Summary
The rapid turnover of the mammalian intestinal epithelium is supported by stem cells located around the base of the crypt1. Alongside Lgr5, intestinal stem cells have been associated with various markers, which are expressed heterogeneously within the crypt base region1-6. Previous quantitative clonal fate analyses have proposed that homeostasis occurs as the consequence of neutral competition between dividing stem cells7-9. However, the short-term behaviour of individual Lgr5+ cells positioned at different locations within the crypt base compartment has not been resolved. Here, we established the short-term dynamics of intestinal stem cells using a novel approach of continuous intravital imaging of Lgr5-Confetti mice. We find that Lgr5+ cells in the upper part of the niche (termed ‘border cells’) can be passively displaced into the transit-amplifying (TA) domain, following division of proximate cells, implying that determination of stem cell fate can be uncoupled from division. Through the quantitative analysis of individual clonal lineages, we show that stem cells at the crypt base, termed ‘central cells’, experience a survival advantage over border stem cells. However, through the transfer of stem cells between the border and central regions, all Lgr5+ cells are endowed with long-term self-renewal potential. These findings establish a novel paradigm for stem cell maintenance in which a dynamically heterogeneous cell population is able to function long-term as a single stem cell pool.
In the small intestine, stem cells are associated with Lgr5 expression, which marks around 14-16 proliferative ‘Crypt Base Columnar (CBC)’ cells distributed throughout the crypt base. The stem cell niche is constituted by Paneth cells10,11 and surrounding mesenchyme12. Cells that become displaced from this region enter the TA compartment and lose stemness13. Quiescent or slow-cycling cells, positioned at or near the ‘+4 position’ may constitute a second stem cell type3,5,6,14, although a recent study indicated that some, if not all, of these cells represent secretory precursors that, in common with Dll1+ cells higher in the crypt15, can be recruited back into the stem cell compartment upon damage16. Hierarchy, heterogeneity, and spatial organization of intestinal stem cells remain a subject of debate17-21. Are stem and progenitors organized in an engrained proliferative hierarchy, defined by a signature of molecular markers, or do stem cells transit reversibly between states of variable competence in which they become biased towards renewal or differentiation? If the latter is true, is bias controlled by intrinsic heterogeneity in the expression of fate determinants, or the consequence of spatio-temporal cues associated with niche-derived signals? Although inducible genetic lineage tracing allows to dissect short-term heterogeneity in self-renewal potential, its reliability may be undermined by transient effects due to drug-inducing agents, Cre activity, or non-representativeness of labelling22. Therefore we applied an in vivo live-imaging strategy, allowing measurements to begin several days after drug administration. In common with previous live-imaging approaches used to study stem cells in hair follicle and testis 23,24,25, our approach enables tracing of the fate of individual marked stem cells and their progeny over time in vivo.
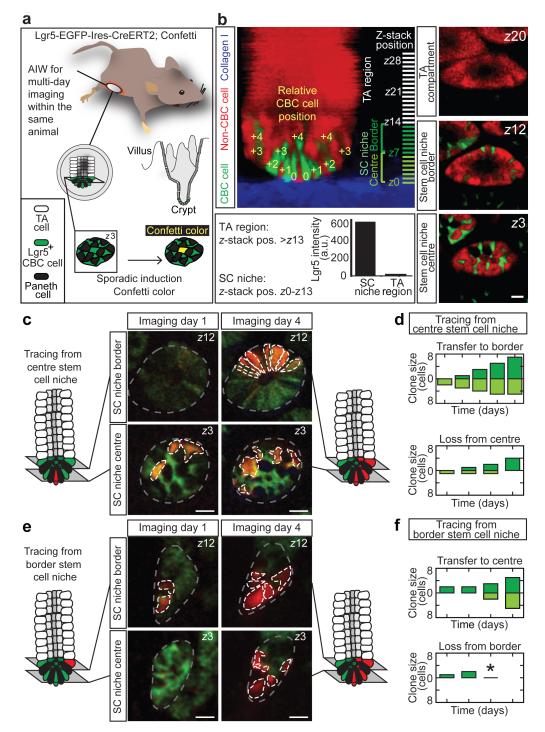
Multiphoton intravital microscopy and surgical implantation of an abdominal imaging window (AIW)26,27 into living Lgr5EGFP-Ires-CreERT2/R26R-Confetti mice were used to obtain visual access to the intestinal stem cell niche (Fig. 1a). Lgr5+ CBC cells and their progeny were lineage traced over time (Extended Data Fig. 1) by activating the expression of one of the Confetti colours (membranous CFP, cytoplasmic YFP and RFP) in individual Lgr5+ cells using Tamoxifen-mediated recombination of the Confetti-construct (Fig. 1a). To characterize fate behaviour of CBC cells we followed lineages of 80 marked cells (n = 4 mice) up to 5 days from the start of time-lapse imaging (Extended Data Fig. 2, for controls see 27 and Extended Data Fig. 3).
Figure 1. Intravital lineage tracing of Lgr5+ cells.
a, Cartoon showing a mouse carrying an abdominal imaging window (AIW) to visualize intestinal Lgr5+ CBC cells and their Confetti progeny over multiple imaging sessions. b, Lateral projection of a Z-stack and representative XY-images of a crypt at indicated Z-stack positions. The stem cell niche (Z0-13) is defined by Lgr5-GFP fluorescence. The relative position of CBC cells to the most basal cell (row 0) determines location in the central (row 0 to +2, which translates to Z0-6) or border region (row +3 to +4, which translates to Z7-13) of the stem cell niche. Scale bar, 20 μm. c-f, Intravital lineage tracing of RFP-expressing Lgr5+ CBC cells located at the centre (c,d) and border (e,f) region. Grey lines indicate crypts, white lines indicate Confetti clones. (d,f) Graphs show time evolution of spatial organization of Confetti clones starting 3 days post-induction. Clone size is divided in central (light green) and border (dark green) CBC cells. Asterisk indicates clones in which all progeny were lost. Scale bar, 20 μm.
Following induction, clonal progeny were observed throughout the stem cell niche. To quantify fate behaviour of Lgr5+ CBC cells, we acquired Z-stacks (Fig. 1b; see Video 1 for the 3D reconstruction) and classified cells based upon their relative position, using the most basal cells (termed ‘row 0’) as a reference (Fig. 1b). Confetti-labelled clones were scored according to cell number, disaggregated by position (Extended Data Fig. 4). In line with predictions of neutral competition7, numbers of marked cells in the stem cell niche varied widely between clones (some expanded in size, others lost attachment to this compartment altogether; Extended Data Figs. 2 and 4). As just 1 of the 28 clones containing a single marked CBC cell at the start of filming remained single after two days of tracing, we chose to neglect the potential impact of lineage committed quiescent Lgr5+ cells, identified previously16.
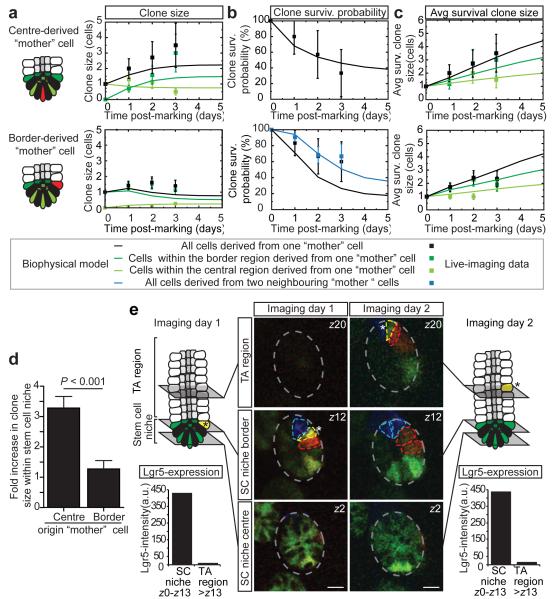
To investigate spatial heterogeneity in self-renewal potential of CBC cells, we defined two regions within the Lgr5+ stem cell niche: a central (rows 0 to +2) and border (+3 and +4) region (Fig. 1b). A ‘mother’ cell in either central or border region could expand and give rise to progeny that extended into both regions (Fig. 1c-f and Extended Data Fig. 5). Further quantitative analysis was necessary to address the potency of CBC cells in these two domains. While the average number of central cells per clone derived from a single central ‘mother’ cell remained approximately constant, consistent with their maintenance over time, the average number of border cells derived from these cells increased to approximately 2 by day 3 (Fig. 2a). Furthermore, maintenance of the average central cell number was achieved through the steady decline in the number of clones retaining at least one central cell (Fig. 2b), compensated by a steady increase in size of those that remain (Fig. 2c). Although clones derived from single border ‘mother’ cells also appeared to approximately maintain their number, they gave rise to a comparatively smaller number of central cells (Fig. 2a). The sustained increase in the number of border cells from a central ‘mother’ cell (Fig. 2a) indicates that these cells typically outcompete cells at the niche border (Fig. 2d).
Figure 2. Central CBC cells experience a short-term positional advantage in self-renewal potential.
a-c, Clonal evolution of a Confetti cell located at the central or border region starting 3 days post-induction. Graphs show: a, average clone size; b, fraction “surviving” clones that contain at least one marked central (top) or border (lower) cell; and c, average size of surviving clones (clones with at least one marked cell). Different colours indicate different regions in the niche. Points show data and lines show fit to the biophysical model (see Fig 3). Error bars represent s.d. d, Fold increase in clone size over three days from a border or central Confetti+ CBC cell. Note that central stem cells have a positional advantage over border stem cells. Error bars represent s.e.m., P <0.001 obtained using a Mann Whitney U test. e, Intravital images of the same crypt at indicated times. Note that the yellow cell is truly expelled from the stem cell niche, since GFP-expression was absent in the TA cell region (see charts at indicated time points). Scale bars, 20 μm.
To investigate the potential basis of this positional advantage, we studied the development of clones with finer time resolution. Every two hours, we acquired multiphoton images of crypts, followed the location of all GFP-labelled cells over time (Extended Data Fig. 6; Video 2-4) and found that division of single Lgr5+ cells coincides with displacement of proximate CBC cells. This suggests that cell proliferation creates competition for space leading to an adjustment of cell positions. Through this rearrangement, and independent of their division history, CBC cells located at the border can become passively displaced from the niche following division of a neighbour (Fig. 2e and Extended Data Fig. 4).
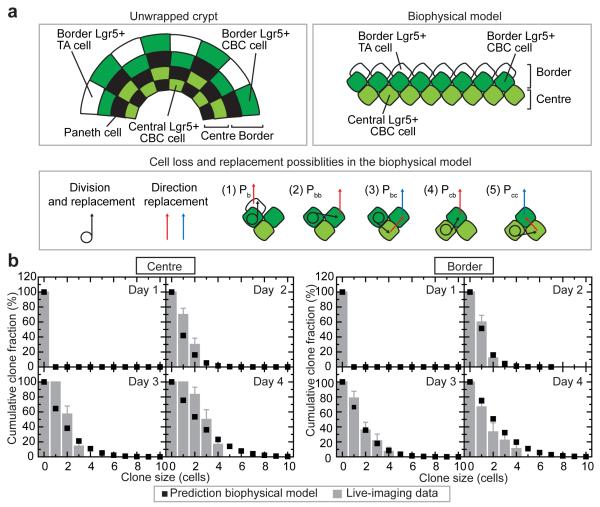
To challenge this conclusion and address the potency of the Lgr5+ CBC stem cell population, we aimed to quantitatively capture the variability seen in the lineage potential of individual cells by a biophysical model, involving a revision of the neutral drift dynamics model introduced in [7,8] in which all stem cells were considered functionally equivalent. In this new model, a periodic quasi-one-dimensional arrangement of stem cells mimicked the ‘collar-like’ geometry of the central and border niche regions of the crypt (Fig. 3a). To account for the mixed GFP expression profile seen at rows +3 and +4 (Fig. 1b), the border region was further subdivided into Lgr5+ CBC cells and Lgr5− TA cells. To accommodate the range of observed dynamical behaviours in the stem cell niche, we allowed for five possible ‘channels’ of stem cell loss and replacement (Fig. 3a): Following division of a border stem cell, one daughter cell remains at its position while the other either (1), displaces a border TA cell out of the niche; (2), displaces a border stem cell which in turn displaces a border TA cell out of the niche; or (3), displaces a central stem cell which in turn displaces a border stem cell into the border TA cell domain. Similarly, after division of a central stem cell, one daughter remains at its position while the other either (4), displaces a border stem cell into the border TA cell region; or (5), displaces a central cell which in turn displaces a border stem cell into the TA cell region. If we define as λ the rate of transfer of border TA cells out of the niche, each of these 5 processes occur at rates Pbλ, Pbbλ, Pbcλ, Pcbλ, and Pccλ, respectively, with Pb+Pbb+Pbc+Pcb+Pcc=1 (Supplementary Notes).
Figure 3. Biophysical model of intestinal stem cell dynamics.
a, From the unfolded crypt caricature (left), we synthesize a quasi-one-dimensional biophysical model of the niche region (right) consisting of two domains: border and centre. To conserve cell number, cell rearrangements following stem cell division displace precisely one cell from the border. To capture the range of lineage data, we include 5 channels of stem cell loss/replacement (1-5) defined in the main text. b, Cumulative size distributions of clones derived from a single cell in the centre (left) or border (right). Clone size is defined in both cases by total number of constituent cells in centre and border. Error bars represent s.e.m. Points represent predictions of the model using the same parameters as that inferred from the average dependences (Supplementary Notes).
By fixing the relative rates of stem cell division and displacement by the observed average clone size dependences and independent estimates of the average cell division rate, we found that the biophysical model can accurately predict clone size distribution and spatial dependencies observed in live-imaging (Fig. 2a-c and Fig. 3b, and Extended Data Fig. 7). More significantly, with the same parameters, the model describes quantitatively convergence onto the hallmark scaling behaviour reported using static lineage tracing assays at intermediate times7 (7 and 14 days post-induction), as well as the predicted progression towards crypt monoclonality at long-times8 (Extended Data Fig. 8 and 9; Supplementary Notes).
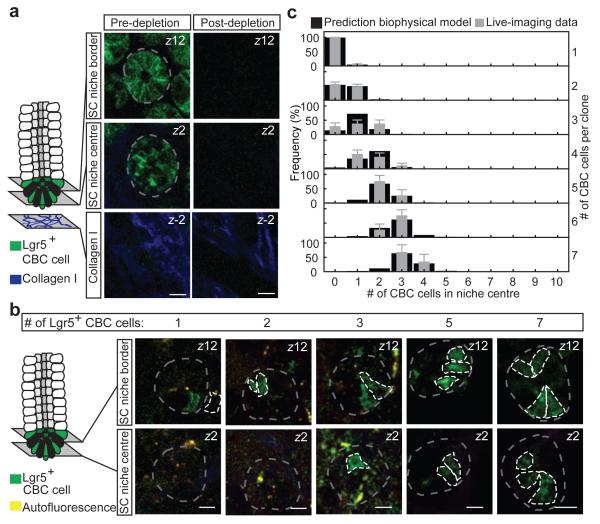
To further challenge the model, we traced the recovery of stem cells following targeted ablation of Lgr5+ cells using diphtheria toxin (DT) injection in mice where the human DT receptor (DTR) fused to EGFP was knocked in the Lgr5 locus (Lgr5DTR:EGFP)28 (Fig. 4a). In these mice, recovered Lgr5+ cells are derived from a TA-lineage28. Following complete depletion (Fig. 4a), we observed a low frequency of initiation and a heterogeneous pattern of recovery (Fig. 4b and Supplementary Video 5), suggesting sporadic transfer of cells from the TA zone into the stem cell niche border. The cohesion of these recovered cell clusters (Supplementary Video 6) suggests clonal expansion of individual TA cells. Intriguingly, by allowing individual border stem cells to recolonize a depleted stem cell niche through cell division uncompensated by loss, our biophysical model provided a quantitative prediction of cluster composition (border versus central) by size, with the same relative rates of stem cell division as those found in steady-state (Fig. 4c and Supplementary Notes).
Figure 4. Recovery of stem cell compartment following ablation of Lgr5+ cells challenges model.
a, Targeted ablation of Lgr5+ cells in Lgr5DTR:EGFP mice was induced by injection of diphtheria toxin. Shown are representative images pre- and post-ablation. Scale bars, 20 μm. b, Recovery of Lgr5+ CBC cells was monitored only in mice where full depletion was confirmed 24 hours after diphtheria toxin injection. Images taken at 72 hours after depletion show representative crypts containing clonal clusters of different sizes (n = 108 crypts in 3 mice). Scale bars, 20 μm c, For all various clone sizes, measured spatial composition (border versus centre) of Lgr5+ CBC cells in clusters (grey) were accurately predicted by the biophysical model (black). Error bars represent s.d.
Our data shows that intestinal stem cell maintenance follows from competition between proximate CBC stem cells for limited niche access and stem cells positioned near the niche boundary experience a bias towards loss and replacement, while stem cells remote from the boundary are biased towards survival. Intriguingly, a similar dependence of self-renewal potential on proximity to the niche border was reported in a recent in vivo live-imaging study of mouse hair follicle29, suggesting that such heterogeneity may be a ubiquitous feature of adult stem cell populations. A recent lineage tracing study based on the continuous and sporadic acquisition of mutations during DNA replication, concluded that only a subfraction of putative intestinal stem cells are ‘functional’30. Our quantitative analysis of live-imaging data shows that central stem cells are about 3 times more likely than border cells to fully colonize a crypt in steady-state, explaining why only a fraction of Lgr5+ cells appears to retain long-term self-renewal potential (Supplementary Notes). Through the transfer of cells between the central and border regions of the niche, the dynamic and heterogeneous population of intestinal stem cells is able to function long-term as a single equipotent pool.
Online-only Methods
Mice
All experiments were carried out in accordance with the guidelines of the Animal Welfare Committee of the Royal Netherlands Academy of Arts and Sciences, the Netherlands. To obtain R26R-Confetti; Lgr5-EGFP-Ires-CreERT2 mice, R26R-Confetti7 mice were crossed with Lgr5-EGFP-Ires-CreERT21. Random double heterozygous male mice between 10-22 weeks old were used for experiments. Three days before imaging, mice were injected with 2.5-5 mg Tamoxifen (single injection; Sigma Aldrich) to induce activation of Cre recombinase to induce expression of one of the Confetti colours (membranous CFP, cytoplasmic YFP and RFP). Nuclear GFP was also activated, but that subset of confetti-labelled cells was not followed. For the targeted ablation studies, 4 male Lgr5DTR:EGFP mice carrying an AIW received 50 μg kg−1 diphtheria toxin (DT) through intraperitoneal injections. Depletion of Lgr5+ cells was confirmed by intravital imaging. Mice in which Lgr5+ cells were not completely depleted after 24 hours received a second DT injection. Mice were housed under standard laboratory conditions and received food and water ad libitum.
AIW surgery
The AIW surgery was performed as described in reference26. In short, all surgical procedures were performed under 2% isoflurane (v/v) inhalation anaesthesia. Before surgery, buprenorphine (3 μg per mouse; Temgesic©, BD pharmaceutical limited) was administered intramuscularly. The left lateral flank of the mice was shaved and the skin was disinfected with 70% (v/v) ethanol. Next, a left lateral flank incision was made through skin and abdominal wall and a purse string suture was placed along the wound edge. A disinfected AIW (> 1 hour in 70% (v/v) ethanol) was placed glass side down next to the mice and the ileum was placed on top. 3M™ Vetbond™ Tissue Adhesive (n-butyl cyanoacrylate; 3M) was used to fix the ileum to the cover glass of the AIW and CyGel (BioStatus Limited) was added to diminish peristaltic movement. After 5 min the AIW was inverted and placed in the mouse, with the skin and abdominal wall placed inside the AIW groove. Then sutures were tightened to stably secure the window into the animal. After surgery the mice were provided food and water ad libitum. Furthermore, mice were closely monitored once a day before imaging for behaviour, reactivity, appearance and defecation.
Equipment and settings
Intravital imaging was performed on an inverted Leica TCS SP5 AOBS two-photon microscope with a chameleon Ti:Sapphire pumped optical parametric oscillator (Coherent Inc.) equipped with a 25x (HCX IRAPO NA0.95 WD 2.5mm) water objective and four non-descanned detectors (NDDs). The NDDs collect the following wavelengths: NDD1 <455 nm, NDD2 455-490 nm, NDD3 500-550 nm, NDD4 560-650 nm. Sequential scanning was performed, exiting the tissue with 860 and 960 nm wavelengths. The Confetti colours were detected as follows: 860 nm: NDD2 (CFP and eGFP), 960 nm: NDD3 (eGFP and YFP), NDD4 (YFP and RFP). Second harmonic generation (SHG) signal is generated by 960 nm excitation at collagen I and detected in NDD2. Scanning was performed in a bidirectional mode at 700 Hz and 12 bit, with a zoom of 1.7, 512×512 pixels. Z-stacks with 2.5 μm z-steps of typically 70-80 images were acquired. Re-identification of the same crypts over multiple days was accomplished by storing the xy coordinates of the imaged regions using the ‘multiple position’ function in the LAS-AF software and using the vasculature and the typical (Confetti) Lgr5+ crypt pattern as visual landmarks.
Multi-day intestinal stem cell imaging
After placing the AIW the mice were kept under anaesthesia and placed face-down in a custom-designed imaging box in which isoflurane (1% (v/v) was administered through a facemask as described before26. For the multi-day imaging sessions (all imaging figures except Extended Data Figure 6), mice were imaged once a day for a maximum of 3 hours during which the climate chamber surrounding the microscope was kept at 32°C. After the imaging session the mice were allowed to wake up to maintain their body temperature. After imaging, acquired z-stacks were corrected for z and xy shifts using a custom-designed Visual Basic software program and further processed and analysed using basic functions in ImageJ software (linear contrasting, blurring, median filtering).
Short-term intestinal stem cell imaging
Mice were anesthetized using isoflurane (2% v/v). The left lateral flank was shaved and the skin was disinfected using 70% (v/v) ethanol. Next, a left lateral flank incision was made through skin and abdominal wall and the ileum was extracorporated using in PBS-drowned cotton swabs. The ileum was placed on a custom-designed inset containing a coverslip fitting the custom-designed imaging box. The ileum was secured to the cover slip using Vetbond and CyGel. The mouse was placed on top of the intestine and in PBS-drowned sterile cotton gauzes were placed next to the animal to prevent dehydration. Parafilm®M (Sigma-Aldrich) was used to cover the mouse and a subcutaneous infusion system was used to provide 100 μl of sterile PBS per hour. The inset was placed within the custom-designed imaging box in which isoflurane (1%) v/v) was administered through a facemask as described above. The temperature of the mouse was monitored during imaging using a rectal probe and was kept between 36 and 37°C by adjusting the temperature of the surrounding climate chamber. Imaging was performed every 2 hours for 14 hours. Z-stacks with a z-step of 2.5 μm of 12 regions with on average 6 crypts were made. Acquired z-stacks were analysed using ImageJ plugins (TurboReg, 3D visualization, 3D viewer).
Intravital imaging of Lgr5+ depleted mice
Mice carrying an AIW received 50 μg kg−1 diphtheria toxin (DT) through intraperitoneal injections. Depletion of Lgr5+ cells was confirmed by intravital imaging. Mice in which Lgr5+ cells were not completely depleted after 24 hours received a second DT injection. Only mice in which full depletion was confirmed by intravital imaging 24 hours after the last DT injection were analyzed. The number of Lgr5+-GFP cells within the stem cell niche border and centre was determined as described in Fig. 1b.
Real-time lineage tracing – clonal competition strength and positional effect on clone size
The data from the lineage tracing was collected at random, and all clones that were imaged with a 3-day interval were included. The strength of a Confetti-labelled Lgr5+ CBC cell to produce offspring was expressed as the fold increase in Confetti-labelled Lgr5+ CBC cell number three days after the first imaging session. A Mann Whitney U test was performed because the data was not normally distributed.
Quantitative data analysis - Multi-day lineage tracing of Lgr5+ CBC cells
Lineage tracing was performed for 80 clones in 80 crypts from 4 mice. No sample size estimate was calculated before the study was executed. Only data from mice from which high enough quality images were acquired were included in the study. The number of Confetti-labelled cells per crypt position (centre (rows 0 to +2), border (rows +3 to +4), TA (rows >4)) was scored. From the 80 lineages, we obtained 33 sublineages originating from the central region (Fig. 4A), and 47 sublineages from the border (Fig 4.B).
Immune cell analysis on intestinal tissue to test potential side-effects of AIW
Six E-Cadherin-CFP/Lgr5EGFP-Ires-CreERT2 mice, 22 weeks of age, were randomly divided into two groups: a control and a window group. AIWs were implanted on top of the small intestines of mice from the window group whereas mice from the control group did not undergo surgery. After 24 hours all mice were sacrificed and the small intestines were harvested. Note that in the window group the part of the small intestine that was located directly behind the window was harvested. The small intestines were fixed for 1 day in fixation mix (1% paraformaldehyde, 0.2% NaIO4, 61mM Na2HPO4, 75mM L-Lysine and 14 mM NaH2PO4 in H2O). After fixation, the tissues were placed for 6 hours into 30% sucrose after which the tissues were snap-frozen using Tissue Freezing Medium (Leice Microsystems Nussloch GmbH). 16 μm sections were cut using a Leica CM3050 cryotome. A standard immunohistochemistry protocol was used to stain the sections with CD45 antibodies (BD Pharmingen™, 553078, Clone 30-F11) and random areas were imaged. For analysis, 10 areas within the imaged regions were selected and analysed in a blinded manner. The number of CD45 positive cells within a region was counted manually and an averaged number for each mouse was calculated. Next, the average of the 3 mice per group was calculated. A Mann-whitney U test was performed because the sample was not distributed normally, and no significant differences were found. The variance between the groups was tested with an F-test, and was not different.
Clone frequency window versus control mice to test potential side-effects of AIW
Eight Lgr5-EGFP-Ires-CreERT2 mice, 22 weeks of age, were divided into two groups: a control and a window group. All mice received 5 mg Tamoxifen by intraperitoneal injection. Three days later, AIWs were implanted on top of the small intestines of mice from the window group whereas mice from the control group did not undergo surgery at this point. Two days after the surgery (five days after Tamoxifen injection) all mice were imaged. In the control group the intestine was exteriorized prior to imaging (as described in short-term intestinal imaging). In the window group the mice were imaged through the AIW. Several random areas were imaged. All recorded clones were used for analysis. For a single clone the number of cells within the stem cell compartment was determined and a frequency distribution was made for the two groups.
Supplementary Material
Extended Data Figure 1 | Retracing of the intravital imaging fields. a The coordinates of the imaging fields within the imaging window (which always has a fixed position within the stage) were stored. By applying these stored coordinates in the subsequent imaging sessions, we recovered the same positions. b The vasculature was used to refine the retracing. c The retraced imaging areas were validated based on the relative position of the colored crypts. Scale bars, 20 μm.
Extended Data Figure 2 | Real-time lineage tracing of individual clones. a, Expression of a confetti colour was induced in Lgr5+ CBC cells, and their progeny was followed over time. Two examples of Confetti-labelled Lgr5+ CBC cells expanding over time to illustrate clonal competition at indicated times. The top images show the continuous expansion of a YFP-expressing Lgr5+ CBC cell (yellow line) in the central region of the crypt. The lower images shows the size of a clone derived from an RFP-expressing Lgr5+ CBC cell in the central region of the crypt, which first increases and then declines (red line). The Confetti-labelled cells are outlined by a white dashed line and the crypts by a grey dashed line. Scale bars, 20 μm. Right, graph shows the increase in the number of YFP− or RFP-expressing Lgr5+ CBC cells in the entire stem cell compartment of the crypts shown in the images on the left (yellow and red line respectively). b, A total of 80 crypts were imaged in 4 mice. The total clone size (border and central) of Confetti+ cells present in the stem cell niche independent of Lgr5-expression (row 0 to +4, which translates to Z0 to Z13 of the Z-stack) was analysed at indicated times (n = 4 mice) for all 80 lineages. The first time point translates to several days post-induction. Every graph represents one crypt.
Extended Data Figure 3 | Clonal dynamics are unaffected by AIW surgery. a Abdominal imaging windows (AIW) were surgically implanted into the abdominal wall of E-Cadherin-CFP/Lgr5EGFP-Ires-CreERT2 mice. To detect CD45+ immune cells, tissue sections of the small intestine of these and control (no AIW) mice were stained with CD45-antibodies. Scale bar, 20 μm. b Quantification of images in a. Regions of interest within the stromal area of the small intestines were measured and the number of CD45+ cells was counted within those regions. The bars show averages and SEM (n = 10 regions per mouse performed in 3 mice per condition). c Five days after tamoxifen injection, the clones of Lgr5EGFP-Ires-CreERT2/R26R-Confetti mice were imaged intravitally. In “control mice”, the intestine was exteriorized prior to imaging and, in “window mice”, a window was placed two days prior to imaging. The frequency of clones with a certain size is plotted in the graph. The lines show the mean and SEM (n = 4 mice per condition).
Extended Data Figure 4 | The spatial distribution of confetti clone expansion within the stem cell niche. a-b, Spatial distribution of confetti clones, subdivided into clones starting in the central (a) or border (b) region (n = 4 mice) Top left panel (a) shows the relative confetti+ cell position within the stem cell niche, where row 0 to +4 translates to Z0 to Z13 of the Z-stack. Every table shows the number of Confetti labelled cells that are present at the different positions in the crypt, independent of the Lgr5-expression. The position within the central and border region of the stem cell niche and TA compartment are colour coded. The hash symbol indicates the presence of Confetti-labelled cells in the TA compartment, and “na” denotes timepoints for which data was not available due crypts that were not retraceable at that specific time point.
Extended Data Figure 5 | Average clone size and survival probability. Cartoon shows the division of the intestinal stem cell niche into a central and border region; The central region contains row 0 to +2, which translates to Z0 to Z6 of the Z-stack and border region contains row +3 to +4, which translates to Z7 to Z13 of the Z-stack (see Fig. 1b). Confetti-expression was induced in Lgr5+ CBC cells. The panels show, over the indicated time, the spatial organization of the 80 lineages of progeny of the confetti-labelled CBC cells (same clones as the Extended Data Figure 2 and 4) (n = 4 mice). For each lineage, we recorded the number of cells per clone in the central and border regions of the stem cell niche at the indicated times. The first time point translates to several days post-induction. The asterisks indicate clones in which all progeny were lost from the niche region.
Extended Data Figure 6 | Lgr5+ CBC cells move and can become expelled from the stem cell niche. Up, maximum projection images (Z2 to Z5) of a time series of a crypt. In the lower cartoons, the Lgr5+ CBC cells are highlighted. The moving cells are indicated with a different colour. The location of the cells at earlier time points are shown by a lighter colour and arrows indicate the direction of movement. Scale bars, 20 μm.
Extended Data Figure 7 | Cumulative clone size distributions derived from 2 border mother cells. Cumulative clone size distributions of clones derived from two neighboring cells in the border region. In each case, the size of the clone is defined by the total number of constituent cells in the central and border regions (rows 0 to +4), independent of GFP expression. Note that the cumulative clone size distribution records the fraction of clones that have a size larger than the given value. The bars represent measurements from individual lineages reconstructed from live-imaging at days 1 (a; n=13, b; n =35, c; n =35), 2 (a; n =10, b; n =25, c; n =28), 3 (a; n =7, b; n =14, c; n =14) and 4 (a; n =6, b; n =8, c; n =6). Error bars denote SEM. The points represent the predictions of the biophysical model using the same parameters as that inferred from the short-term live-imaging assay (for details, see Fig. 3a and the Supplementary Notes).
Extended Data Figure 8 | Longer-term clonal evolution and the approach to scaling behaviour. a-b, Cumulative clone size distribution at 7 days (a) and 14 days (b) post-induction showing the percentage of clones which have a constituent number of Lgr5+ CBC cells larger than the given value. For example, the data point at around (4 cells, 40%) in (a) shows that, at 7 days post-induction, some 40% of clones have a size larger than 4 Lgr5+ cells, etc. The bars reproduce the findings of a fixed clonal assay using the same Lgr5-Confetti mouse construct used in the present study and first presented in Ref. [7]. Error bars denote SEM. Points represent the predictions of the biophysical model using the same parameters as that inferred from the short-term live-imaging assay (see Fig. 3 and Supplementary Notes). To account for the prolonged activity of the Cre recombinase in the static clonal assay, in both cases we have introduced a one-day time delay. At the 14 day time point, the measured clone size distribution and model prediction are beginning to converge onto the universal scaling behaviour characteristic of a strictly one-dimensional neutral drift dynamics (line) (for details, see Supplementary Notes).
Extended Data Figure 9 | Long-term drift towards monoclonality of labeled crypts. Predicted frequency of monoclonal crypts over time expressed as a percentage of surviving clones following pulse-labelling of stem cells at clonal density. The points show the predictions of the biophysical model defined in the main text and Supplementary Notes using the same parameters as that inferred from the short-term live-imaging assay (see Fig. 3 and supplementary notes) following the representative marking of stem cells at the crypt base, and the line shows the predictions of the strictly one-dimensional neutral drift dynamics model introduced in Ref. [8] with a stem cell loss replacement rate of 0.24 per day and a total of 8 stem cells. The convergence of these two model predictions at longer times shows that, first, the behaviour of the quasi one-dimensional model approaches that of the strictly one-dimensional model at longer times and, second, that the effective loss/replacement rate and stem cell number in the new model is essentially fixed by the rate λpcc=0.24 per day and the 8 stem cells that occupy the central region. Significantly, these parameters translate to the ratio λpcc/Nstem2 =0.026 per week, very close to the figure of 0.025 per week obtained from a fit of the measured monoclonal crypt fraction to the one-dimensional neutral drift dynamics model in Ref. [8]. (For the labeling protocol and the experimental data points, we refer to the original reference).
Acknowledgements
The authors would like to thank Anko de Graaff from the Hubrecht Imaging Center for imaging support, all members of the van Rheenen group for useful discussions and the Hubrecht Institute animal caretakers for animal support. This work was supported by a Vidi fellowship (91710330; J.v.R.) and equipment grants (175.010.2007.00 and 834.11.002; J.v.R.) from the Dutch organization of scientific research (NWO), a grant from the Dutch cancer society (KWF; HUBR 2009-4621; J.v.R.), a grant from the Association for International Cancer Research (AICR; 13-0297; J.v.R.), and the Wellcome Trust (grant number 098357/Z/12/Z; B.D.S.).
Footnotes
Full Methods and any associated references are available in the online version of the paper.
Reprints and permissions information is available at ww.nature.com/reprints.
The authors declare no competing financial interests Readers are welcome to comment on the online version of the paper.
References
- 1.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 2.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeda N, et al. Interconversion Between Intestinal Stem Cell Populations in Distinct Niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montgomery RK, et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proceedings of the National Academy of Sciences. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powell Anne E., et al. The Pan-ErbB Negative Regulator Lrig1 Is an Intestinal Stem Cell Marker that Functions as a Tumor Suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong VWY, et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol. 2012;14:401–408. doi: 10.1038/ncb2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snippert HJ, et al. Intestinal Crypt Homeostasis Results from Neutral Competition between Symmetrically Dividing Lgr5 Stem Cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. Intestinal Stem Cell Replacement Follows a Pattern of Neutral Drift. Science. 2010;330:822–825. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- 9.Snippert HJ, Clevers H. Tracking adult stem cells. EMBO Rep. 2011;12:113–122. doi: 10.1038/embor.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.VanDussen KL, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farin HF, Van Es JH, Clevers H. Redundant Sources of Wnt Regulate Intestinal Stem Cells and Promote Formation of Paneth Cells. Gastroenterology. 2012;143:1518–1529.e1517. doi: 10.1053/j.gastro.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 13.van der Flier LG, Clevers H. Stem Cells, Self-Renewal, and Differentiation in the Intestinal Epithelium. Annual Review of Physiology. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 14.Sangiorgi E, Capecchi MR. Bmi1 lineage tracing identifies a self-renewing pancreatic acinar cell subpopulation capable of maintaining pancreatic organ homeostasis. Proceedings of the National Academy of Sciences. 2009;106:7101–7106. doi: 10.1073/pnas.0902508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Es JH, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buczacki SJA, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 17.Stine RR, Matunis EL. Stem cell competition: finding balance in the niche. Trends in Cell Biology. doi: 10.1016/j.tcb.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simons Benjamin D., Clevers H. Strategies for Homeostatic Stem Cell Self-Renewal in Adult Tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 19.Morrison SJ, Spradling AC. Stem Cells and Niches: Mechanisms That Promote Stem Cell Maintenance throughout Life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goulas S, Conder R, Knoblich Juergen A. The Par Complex and Integrins Direct Asymmetric Cell Division in Adult Intestinal Stem Cells. Cell stem cell. 2012;11:529–540. doi: 10.1016/j.stem.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheng XR, Matunis E. Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development. 2011;138:3367–3376. doi: 10.1242/dev.065797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Y, Huang Y-F, Kek C, Bulavin Dmitry V. Apoptosis Differently Affects Lineage Tracing of Lgr5 and Bmi1 Intestinal Stem Cell Populations. Cell stem cell. 2013;12:298–303. doi: 10.1016/j.stem.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa T, Sharma M, Nabeshima Y.-i., Braun RE, Yoshida S. Functional Hierarchy and Reversibility Within the Murine Spermatogenic Stem Cell Compartment. Science. 2010;328:62–67. doi: 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein AM, Nakagawa T, Ichikawa R, Yoshida S, Simons BD. Mouse Germ Line Stem Cells Undergo Rapid and Stochastic Turnover. Cell stem cell. 2010;7:214–224. doi: 10.1016/j.stem.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 25.Rompolas P, et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature. 2012;487:496–499. doi: 10.1038/nature11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritsma L, et al. Surgical implantation of an abdominal imaging window for intravital microscopy. Nat. Protocols. 2013;8:583–594. doi: 10.1038/nprot.2013.026. [DOI] [PubMed] [Google Scholar]
- 27.Ritsma L, et al. Intravital Microscopy Through an Abdominal Imaging Window Reveals a Pre-Micrometastasis Stage During Liver Metastasis. Science Translational Medicine. 2012;4:158ra145. doi: 10.1126/scitranslmed.3004394. [DOI] [PubMed] [Google Scholar]
- 28.Tian H, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rompolas P, Mesa KR, Greco V. Spatial organization within a niche as a determinant of stem-cell fate. Nature. 2013;502:513–518. doi: 10.1038/nature12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozar S, et al. Continuous Clonal Labeling Reveals Small Numbers of Functional Stem Cells in Intestinal Crypts and Adenomas. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Figure 1 | Retracing of the intravital imaging fields. a The coordinates of the imaging fields within the imaging window (which always has a fixed position within the stage) were stored. By applying these stored coordinates in the subsequent imaging sessions, we recovered the same positions. b The vasculature was used to refine the retracing. c The retraced imaging areas were validated based on the relative position of the colored crypts. Scale bars, 20 μm.
Extended Data Figure 2 | Real-time lineage tracing of individual clones. a, Expression of a confetti colour was induced in Lgr5+ CBC cells, and their progeny was followed over time. Two examples of Confetti-labelled Lgr5+ CBC cells expanding over time to illustrate clonal competition at indicated times. The top images show the continuous expansion of a YFP-expressing Lgr5+ CBC cell (yellow line) in the central region of the crypt. The lower images shows the size of a clone derived from an RFP-expressing Lgr5+ CBC cell in the central region of the crypt, which first increases and then declines (red line). The Confetti-labelled cells are outlined by a white dashed line and the crypts by a grey dashed line. Scale bars, 20 μm. Right, graph shows the increase in the number of YFP− or RFP-expressing Lgr5+ CBC cells in the entire stem cell compartment of the crypts shown in the images on the left (yellow and red line respectively). b, A total of 80 crypts were imaged in 4 mice. The total clone size (border and central) of Confetti+ cells present in the stem cell niche independent of Lgr5-expression (row 0 to +4, which translates to Z0 to Z13 of the Z-stack) was analysed at indicated times (n = 4 mice) for all 80 lineages. The first time point translates to several days post-induction. Every graph represents one crypt.
Extended Data Figure 3 | Clonal dynamics are unaffected by AIW surgery. a Abdominal imaging windows (AIW) were surgically implanted into the abdominal wall of E-Cadherin-CFP/Lgr5EGFP-Ires-CreERT2 mice. To detect CD45+ immune cells, tissue sections of the small intestine of these and control (no AIW) mice were stained with CD45-antibodies. Scale bar, 20 μm. b Quantification of images in a. Regions of interest within the stromal area of the small intestines were measured and the number of CD45+ cells was counted within those regions. The bars show averages and SEM (n = 10 regions per mouse performed in 3 mice per condition). c Five days after tamoxifen injection, the clones of Lgr5EGFP-Ires-CreERT2/R26R-Confetti mice were imaged intravitally. In “control mice”, the intestine was exteriorized prior to imaging and, in “window mice”, a window was placed two days prior to imaging. The frequency of clones with a certain size is plotted in the graph. The lines show the mean and SEM (n = 4 mice per condition).
Extended Data Figure 4 | The spatial distribution of confetti clone expansion within the stem cell niche. a-b, Spatial distribution of confetti clones, subdivided into clones starting in the central (a) or border (b) region (n = 4 mice) Top left panel (a) shows the relative confetti+ cell position within the stem cell niche, where row 0 to +4 translates to Z0 to Z13 of the Z-stack. Every table shows the number of Confetti labelled cells that are present at the different positions in the crypt, independent of the Lgr5-expression. The position within the central and border region of the stem cell niche and TA compartment are colour coded. The hash symbol indicates the presence of Confetti-labelled cells in the TA compartment, and “na” denotes timepoints for which data was not available due crypts that were not retraceable at that specific time point.
Extended Data Figure 5 | Average clone size and survival probability. Cartoon shows the division of the intestinal stem cell niche into a central and border region; The central region contains row 0 to +2, which translates to Z0 to Z6 of the Z-stack and border region contains row +3 to +4, which translates to Z7 to Z13 of the Z-stack (see Fig. 1b). Confetti-expression was induced in Lgr5+ CBC cells. The panels show, over the indicated time, the spatial organization of the 80 lineages of progeny of the confetti-labelled CBC cells (same clones as the Extended Data Figure 2 and 4) (n = 4 mice). For each lineage, we recorded the number of cells per clone in the central and border regions of the stem cell niche at the indicated times. The first time point translates to several days post-induction. The asterisks indicate clones in which all progeny were lost from the niche region.
Extended Data Figure 6 | Lgr5+ CBC cells move and can become expelled from the stem cell niche. Up, maximum projection images (Z2 to Z5) of a time series of a crypt. In the lower cartoons, the Lgr5+ CBC cells are highlighted. The moving cells are indicated with a different colour. The location of the cells at earlier time points are shown by a lighter colour and arrows indicate the direction of movement. Scale bars, 20 μm.
Extended Data Figure 7 | Cumulative clone size distributions derived from 2 border mother cells. Cumulative clone size distributions of clones derived from two neighboring cells in the border region. In each case, the size of the clone is defined by the total number of constituent cells in the central and border regions (rows 0 to +4), independent of GFP expression. Note that the cumulative clone size distribution records the fraction of clones that have a size larger than the given value. The bars represent measurements from individual lineages reconstructed from live-imaging at days 1 (a; n=13, b; n =35, c; n =35), 2 (a; n =10, b; n =25, c; n =28), 3 (a; n =7, b; n =14, c; n =14) and 4 (a; n =6, b; n =8, c; n =6). Error bars denote SEM. The points represent the predictions of the biophysical model using the same parameters as that inferred from the short-term live-imaging assay (for details, see Fig. 3a and the Supplementary Notes).
Extended Data Figure 8 | Longer-term clonal evolution and the approach to scaling behaviour. a-b, Cumulative clone size distribution at 7 days (a) and 14 days (b) post-induction showing the percentage of clones which have a constituent number of Lgr5+ CBC cells larger than the given value. For example, the data point at around (4 cells, 40%) in (a) shows that, at 7 days post-induction, some 40% of clones have a size larger than 4 Lgr5+ cells, etc. The bars reproduce the findings of a fixed clonal assay using the same Lgr5-Confetti mouse construct used in the present study and first presented in Ref. [7]. Error bars denote SEM. Points represent the predictions of the biophysical model using the same parameters as that inferred from the short-term live-imaging assay (see Fig. 3 and Supplementary Notes). To account for the prolonged activity of the Cre recombinase in the static clonal assay, in both cases we have introduced a one-day time delay. At the 14 day time point, the measured clone size distribution and model prediction are beginning to converge onto the universal scaling behaviour characteristic of a strictly one-dimensional neutral drift dynamics (line) (for details, see Supplementary Notes).
Extended Data Figure 9 | Long-term drift towards monoclonality of labeled crypts. Predicted frequency of monoclonal crypts over time expressed as a percentage of surviving clones following pulse-labelling of stem cells at clonal density. The points show the predictions of the biophysical model defined in the main text and Supplementary Notes using the same parameters as that inferred from the short-term live-imaging assay (see Fig. 3 and supplementary notes) following the representative marking of stem cells at the crypt base, and the line shows the predictions of the strictly one-dimensional neutral drift dynamics model introduced in Ref. [8] with a stem cell loss replacement rate of 0.24 per day and a total of 8 stem cells. The convergence of these two model predictions at longer times shows that, first, the behaviour of the quasi one-dimensional model approaches that of the strictly one-dimensional model at longer times and, second, that the effective loss/replacement rate and stem cell number in the new model is essentially fixed by the rate λpcc=0.24 per day and the 8 stem cells that occupy the central region. Significantly, these parameters translate to the ratio λpcc/Nstem2 =0.026 per week, very close to the figure of 0.025 per week obtained from a fit of the measured monoclonal crypt fraction to the one-dimensional neutral drift dynamics model in Ref. [8]. (For the labeling protocol and the experimental data points, we refer to the original reference).