochondral ossification is unique to bony vertebrates. The C. milii genome sequence provided a unique opportunity to address the question of why the endoskeleton of chondrichthyans is not ossified.
Figure 3. Genetic events underlying the emergence of bone formation in vertebrates.
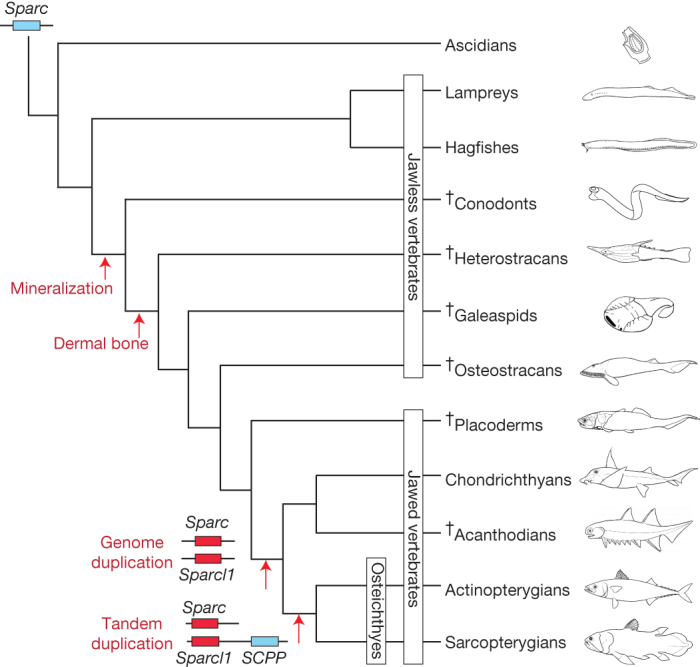
Duplication of Sparc by whole-genome duplication initially gave rise to Sparcl1, and the subsequent tandem duplication of Sparcl1 gave rise to the SCPP gene family responsible for endochondral ossification. Because the sea lamprey genome contains only Sparc but no Sparcl1 (ref. 22), we have placed the genome duplication event that gave rise to Sparcl1 after the divergence of jawless vertebrates from the jawed vertebrate ancestor. The sister relationship of chondrichthyans and acanthodians is based on ref. 37. SCPP, secretory calcium-binding phosphoprotein gene family member. †, extinct.
We searched the C. milii genome assembly and transcriptomes for genes known to be involved in bone formation in osteichthyans (Supplementary Note X). All gene family members involved in bone formation were present, except the secretory calcium-binding phosphoprotein (SCPP) gene family (Supplementary Table X.1). This gene family encodes a diverse array of secreted phosphoproteins that arose from the gene Sparc-like 1 (Sparcl1) through tandem duplication, and Sparcl1 itself arose from an ancient metazoan gene, Sparc, through whole-genome duplication22. There are two main categories of SCPP genes: one group encodes acidic proteins and the other encodes proline- and glutamine-rich (P/Q-rich) proteins. In the human genome, the two groups are found in two different clusters on chromosome 4; the acidic SCPP genes (SPP1, MEPE, IBSP, DMP1 and DSPP, collectively known as SIBLING genes) occur between PKD2 and SPARCL1, whereas the P/Q-rich SCPP genes are found in the enamel matrix protein-SCPP cluster ∼17 megabases downstream of SPARCL1 (Supplementary Fig. X.2). Acidic SCPP or SIBLING genes are involved in the ossification of collagenous matrix in bone and dentine, and P/Q-rich SCPP genes are involved in the production of enamel, milk, tears and saliva. Although there are variable numbers of P/Q-rich SCPP genes in teleosts23, there is a single SIBLING gene, spp1, in zebrafish and medaka. Zebrafish spp1 (also known as osteopontin) is expressed specifically in osteoblasts24 and has therefore been proposed to have a primary function in bone formation similar to its mammalian orthologue23.
The C. milii genome contains both Sparc and Sparcl1 on different scaffolds that show extensive conserved synteny with orthologous loci in human and other bony vertebrates (Supplementary Figs X.1 and X.2). However, there is no SCPP gene cluster in the intergenic region between Pkd2 and Sparcl1 or elsewhere in the genome (Supplementary Fig. X.2). The genomic and transcriptomic resources available for other cartilaginous fishes such as the little skate (Leucoraja erinacea) and the small-spotted catshark (Scyliorhinus canicula) as well as the genome assembly of the sea lamprey, a jawless vertebrate, also do not contain any SCPP genes (Supplementary Note X). These findings suggest that the tandem duplication of Sparcl1 that gave rise to SCPP genes occurred in the common ancestor of osteichthyans after this lineage split from the chondrichthyan lineage (Fig. 3). Because SCPP genes have a crucial role in the formation of bone, we propose that their absence from C. milii explains the absence of bone from the endoskeleton of cartilaginous fishes.
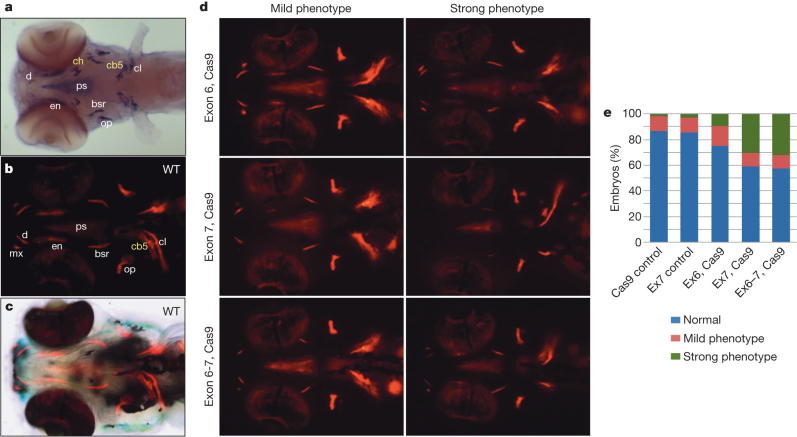
To test this hypothesis, we used two different methods to disrupt the function of the single bone-specific SIBLING gene spp1 in zebrafish. The knockdown of spp1 using two gene-specific morpholinos (ATG MO and E2-I2 MO) resulted in a significant reduction in endochondral and dermal bone formation by comparison with embryos injected with 5-base-pair-mismatch control morpholinos (Supplementary Fig. X.4). Unlike the transient effects exerted by morpholinos, the genetic interference afforded by the CRISPR/Cas9 system25 results in heritable genomic modifications; indeed, targeting exons 6 and 7 of the spp1 gene using the CRISPR/Cas9 system resulted in the generation of specific insertion/deletion mutations at the target sites, including a ∼2.6-kilobase deletion when exons 6 and 7 were simultaneously targeted (Supplementary Fig. X.6). Embryos 5 days post-fertilization (dpf) with deletions in exon 7 alone or in both exon 6 and exon 7 of spp1 showed a significant reduction in the formation of endochondral and dermal bone (Fig. 4), with the defect in bone formation persisting in 15-dpf mutant embryos (Supplementary Fig. X.9). The similar phenotypes obtained using two different methods of manipulation indicate that the effects on bone formation are specific, and strongly suggest that spp1 has an essential role in the modulation of bone formation in zebrafish.
Figure 4. Targeted mutagenesis of zebrafish spp1 by sgRNA:Cas9 results in reduced bone formation.
a, spp1 is specifically expressed in cells surrounding the bone matrix. Ventral view of a 5-dpf embryo hybridized with a spp1-specific RNA probe. Yellow labels, endochondral bones (cb5, ceratobranchial 5; ch, ceratohyal); white labels, dermal bones (bsr, branchiostegal ray; cl, cleithrum; d, dentary; en, entopterygoid; op, operculum; ps, parasphenoid). b, Ventral view of a 5-dpf wild-type (WT) embryo stained with alizarin red to reveal sites of bone deposition (red fluorescence). mx, maxilla. c, Bright-field image merged with b to visualize anatomical structures and locations of bone deposition simultaneously. d, Ventral views of 5-dpf embryos injected with Cas9 mRNA together with single guide RNA (sgRNA) targeting spp1 exon 6, exon 7 or both (alizarin red staining). The embryos were scored as normal (resembling wild type), mild or strong bone phenotypes, with the latter showing the greatest reduction in bone formation. The variations in the extent of bone reduction are presumably due to somatic chimaerism with regard to spp1 disruption. e, Proportions of mild and strong bone phenotypes induced by disruption of spp1 by sgRNA/Cas9. Targeting of exon 7 (n = 206 embryos) or both exons 6 and 7 (n = 143) resulted in significantly higher proportions of strong bone phenotype (P < 0.01, Fisher’s exact test) compared with control injections of Cas9 mRNA (n = 190) and exon 7 sgRNA (n = 143) (Ex6, Cas9: n = 72).
The results of the zebrafish spp1 knockdown experiments provide strong support for our hypothesis that the absence of SCPP genes from cartilaginous fishes is related to the unossified nature of their endoskeleton. In turn, the absence of SCPP genes from chondrichthyans raises questions about the genetic basis of dermal and perichondral bone formation in chondrichthyans, placoderms, acanthodians and extinct jawless vertebrates. We speculate that one or more SCPP-related genes, probably Sparc, Sparcl1 or both, mediate the mineralization of skeleton in these vertebrates.
Primordial adaptive immune system
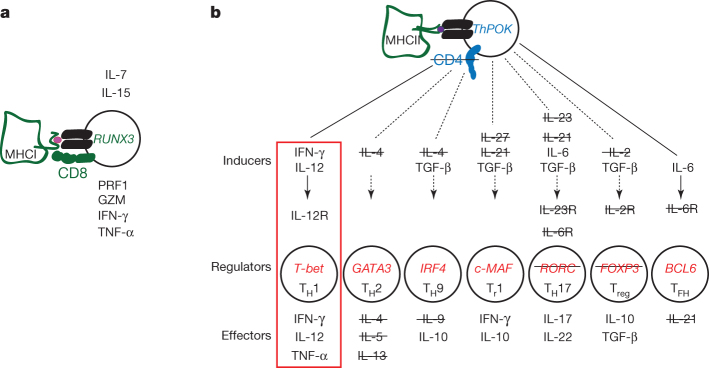
The chondrichthyan immune system shares many features of the innate and adaptive immune systems of mammals9,26. However, several important differences, confirmed by transcriptome analysis of an elasmobranch cartilaginous fish, the nurse shark (Ginglymostoma cirratum), which diverged from the C. milii lineage ∼420 Myr ago6, highlight several unexpected features of the primordial state of gnathostome immune systems, especially for adaptive immunity (Supplementary Note XI, Supplementary Figs XI.1–XI.10 and Supplementary Tables XI.1–XI.13). First, the genome assembly suggests a close linkage of immunoglobulin and T-cell receptor genes, compatible with the notion that the somatically diversifying antigen receptor genes evolved from a common ancestor via en bloc duplications27. Indeed, the presence of the variant antigen receptor NAR–TCR, but lack of other specialized forms of antigen receptors, such as IgW and IgNAR in the C. milii genome suggest that the single-domain V region, subjected to somatic diversification by Rag proteins, was at first part of a T-cell receptor (TCR)-like structure before being co-opted by immunoglobulins. Second, the linkage of antigen receptor genes with certain MHC genes, whose products functionally interact in regulating the immune response, supports the co-evolutionary origin of antigen presentation and recognition28,29. Third, the immunogenome of C. milii is compatible with the presence of cytotoxic natural-killer and CD8+ T cells; by contrast, the absence of the canonical CD4 co-receptor, the transcription factor RORC, several cytokines and cytokine receptors that are associated with helper and regulatory functions of CD4+ T cells in mammals (Fig. 5) suggests the presence of a primordial type of helper function in cartilaginous fishes. Restricted helper functions (exemplified by the lack of the T-follicular helper lineage) in cartilaginous fishes may explain the long lag time required to generate humoral immunity (affinity maturation and memory) in sharks30. Thus, the emerging adaptive immune system seems to have been characterized by the presence of a full-blown cytotoxic system and a primordial helper system that was geared towards a TH1-type response.
Figure 5. T-cell lineages in mammals and cartilaginous fishes.
a, Schematic diagram of mammalian CD8+ cytotoxic cells producing various effector molecules, such as PRF1 (perforin), GZM (granzymes), INF-γ (interferon-γ) and TNF-α (tumour necrosis factor-α). Interleukins 7 (IL-7) and 15 (IL-15) are indispensable for induction of the transcription factor RUNX3, which controls expression of the genes encoding the CD8 co-receptor that interacts with the TCR and MHC class I molecules. All key elements of this T-cell lineage are present in the genomes of cartilaginous fishes. b, Diversification of CD4 lineages in mammals. The transcription factor ThPOK controls expression of the co-receptor CD4, which interacts with the TCR and MHC class II molecules and represses the CD8 lineage programme. The listed inducer cytokines activate key transcriptional regulators that define the multiple CD4+ lineages specialized for producing various effector molecules; genes not found in cartilaginous fishes are struck through. Cartilaginous fishes probably possess only TH1 cells (red rectangle) and lack other helper subsets. TGF-β, transforming growth factor-β.
Despite the apparent lack of Treg cells in cartilaginous fish, the elimination of self-reactive lymphocytes during the process of central tolerance seems to be a primordial vertebrate feature, as demonstrated by the presence of AIRE, which is responsible for the expression of peripheral self-antigens for presentation via MHC to T cells in the medulla of the thymus31. The absence of the gene encoding a bona fide CD4 co-receptor despite the presence of polymorphic MHC class II genes suggests the presence of unusual types of CD8− helper T cells, capable of exerting TH1-like activities through IFN-γ, IL-12 and TNF-α (Fig. 5). We suggest that such cells are capable of recognizing exogenous antigens presented by MHC class II molecules but might also interact with other antigens in an MHC-independent, antibody-like recognition mode, as recently demonstrated in mice deficient in both CD8 and CD4 co-receptors32. It is to be noted that the secondary loss of CD4, MHC class II and invariant chain genes in a teleost, the Atlantic cod (Gadus morhua), is accompanied by compensatory changes, such as amplification of MHC class I and Toll-like receptor genes33, most probably related to the disappearance of T-helper lineages only in this species. In stark contrast, the lack of such compensatory features in cartilaginous fishes, demonstrated in two highly divergent species, C. milii and G. cirratum, supports the hypothesis that the features described here are primordial. Our data also suggest that CD8 and CD4 were co-opted as co-receptors for the TCR at different times in evolution, and that T cells recognizing MHC class II molecules in cartilaginous fish recruit the downstream signalling components to the immunological synapse entirely through the TCR complex.
Conclusion
Among the three living lineages of vertebrates (cyclostomes, cartilaginous fishes and bony vertebrates), bony vertebrates are the largest and most diverse group of vertebrates. Because cartilaginous fishes are the sister group of bony vertebrates, they constitute a critical outgroup for understanding the evolution and diversity of bony vertebrates. The whole-genome analysis of C. milii, a holocephalan cartilaginous fish, shows that the C. milii genome is evolving significantly slower than other vertebrates, including the coelacanth, which is considered a ‘living fossil’. Although several physiological and environmental factors have been proposed to explain the interspecific variation in molecular evolutionary rates34,35, the factors contributing to the lower evolutionary rate of C. milii are not known. Overall, the C. milii genome is the least derived among known vertebrates and is therefore a good model for inferring the state of the ancestral chondrichthyan and gnathostome genomes. Its value for comparative genomic studies is illustrated by our analysis of genetic events that led to the ossification of endoskeleton in bony vertebrates. Unexpected was our finding that the adaptive immune system of cartilaginous fishes possesses highly restricted subsets of T helper cells (perhaps only one) with unconventional antigen-binding properties; this suggests that helper and regulatory functions of T cells that recognize MHC class II molecules became more elaborate in the ancestor of bony vertebrates through the acquisition of transcription factors such as RORC, the CD4 co-receptor, conventional FOXP3 and a host of CD4-lineage-specific cytokines and cytokine receptors. Thus, the whole-genome analysis of C. milii provides fresh insight into the mechanism of bone formation and the origin of adaptive immunity of gnathostomes.
Methods Summary
Genomic DNA was obtained from the testis of a single C. milii caught in Tasmania, Australia, and used to prepare libraries with inserts of different sizes. Sequencing was conducted on the Roche 454 GS FLX Titanium platform (for shotgun fragments and 3-kilobase and 8-kilobase inserts) and the ABI3730 instrument (for plasmid, fosmid and BAC ends). The C. milii genome was assembled using CABOG v6.1. PolyA-selected RNA from ten tissues of C. milii (brain, gills, heart, intestine, kidney, liver, muscle, ovary, spleen and testis) and the spleen and thymus of G. cirratum were sequenced on the Illumina GAIIx platform. Transcripts were assembled de novo using Trinity r2011-07-13. MicroRNA genes were identified by deep sequencing of small RNA from 16 tissues (brain, blood, eye, gills, heart, intestine, kidney, liver, muscle, ovary, pancreas, rectal gland, skin, spleen, testis and uterus) on the Illumina GAIIx platform. Annotation of the genome was carried out using the Ensembl gene annotation pipeline which integrated ab initio gene predictions and evidence-based gene models. The gene set can be viewed at http://esharkgenome.imcb.a-star.edu.sg/. Annotation of protein domains in the C. milii proteome was carried out by searching it against the Pfam v26 database using HMMER v3.0. Zebrafish spp1 was knocked down using morpholinos or the CRISPR/Cas9 system. Methods are described in detail in individual sections of the Supplementary Information.
All animals were cared for in strict accordance with National Institutes of Health (USA) guidelines. The protocol was approved by the Institutional Animal Care and Use Committee of the Biological Resource Centre, A*STAR (protocol #100520).
Supplementary information
This file contains Supplementary Notes 1-11, Supplementary Figures 1-11 and Supplementary Table 1-11 (please note that Supplementary Tables 3, 4, 8 and 9 are in a separate excel file) - see pages 1-7 for more details of the Content. (PDF 18504 kb)
This zipped file contains Supplementary Tables 3, 4, 8 and 9 (see the Supplementary Information file for more details. (ZIP 3143 kb)
Acknowledgements
Generation of C. milii genome sequence at the Genome Institute at Washington University was supported by grants from the National Human Genome Research Institute, USA. We thank members of the Production Sequencing group at the Genome Institute at Washington University. Sequencing of messenger RNA, small RNA and BAC ends was supported by grants from the Biomedical Research Council of A*STAR, Singapore. This work was supported by the A*STAR Computational Resource Centre through the use of its high-performance computing facilities. We would like to thank J. Danks, J. Bell and J. G. Patil for their help in collecting C. milii samples, and J. K. Joung for CRISPR and Cas9 plasmids. We also thank the following funding agencies: the Max Planck Society (T.B.); NIH grants RR006603 and AI27877 (M.F.F.); the Ministry of Education, Culture, Sports, Science and Technology, Japan (M.K.); the Human Frontiers Science Program Organization (M.I.); ERC Starting Grant (260372) and MICINN (Spain) BFU2011-28549 (T.M.-B.); and the Biomedical Research Council of A*STAR, Singapore (B.V., P.W.I., S. Hoon and V.K.).
PowerPoint slides
Author Contributions
B.V., R.K.W., S.B. and W.C.W. planned and oversaw the project. P.M., L.W.H. and W.C.W. sequenced and assembled the genome, and evaluated assembly coverage and quality. A.P.L., M.M.L. and B.V. annotated the genome. A.P.L. characterized repetitive sequences, GC content, isochores and CNEs. B.J.R. generated whole-genome alignments. B.L.-G., J.Q. and T.M.-B. analysed gene duplication and heterozygosity. S. Hoon, V.G., K.W.K. and S. Ho generated RNA-seq data and analysed miRNA families and protein domains. B.-H.T., S.T. and A.T. sequenced BAC ends and RNA-seq. M.M.L. and A.P.L. assembled and annotated RNA-seq data. V.R. performed phylogenomic and molecular evolutionary rate analyses. S.W.R. and M.I. investigated intron evolution. A.K.M., A.P.L., V.R., P.W.I. and B.V. analysed genes involved in bone formation. A.K.M. and Z.W.L. carried out functional analysis of zebrafish spp1. T.B., J.B.S., Y.O., M.F.F., Y.S., M.K., A.P.L., M.M.L. and B.V. analysed the immunogenome. V.K., V.R. and I.K. analysed evolution of protein-coding gene families. M.M.L., A.P.L. and B.V. analysed large-scale synteny. B.V., T.B. and W.C.W. wrote the manuscript with input from other authors.
Accession codes
Accessions
GenBank/EMBL/DDBJ
Sequence Read Archive
Data deposits
The C. milii genome assembly has been deposited at DDBJ/EMBL/GenBank under the accession number AAVX00000000. The version described in this paper is the second version, AAVX02000000. The C. milii and G. cirratum RNA-seq data have been deposited at SRA under accession numbers SRA054255 and SRA062964, respectively. The transcripts have been deposited into GenBank under accession numbers JW861113–JW881738, KA353634–KA353668; and the miRNA sequences under accession numbers JX994303–JX994995.
Competing interests
The authors declare no competing financial interests.
Contributor Information
Byrappa Venkatesh, Email: mcbbv@imcb.a-star.edu.sg.
Wesley C. Warren, Email: wwarren@genome.wustl.edu
References
- 1.Shimeld SM, Holland PW. Vertebrate innovations. Proc. Natl Acad. Sci. USA. 2000;97:4449–4452. doi: 10.1073/pnas.97.9.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gans C, Northcutt RG. Neural crest and the origin of vertebrates: a new head. Science. 1983;220:268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- 3.Young GC. Placoderms (armored fish): dominant vertebrates of the Devonian period. Annu. Rev. Earth Planet. Sci. 2010;38:523–550. doi: 10.1146/annurev-earth-040809-152507. [DOI] [Google Scholar]
- 4.Boehm T. Evolution of vertebrate immunity. Curr. Biol. 2012;22:R722–R732. doi: 10.1016/j.cub.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Janvier P. Early Vertebrates. 1996. [Google Scholar]
- 6.Inoue JG, et al. Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): a mitogenomic perspective. Mol. Biol. Evol. 2010;27:2576–2586. doi: 10.1093/molbev/msq147. [DOI] [PubMed] [Google Scholar]
- 7.Smith JJ, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nature Genet. 2013;45:415–421. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkatesh B, Tay A, Dandona N, Patil JG, Brenner S. A compact cartilaginous fish model genome. Curr. Biol. 2005;15:R82–R83. doi: 10.1016/j.cub.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 9.Venkatesh B, et al. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 2007;5:e101. doi: 10.1371/journal.pbio.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson JS. Fishes of the World. 2006. [Google Scholar]
- 11.Last PR, Stevens JD. Sharks and Rays of Australia. 1994. [Google Scholar]
- 12.Flicek P, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasmussen AS, Arnason U. Molecular studies suggest that cartilaginous fishes have a terminal position in the piscine tree. Proc. Natl Acad. Sci. USA. 1999;96:2177–2182. doi: 10.1073/pnas.96.5.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol. Biol. Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- 15.Hallstrom BM, Janke A. Gnathostome phylogenomics utilizing lungfish EST sequences. Mol. Biol. Evol. 2009;26:463–471. doi: 10.1093/molbev/msn271. [DOI] [PubMed] [Google Scholar]
- 16.Martin AP, Naylor GJ, Palumbi SR. Rates of mitochondrial DNA evolution in sharks are slow compared with mammals. Nature. 1992;357:153–155. doi: 10.1038/357153a0. [DOI] [PubMed] [Google Scholar]
- 17.Martin AP. Substitution rates of organelle and nuclear genes in sharks: implicating metabolic rate (again) Mol. Biol. Evol. 1999;16:996–1002. doi: 10.1093/oxfordjournals.molbev.a026189. [DOI] [PubMed] [Google Scholar]
- 18.Amemiya CT, et al. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. doi: 10.1038/nature12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burt DW. Origin and evolution of avian microchromosomes. Cytogenet. Genome Res. 2002;96:97–112. doi: 10.1159/000063018. [DOI] [PubMed] [Google Scholar]
- 20.Ellegren H. Evolutionary stasis: the stable chromosomes of birds. Trends Ecol. Evol. 2010;25:283–291. doi: 10.1016/j.tree.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Donoghue PC, Sansom IJ, Downs JP. Early evolution of vertebrate skeletal tissues and cellular interactions, and the canalization of skeletal development. J. Exp. Zoolog. B. 2006;306B:278–294. doi: 10.1002/jez.b.21090. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki K, Buchanan AV, Weiss KM. Gene duplication and the evolution of vertebrate skeletal mineralization. Cells Tissues Organs. 2007;186:7–24. doi: 10.1159/000102678. [DOI] [PubMed] [Google Scholar]
- 23.Kawasaki K. The SCPP gene repertoire in bony vertebrates and graded differences in mineralized tissues. Dev. Genes Evol. 2009;219:147–157. doi: 10.1007/s00427-009-0276-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laue K, Jänicke M, Plaster N, Sonntag C, Hammerschmidt M. Restriction of retinoic acid activity by Cyp26b1 is required for proper timing and patterning of osteogenesis during zebrafish development. Development. 2008;135:3775–3787. doi: 10.1242/dev.021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- 26.Dooley H, Flajnik MF. Antibody repertoire development in cartilaginous fish. Dev. Comp. Immunol. 2006;30:43–56. doi: 10.1016/j.dci.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 27.Glusman G, et al. Comparative genomics of the human and mouse T cell receptor loci. Immunity. 2001;15:337–349. doi: 10.1016/S1074-7613(01)00200-X. [DOI] [PubMed] [Google Scholar]
- 28.Du Pasquier L, Zucchetti I, De Santis R. Immunoglobulin superfamily receptors in protochordates: before RAG time. Immunol. Rev. 2004;198:233–248. doi: 10.1111/j.0105-2896.2004.00122.x. [DOI] [PubMed] [Google Scholar]
- 29.Flajnik MF, Tlapakova T, Criscitiello MF, Krylov V, Ohta Y. Evolution of the B7 family: co-evolution of B7H6 and NKp30, identification of a new B7 family member, B7H7, and of B7’s historical relationship with the MHC. Immunogenetics. 2012;64:571–590. doi: 10.1007/s00251-012-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dooley H, Flajnik MF. Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur. J. Immunol. 2005;35:936–945. doi: 10.1002/eji.200425760. [DOI] [PubMed] [Google Scholar]
- 31.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 32.Tikhonova AN, et al. αβ T cell receptors that do not undergo major histocompatibility complex-specific thymic selection possess antibody-like recognition specificities. Immunity. 2012;36:79–91. doi: 10.1016/j.immuni.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Star B, et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011;477:207–210. doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin AP, Palumbi SR. Body size, metabolic rate, generation time, and the molecular clock. Proc. Natl Acad. Sci. USA. 1993;90:4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlötterer C, Amos B, Tautz D. Conservation of polymorphic simple sequence loci in cetacean species. Nature. 1991;354:63–65. doi: 10.1038/354063a0. [DOI] [PubMed] [Google Scholar]
- 36.Janvier P. Palaeontology: modern look for ancient lamprey. Nature. 2006;443:921–924. doi: 10.1038/443921a. [DOI] [PubMed] [Google Scholar]
- 37.Zhu M, et al. A Silurian placoderm with osteichthyan-like marginal jaw bones. Nature. 2013;502:188–193. doi: 10.1038/nature12617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Notes 1-11, Supplementary Figures 1-11 and Supplementary Table 1-11 (please note that Supplementary Tables 3, 4, 8 and 9 are in a separate excel file) - see pages 1-7 for more details of the Content. (PDF 18504 kb)
This zipped file contains Supplementary Tables 3, 4, 8 and 9 (see the Supplementary Information file for more details. (ZIP 3143 kb)
Data Availability Statement
Accessions
GenBank/EMBL/DDBJ
Sequence Read Archive
Data deposits
The C. milii genome assembly has been deposited at DDBJ/EMBL/GenBank under the accession number AAVX00000000. The version described in this paper is the second version, AAVX02000000. The C. milii and G. cirratum RNA-seq data have been deposited at SRA under accession numbers SRA054255 and SRA062964, respectively. The transcripts have been deposited into GenBank under accession numbers JW861113–JW881738, KA353634–KA353668; and the miRNA sequences under accession numbers JX994303–JX994995.