Abstract
Structure–activity profiles for the phytohormone auxin have been collected for over 70 years, and a number of synthetic auxins are used in agriculture. Auxin classification schemes and binding models followed from understanding auxin structures. However, all of the data came from whole plant bioassays, meaning the output was the integral of many different processes. The discovery of Transport Inhibitor-Response 1 (TIR1) and the Auxin F-Box (AFB) proteins as sites of auxin perception and the role of auxin as molecular glue in the assembly of co-receptor complexes has allowed the development of a definitive quantitative structure–activity relationship for TIR1 and AFB5. Factorial analysis of binding activities offered two uncorrelated factors associated with binding efficiency and binding selectivity. The six maximum-likelihood estimators of Efficiency are changes in the overlap matrixes, inferring that Efficiency is related to the volume of the electronic system. Using the subset of compounds that bound strongly, chemometric analyses based on quantum chemical calculations and similarity and self-similarity indices yielded three classes of Specificity that relate to differential binding. Specificity may not be defined by any one specific atom or position and is influenced by coulomb matrixes, suggesting that it is driven by electrostatic forces. These analyses give the first receptor-specific classification of auxins and indicate that AFB5 is the preferred site for a number of auxinic herbicides by allowing interactions with analogues having van der Waals surfaces larger than that of indole-3-acetic acid. The quality factors are also examined in terms of long-standing models for the mechanism of auxin binding.
The identification of Transport Inhibitor Response 1 (TIR1) as a receptor for the small hormonal ligands in the auxin family1,2 was a landmark advance for both ubiquitin biochemistry and auxin physiology. TIR1 is an F-box protein and forms the substrate binding platform of an ubiquitin E3 ligase complex of the Skp1-Cullin-F-box protein class, hence SCFTIR1. Previous genetic and pull-down experiments had suggested that the endogenous auxin indole-3-acetic acid (IAA) activated either TIR1 or its substrates, the Aux/IAA proteins.3 This activation induced ubiquitination of the Aux/IAA proteins, which were known to be transcriptional regulators.4 Dharmasiri et al.1 and Kepinski and Leyser2 showed that the F-box protein itself was necessary for ligand binding. Shortly afterward the crystal structure of the receptor–ligand complex was published,5 giving a detailed crystal structure of the ligand-binding pocket and the three-component complex that constitutes the activated receptor. The crystallography data also showed that the activated TIR1 complex was a new paradigm for receptor binding because the ligand was shown to be acting as “molecular glue”, participating in substrate binding by completing the nascent recognition pocket. More recently TIR1 and substrate Aux/IAA proteins have been described as co-receptors because both appear to be necessary for ligand binding,6 although the crystallography implies that the leading interaction is the binding of auxin to TIR1.
Auxins have been studied for many decades, and long before receptor candidates were identified, bioassays were in use to generate structure–activity relationships (SARs).7,8 From the early bioassay data sets, a string of chemical hypotheses9,10 and virtual models11 of the receptor binding site have been generated. Auxins have been classified according to chemical scaffold (phenoxyacetic acid, picolinate, etc.)7 and molecular interaction fields resembling the virtual model11 and based on quantum chemical similarity measures defining independent biologically active chemical spaces.12 At the same time, academic and commercial groups have continued screening compound libraries for new synthetic auxins driven by the growing importance of such analogues in agriculture, particularly as herbicides, and the growth of chemical genomics.13−15
Compound screens continue to be based on whole plant bioassays, and novel active compounds such as DAS534 continue to be discovered.16 Auxin bioassays are, clearly, fit for the purpose but, given that bioassay output is the sum of compound uptake, transport, and receptor activation, may not give activity profiles that reflect selectivity in receptor binding. The recent description of the TIR1 complex offers, for the first time, the opportunity of a direct survey of co-receptor structural selectivity. In Arabidopsis the TIR1 family also contains orthologues AFB1, AFB2, AFB3, AFB4, and AFB5.17 The subgroup of AFB4 and AFB5 is the most distinct from the prototypical TIR1. AFB5 has been shown to be fully functional as a receptor for auxin and, notably, the site preferred by the herbicidal auxin Picloram.16,6 In this paper TIR1 and its close orthologue AFB5 have been used as templates for a mixed, high-throughput screen for a selection of active auxins and other auxin analogues in order to build accurate, receptor-specific structure–activity profiles for each.
Surface plasmon resonance (SPR) has proved a reliable and very versatile technology for label-free immunological and pharmacological screening.18,19 The technique requires little protein, shows interactions in real time, and has robust evaluation software to allow both detailed kinetic and rapid, high-throughput binding analyses. In most cases the ligand (frequently this is the protein receptor) is immobilized on the chip surface and binding is followed for the analyte (non-protein small molecule) in solution as it is injected over the receptor on the chip. The newest generation of SPR instruments has sensitivity sufficient to record binding of analytes as small as 100 Da, but previous generation instruments are less sensitive and are still widely used. In such cases the assay may sometimes be inverted to immobilize the small analyte and pass the receptor across the chip, recording the binding of the larger partner. However, many small ligands may not be immobilized without losing activity. For example, the biological activity of auxin IAA (Mr = 172) is compromised if it is derivatized. The knowledge of the TIR1-based three-way co-receptor complex offers an additional binding format, allowing auxin and other small molecules to be screened as analytes in solution without compromising their activity.
The utility of structure–activity relationships may be developed in many ways using chemical and physical properties of the ligand, often based on side groups, charge distributions, and conformations. Statistical analysis then allows grouping or classification of ligands based on quantum molecular similarity measures. As shown in this report, direct quantitations of co-receptor assembly coupled with quantum chemical mapping of a ligand library offers new insights into the molecular properties of auxins behind their affinity.
Results and Discussion
Measuring Co-receptor Assembly and Dissociation Using Surface Plasmon Resonance
In order to examine the assembly requirements of the TIR1 receptor complex a binding assay was established using SPR. Peptides representing the degron domain,20 biotinylated at the N-terminus, were used in place of whole Aux/IAA proteins. Aux/IAA7 (IAA7) was the default sequence unless specified otherwise (Table 1). Channel 1 of a streptavidin-coated chip was blocked with biocytin, channels 2 and 4 were loaded with peptide IAA7, and channel 3 was loaded with a mutant version of the same peptide synthesized with four substitutions around the core degron motif (IAAm7). Purified TIR1 was passed across all four channels. In the absence of auxin, little binding was seen (Figure 1a). Mixing the auxin IAA with TIR1 protein before injection induced binding of TIR1 to peptide IAA7. There was little or no binding to the in-chip control, the mutated degron peptide IAAm7, even in the presence of IAA (Figure 1b).
Table 1. Degron Peptide Sequences; Residues varying from IAA7 are Underscored.
| peptide | sequence |
|---|---|
| IAA7 | biot-AKAQVVGWPPVRNYRKN |
| IAAm7 | biot-AKAQVVEWSSGRNYRKN |
| IAA9 | biot-AKAQIVGWPPVRNYRKN |
| IAA28 | biot-EVAPVVGWPPVRSSRRN |
| IAA31 | biot-QREARQDWPPIKSRLRD |
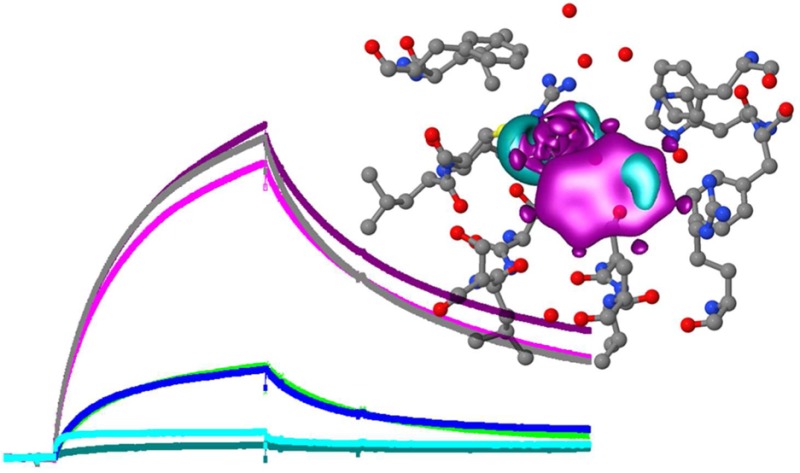
Figure 1.
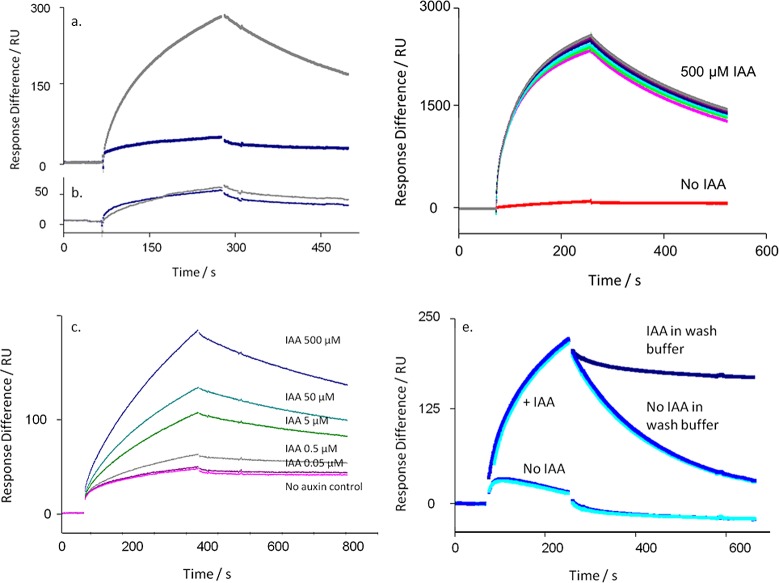
The co-receptor complex is specific for auxin. (a) Sensorgram showing the binding of TIR1 to degron peptide IAA7 on the chip in the absence (blue) and presence of IAA (500 μM; gray). (b) As in panel a, but showing the absence of binding even to a mutagenized IAA7 peptide (IAAm7). (c) The assembly of the TIR1 co-receptor complex is not dependent on exogenous IP6. (d) The response to auxin is dose-dependent. Sensorgrams in panels c and d show ΔRU using channels 4-3 (binding to peptide IAA7 minus peptide IAAm7). (e) The co-receptor complex remains stable in the presence of IAA. The dissociation of the complex is markedly reduced with IAA in the wash buffer (dark blue) compared to dissociation without auxin in the wash buffer (mid and light blue, repeats). Calculated kd with IAA = 1.09 × 10–3 s–1; kd without IAA = 5.58 × 10–3 s–1; X2 = 1.07.
The amplitude of binding was highest at mildly acidic pH (pH 6.8), declining by 15% at pH 7.2 and a further 20% up to pH 8.2 (Supplementary Figure S2). The co-receptor complex assembles in the nucleus,21 and so subsequent experiments were carried out at pH 7.4.22
The crystal structure for TIR1 showed that the active, folded protein held inositol-6-phosphate (IP6) as a cofactor.5 We tested the requirement for IP6 on the binding of TIR1 to immobilized peptide by adding IP6 to [TIR1 + IAA] before injection, using a series of concentrations up to 100 μM (Figure 1c). No significant change in binding or dissociation was observed, suggesting that the expression and purification steps produced IP6-competent TIR1, as had been the case for crystallization work.5 Consequently, IP6 was not routinely added to the reaction mixes. In contrast, binding of TIR1 was strongly dependent on the concentration of IAA (Figure 1d), with half maximal binding data suggesting an affinity of around 5 μM for IAA.
Auxin is unlikely to be removed abruptly in vivo as it is in the SPR experiments. If IAA is retained in the wash buffer after the association phase (but TIR1 is no longer being injected), dissociation of the complex is markedly slowed (Figure 1e). Dissociation off-rate constants assuming first-order 1:1 Langmuir binding are calculated as kd with IAA = 1.09 × 10–3 s–1, and kd without IAA = 5.58 × 10–3 s–1 (X2 = 1.07).
Establishing Preferences for IAA/Aux Co-receptors
There are 29 members of the Aux/IAA family of transcriptional regulators in Arabidopsis.23 Variation within the degron is somewhat lower, but there remains a diversity of degron sequences. A set peptides was selected to represent major degron clades, IAA7 (identical to IAA14), IAA9 (identical to IAA3 and IAA4), IAA28 and IAA31 (Table 1).
There is high selectivity for degron sequence and little qualitative difference in selectivity between TIR1 and AFB5 (Figure 2). Residue substitutions affected the amplitude of binding, although this was position-dependent (Table 1 and Figure 2). The conservative substitution between IAA7 and IAA9 [V/I] three residues upstream from the core degron motif WPPVRN (Table 1) did not affect binding kinetics (Figure 2a and b). Changes both C-terminal and N-terminal to this core (IAA28) reduced binding appreciably, with similar effects in both TIR1 and AFB5. The very different IAA31 showed little auxin-dependent binding.
Figure 2.
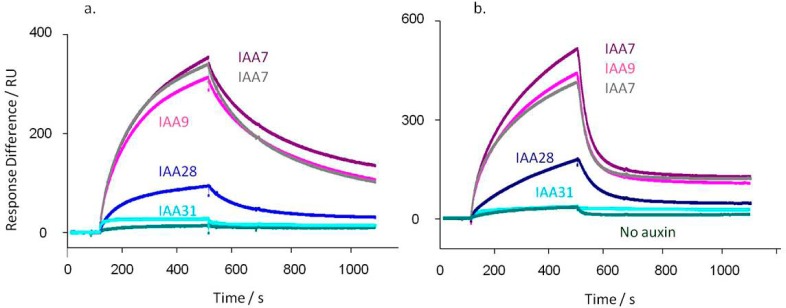
TIR1 (a) and AFB5 (b) co-receptor assembly is dependent on degron sequence. In each case TIR1 or AFB5 protein was mixed prior to injection with IAA at 50 μM, except for the control (no auxin). (a) Calculated kd’s for TIR1: IAA7 kd = 3.3 × 10–3 s–1; IAA7 repeat kd = 4.1 × 10–3 s–1; IAA9 kd = 3.5 × 10–3 s–1; IAA28 kd = 5.8 × 10–3 s–1; IAA31 kd = 3.6 × 10–3 s–1. X2 for the set = 1.24. (b) AFB5 dissociation rates are more rapid than for TIR1: IAA7 kd = 0.019 s–1; IAA7 repeat kd = 0.019 s–1; IAA9 kd = 0.020 s–1; IAA28 kd = 0.011 s–1; IAA31 kd = 0.002 s–1. X2 for the set = 0.70. Assays were set up with channel 1 blocked with biocytin, channel 2 coated with IAA7, and two other peptides on channels 3 and 4. Channel surfaces were saturated with biotinylated peptide in all cases. The IAA7 signal was available to normalize responses between chips, although within batches of protein this was not found necessary. The binding assays were done using a series of auxins. Only data collected at 50 μM IAA are shown, plus one of the series of control injections without auxin.
The kinetic off-rates were calculated in each case. For TIR1 and IAA7, 9, and 31 they all fall within the range of 3.3–4.1 × 10–3 s–1. They are marginally higher for IAA17 and 28 at 5.0 and 5.8 × 10–3 s–1, respectively (Figure 2). With AFB5 the order and relative amplitudes of binding were similar to the pattern for TIR1, but all dissociation rates from AFB5 were much more rapid.6 Rates for IAA7, 9, and 28 were between 1.1 and 1.9 × 10–2 s–1, and no binding was measured for IAA 31.
The co-receptor binding preferences reflect phenotypes shown in mutant plant lines and in the measured half-lives of the mutant Aux/IAA proteins.23−25 The Arabidopsis lines axr2 (IAA7), axr3 (IAA17), shy2 (IAA3, sharing the same degron sequence as IAA9), and iaa28 (IAA28) are all gain-of-function mutations with altered degron sequences. Their phenotypes are all consistent with the consequences of disruption in TIR1 binding, inefficient ubiquitination, a longer half-life, and accumulation of these transcriptional repressors.23 The Aux/IAA family member with most distinct degron motif, IAA31, is long-lived23 and shows very poor binding to TIR1 or AFB5.
Establishing Selectivity for Ligand
A range of synthetic auxins were tested in IAA7 peptide-based assays with TIR1 as described above (Figure 3). In each experiment 500 μM IAA was included to saturate binding (Rmax) as a comparator, and all other auxins and compounds were added at 50 μM. Most compounds known to be active auxins did induce binding, although with somewhat differing association and dissociation characteristics. Allowing for the more accelerated dissociation rates from AFB5 (Figure 3b), most of the compounds tested showed the same pattern of binding and dissociation from AFB5 as for TIR1 (Figure 3a). The herbicide Picloram is a known exception and has a higher affinity for AFB5 than for TIR1.6 Some auxins support assembly of the co-receptor complex but demonstrate far more rapid dissociation kinetics than recorded for IAA. An example is seen by comparing the herbicidal auxins Fluroxypyr and Triclopyr (Figure 3). Both bind as actively as IAA to both TIR1 and AFB5, and Fluroxypyr has a similar off-rate (TIR1: kd,IAA = 1.1 × 10–3 s–1, kd,Flu = 0.93 × 10–3 s–1; AFB5: kd,IAA = 3.0 × 10–2 s–1; kd,Flu =3.3 × 10–2 s–1). However, with both receptors Triclopyr off rates are approximately 3-fold faster (TIR1: kd,Tri = 3.4 × 10–3 s–1; AFB5: kd,Tri = 8.1 × 10–2 s–1).
Figure 3.
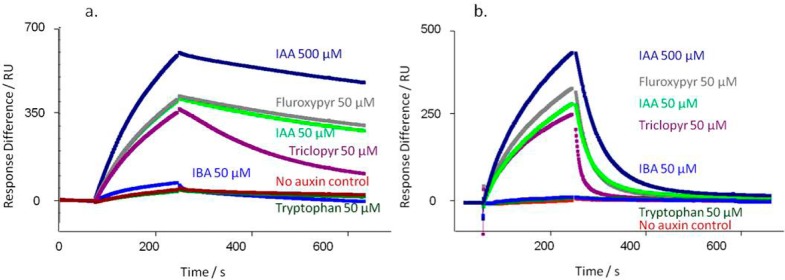
Co-receptor assembly kinetics vary with ligand. A series of commercially relevant synthetic auxins was compared to the IAA response for TIR1 (a) and AFB5 (b). (a) Calculated kd’s for TIR1: IAA500kd = 0.73 × 10–3 s–1; IAA50kd = 1.1 × 10–3 s–1; Fluroxypyr kd = 0.93 × 10–3 s–1; Triclopyr kd = 3.4 × 10–3 s–1; IBA kd = 6.9 × 10–3 s–1. X2 for the set = 0.31. (b) Calculated kd’s for AFB5: IAA500kd = 1.8 × 10–2 s–1; IAA50kd = 3.0 × 10–2 s–1; Fluroxypyr kd = 3.3 × 10–2 s–1; Triclopyr kd = 8.1 × 10–2 s–1; IBA no binding. X2 for the set = 1.81. In all cases a control with no added auxin was included (red).
Domain 1 of Aux/IAA proteins has been shown to contribute toward co-receptor selection.6 Nevertheless, the peptide-based SPR assay appeared to reflect physiological auxin activity well, in addition to providing kinetic details unavailable from radiolabel binding assays6 and pull-down assays.17 It follows that the defined co-receptor assays may be useful for developing a new, more instructive auxin quantitative structure–activity model.
Establishing a Compound Screen
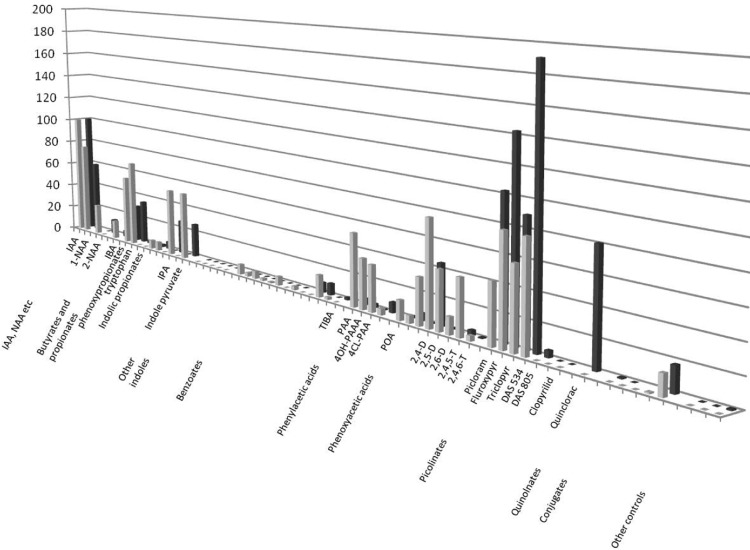
On the basis of results with the training compounds described above, the SPR assay was modified to set up ligand screens for both TIR1 and AFB5. Association times were 180 s, and dissociation times were 300 s. In order to facilitate comparisons between compounds and runs, data have been normalized to the response for 100 μM IAA recorded at the beginning, middle, and end of each data set. All compounds were tested at 100 μM, and report points were introduced19 before injection, 10 s before the end of association, 60 s after dissociation started, and at the end of the dissociation phase. Figure 4 shows the binding data for 58 compounds normalized using the report point at the end of the association phase. The full list of compounds and resonance unit (RU) values are given in Supplementary Table S1.
Figure 4.
Screening analogue libraries for co-receptor ligand specificity. Assembly of the co-receptor varies with ligand and with F-box partner. Each compound was assayed at 100 μM, and the data are presented normalized to 100 μM IAA, which was run at the start and end of each set of experiments. The report point was taken 10 s before the end of the association phase, and the data are for binding to peptide IAA7 with mIAA7 as reference.
Binding Efficiency
All previous classifications of auxins have been based on data collected from growth bioassays.10−12 Early work classified auxins by chemical structure.26,10 Later, molecular interaction energy fields correlated with biological activity were used to group 53 compounds into 4 classes.11 Most recently, a suite of analyses defined 11 quantum chemical classes associated with five levels of biological function using 241 compounds.12 Further calculations of electron density using a molecular harness coupled with similarity indices offered additional theoretical and experimental insights.27 The earlier work assumed activity was associated with activation of a single receptor. The more recent works have accepted that there may be multiple components to molecular efficacy, including uptake, efflux, and catabolism as well as receptor activation. The most recent work also accepted that the system needed to account for more than one family of receptors.12,27
The present structure–activity analysis of auxin-like molecules is based directly on experimental binding data for co-receptor assembly (Figure 4). By projecting quantum chemical similarities onto orthogonal (independent) factors, it was possible to uncover parameters of both binding Efficiency and Specificity (Figure 5) to open new perspectives on the molecular properties of auxins. By measuring TIR1 and AFB5 binding in isolation, it was possible to refine the observation that the coulomb matrix (electrostatic interaction surface) plays the major role in specifying auxins.12 The present analysis suggests that binding Efficiency is associated with the overlap matrixes (volumes of the components in the molecular system), whereas receptor Selectivity includes further variables of the coulomb matrixes (electrostatic surfaces). It infers that the recognition and activation reactions of auxins are driven by different reaction mechanisms.
Figure 5.
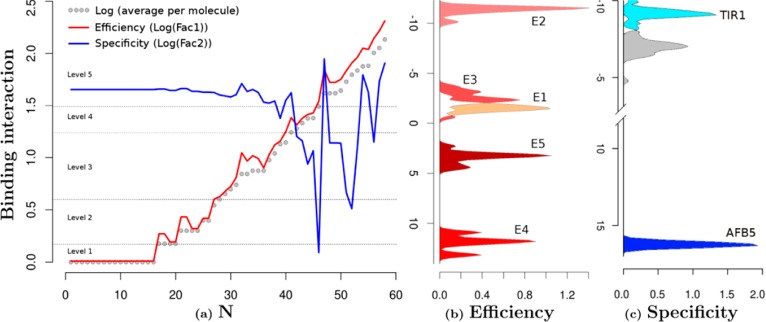
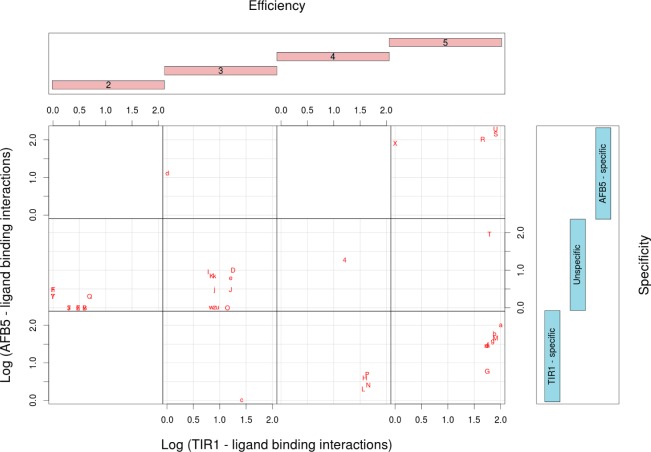
Structure and functional characterization of auxin-like molecules toward their binding with TIR1 and AFB5. (a) Ligand screening data for binding at both receptors (N is the compound reference number in Supplementary Table S2). The black data points are the log values of the mean of the binding per compound for both TIR1 and AFB5 (binding average). The red line represents the first orthogonal factor that is associated with general binding Efficiency of each compound (87.29% of variance). The blue line is the second orthogonal factor, which we associate with binding Specificity (12.71% of variance). Compounds are divided by the analysis into five groups of binding Efficiency along the curve. (b) Quantum chemical classification to predict the membership of the ligands based on their binding Efficiency (E1–E5) and (c) binding specificity, which distinguishes the chemical nature of the binding to TIR1 and AFB5. The statistical membership of both Efficiency and Specificity using quantum chemical variables was predicted with 100% of efficiency. The density of molecules predicted per group is projected on the first lineal discriminant equation for both cases.
Initially molecules were organized in order of increasing binding (binding average in Figure 5). The first result is offered by a factorial analysis of the binding activities with each compound for both TIR1 and AFB5. The analysis offered two uncorrelated factors (Figure 5): the first factor explains 87.29% of the variance and correlated with the average of the binding activity of both TIR1 and AFB5 with r = −1.0 (Factor 1 = Efficiency). The analysis inferred that binding is dominated by a common molecular recognition mechanism. The second component (12.71% of variance) did not correlate with the average of the binding activities (r = −0.08) but did correlate with the differences in binding between TIR1 and AFB5 (r = 0.86). This suggested that structural details on the ligand are driving the small differences in binding specificity toward either TIR1 or AFB5 and that these details may be uncovered by the second factor (Factor 2 = Specificity).
Factor 1 was further analyzed using the equation ((xi – yi)2/(xi + yi)2) categorizing binding Efficiency into five levels (1–5 shown in Supplementary Table S2). Essentially, groups 1–3 are populated by compounds that are inactive in the SPR co-receptor assembly assay or are poor ligands, group 4 compounds are weak ligands, and group 5 compounds are active or highly active against either TIR1 or AFB5 or both.
Four discriminant equations predict correctly 100.00% (Figure 5b) of the changes of binding activity based on an orthogonal arrangement of the overlap and coulomb similarity and self-similarity matrixes. The six maximum-likelihood estimators of binding Efficiency are changes in the overlap matrixes. The overlap self-similarity diagonal of the molecules {Zii(J)}tot is one of these six predictors. This powerful predictor contains electronic structure information, inferring that the fundamental chemical nature of binding Efficiency is related to the volume component of the electronic system.28 The remaining major predictor variables are provided by one component of the total matrix and four components that are a consequence of NH2, fluorine, or chlorine substitutions.
The physicochemical properties that confer specific activity to the natural auxin IAA (labeled as “a” in the scatter plot, Figure 6) can be seen to be mimicked by modifying the structure (in terms of the inter-relationship of a set of basis vectors of a quantum system) of other unsaturated ring systems using an appropriate balance of substitutions using halogens such as F or Cl or an amino group. Halogens may affect the reactivity of the aromatic rings by deactivation, which simultaneously directs electrophilic attack to ortho/para/meta positions. The amino group may activate the electronic structure of aromatic rings according to the classical mesomeric and inductive electronic processes, as well as acting as both a proton donor and acceptor.
Figure 6.
Scatter plot of the binding profiles of the ligands with both TIR1 and AFB5 proteins taking Efficiency and Specificity as independent factors. This classification is inferred from the structural comparison of the ligand molecules by evaluation of the lineal discriminant equations. Compounds are plotted as “Label” according to Supplementary Table S2. Note, Efficiency class 1 does not bind, and hence there are no entries for these compounds.
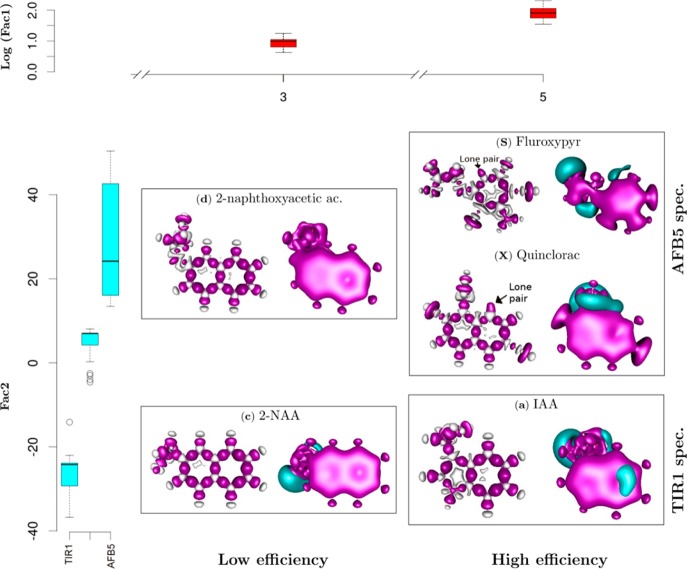
Ring-substituted halogens and N atoms deform the electronic structure of the atomic neighborhood, provoking differences in quadrupole moments and potential energy surfaces of the molecule (Figure 7). The herbicide Fluroxypyr, for example, illustrates the influence of a lone pair on the picolinic ring -N-, which in this case is attracted by the nearby F atom to form a negative potential. The two chlorines are withdrawing electrons from the ring and are connected with the NH2 group to form important quadrupole moments (Figure 7). Quinclorac is another example of an active auxin exhibiting a depletion of ring electron density due to a Cl and a lone pair on the N atom (Figure 7). Interestingly, in this case these atoms are increasing significantly the number of electrons of the system and orienting the π-system. Our analysis infers that halogen substitutions contribute the major direct influence on the electron structure, but further analysis has to be done to explain the details of each specific substitution on auxin-binding interactions.
Figure 7.
Discrimination of auxin-like molecules. Functional dependences were analyzed between the quantum chemical variables of each ligand and binding with TIR1 and AFB5. Box plots show statistical analyses of molecular groupings (Figure 6), relating the feedback between molecular structure and binding Efficiency (log Factor 1 abbreviated to Fac1) and binding Specificity (Fac2). Compounds are organized at two levels of efficiency, low (E3) and high (E5) determined by Fac1, and two levels of specificity, TIR1 and AFB5 (Fac2). Quantum solutions for representative molecules are illustrated. At the right of each panel the blue color represents negative potential at −0.025 atomic units [au], and red the quadrupole moment (0.001 au). These areas are highly likely to contribute hydrogen bond and van der Waals interactions, respectively. The skeletal molecular structure at the left of each panel represents the deformation of the electron density forming intramolecular covalent bonds. The red areas are the positive deformation of the electron density (−0.01 au) or covalent bonds, while the gray areas are the negative deformation (−0.02 au) of the electron density or donor areas. The analysis indicates lone pair electrons on Fluroxypyr and Quinclorac molecules, indicating regions that play a determining role in intermolecular forces.
Unfortunately few commercial auxins have been included in published data sets, and so comparisons to previous classification systems are limited. However, our receptor binding Efficiency ranking agrees well with previous systems, including those based only on biological activity (Supplementary Table S2) and the recent compilation linking tolerance to, e.g., 2,4-D and 2,4,5-T with receptor selectivity,29 adding mechanistic detail.
Co-receptor Selectivity
Picolinates display a higher than average binding activity for AFB5 than IAA along with Quinclorac (Figures 4–7). An important part of the work was to clarify the differences in structural selectivity of the co-receptor complexes TIR1 and AFB5. For this we analyzed the second factor and found that the differences of the binding between receptors were defined by a categorical variable independent of Efficiency. Analysis of the electronic structure of the molecules explained 100.00% of the membership in three levels according to the coefficients (Figure 5c; Specificity). Coefficients close to zero were classified as unspecific (the gray region), high negative coefficients as TIR1-specific (cyan), and high positive as AFB5-specific (blue). The categories are statistically consistent as dependent variables and may help uncover structural features (independent variables) contributing to selective binding activities between auxin molecules and the co-receptors. The independent variables are based on chemometrics analyses of quantum chemical similarity (Zij(Ω)) and self-similarity (own molecular index: Zij(Ω)) indices.12 The solutions were found by using linear discriminant analysis (LDA).
Specificity or unspecificity may not be defined by any one specific atom or position and is influenced by coulomb matrixes, suggesting also that it is driven by electrostatic forces. From the analysis of quantum chemical descriptors, the following are the main factors defining Specificity: (1) double effect of an NH2 group, two orthogonal factors depending on the atomic neighborhood, (2) reduction by iodine of the binding specificity of the molecules, possibly by increasing the van der Waals volumes, (3) electrostatic effects (Coulomb matrix) of fluorine atoms and heterocyclic ring-N substitutions, and (4) the total Coulomb matrix of dissimilarities.
The preference by AFB5 for Picloram, Fluoxypyr, DAS 534, and Quinclorac among others is explained by the Specificity functions. Representative molecules for each of the groupings are shown as quadrupole moment electron densities (Figure 7, right part of each molecule). The upper panel gives class size and illustrates the independence of Specificity factor from Efficiency factor. Global and local 3D electronic structures (Figure 7) show the lone pairs in the nitrogen atom of Quinclorac as well as the corresponding negative potential, most notably for Quinclorac. The quadrupole moment of the electron density represents the deviation of the electron distribution from its ideal, undisturbed spherical cloud around each atom. This will coordinate different multipole–multipole interactions during mutual orientation within the binding pocket of TIR1/AFB5. Further differences of the quadrupole structure will contribute to specific local van der Waals interactions in the binding pocket and with the Aux/IAA degron (compare weak (Low Efficiency) and strong (High Efficiency) ligands in Figure 7).
The analysis suggests overall that electrostatic interactions are dominating the recognition of molecules with greater affinity to AFB5. In comparison, the molecules with greater affinity toward TIR1 present additional negative potential on the aromatic ring structure.
Picolinate Auxins and AFB5
The picolinate and quinolinate auxins provide valuable commercial auxins. Most of the picolinates supported very strong binding to both TIR1 and AFB5, although in general the interactions were stronger with AFB5 than with TIR1 (Figure 4). This is consistent with previous observations identifying AFB5 as the primary target for Picloram.16,6 Fluroxypyr also supported stronger AFB5 co-receptor assembly compared with that of IAA. Of all the compounds tested, DAS534 has the largest van der Waals surface area and was also the strongest by SPR assay with far stronger binding to AFB5 than IAA. Interestingly, DAS534 was found to be among the most active auxins in bioassays.16
The chemometric statistical analysis infers differences of binding activity for sets of compounds against TIR1 and AFB5. However, this inference is based on just 12.64% of the binding information and, so far, a relatively small collection of compounds. Therefore it is noted that it is not possible to define two populations of molecules fitting differentially on these receptors with statistical confidence. Nevertheless, taking into account previous genetic and biochemical evidence,16,6 it is clear that AFB5 is the dominant target for agricultural picolinate auxins and the chemometrics define the factors conferring high Efficiency and high Specificity.
The discovery of a set of compounds with selectivity for AFB5 also raises a question about why this distinction might have developed. The data on selectivity for co-receptor degrons shows no difference between TIR1 and AFB5 (Figure 2), and so the reason for the distinction is likely to lie with ligand binding on TIR1 and AFB5, not co-receptor partnerships. The principle endogenous auxin is IAA, with evidence only of IBA, 4-chloroIAA, and phenylacetic acid as additional active auxins occurring naturally. Of these, no preference was shown for AFB5 (4-chloroIAA was not tested), and chemometrics classifies IBA with no preference, phenylacetic acid with TIR1. Therefore, while IAA does bind strongly to AFB5, selection for more than a single binding geometry suggests that there might be additional endogenous ligands still to be described. So far it is not possible to predict what such additional native ligands might be, and larger chemical libraries will need to be screened to improve statistical definitions.
Antiauxins and Nonbinders
Extensions and elaborations of the indolic side chain have been shown to make effective antiauxins acting as competitive inhibitors.30,31 It remains possible that some of the compounds tested for binding do indeed bind to TIR1/AFB5 but then prevent approach of the Aux/IAA degron and hence block co-receptor assembly. Such antiauxins would give no binding Efficiency in the SPR assay. However, such compounds could record activity in some whole plant bioassays because effective blockade of these receptors would lead to longevity of the Aux/IAA substrates and auxin hyperactivity in the same manner as has been noted for mutations to certain Aux/IAA degrons.24 It is also noted that the naturally occurring IBA supported only minimal binding (Figure 4; Supplementary Tables S1 and S2). It has been reported that IBA may be catabolized to IAA in vivo to become active.32 The data presented (Figure 4) support the hypothesis that IBA is minimally active per se, but it remains to be seen whether this compound has activity in vivo as an antagonist.
To date, the complete co-receptor complex has been found necessary in all binding assay formats.6 Binding to TIR1/AFB5 alone has not yet been recorded, although this must precede co-receptor assembly given that the binding pocket is completely occluded by degron association.5 Until such an assay is devised, it will be necessary to address the distinctions between antiauxins and nonbinders by developing an antiauxin assay by competition.
A few compounds in addition to IBA that are known to be active as auxins in whole plant bioassays showed poor binding in the SPR assay. For example, the benzoate Amiben (also known as Chloramben, 3-amino-2,5-dichlorobenzoic acid), a commercial herbicide, gave little or no binding to either receptor. By quantum chemometrics Amiben is placed in the group of compounds with unspecified target site. We recognize that we have selected the two most extreme receptor proteins for this study and that some selectivity may lie with AFB1–4. If one of these other AFBs proves to be the preferred target for, e.g., benzoate auxins, further routes will open for the development of site-selective compounds and crop resistance to such compounds.
Concluding Comments and Binding Site Models
Co-receptor binding data and chemometric analyses have yielded two independent factors related to auxin activity. The first, which we have labeled Efficiency, defines whether a compound will bind. Five categories of compounds were identified (Efficiency 1–5; Supplementary Table S2). Then, using the compounds that bind well (levels 4 and 5), a second factor was found defining binding Specificity. The data suggest that Efficiency is associated with the overlap matrix (volumes of the components in the molecular system). Specificity is defined by coulomb matrixes, suggesting that it is driven by electrostatic surfaces on the ligand. Larger van der Waals surfaces dominate the recognition of molecules with greater affinity to AFB5 such as the picolinates. Molecules with greater affinity toward TIR1 present additional negative potential on the aromatic ring.
Our results may be compared to previous binding interaction models and SARs. Specific descriptive definitions of auxins have been known to be inadequate for many years (reviewed in 195326), and the focus has been on functional descriptors.26,10,11,8 A molecular interaction energy field model in combination with similarity indices and bioassay data created a useful classification system11 that included antiauxins. Interestingly, this system classed IBA and triiodobenzoic acid (TIBA; no binding, Figure 4) as antiauxins. 2-NAA was moved from an initial classification as an antiauxin to a weak auxin, and this agrees with our data (Figure 4). Indeed, there is general agreement between the classifications assigned by Tomic et al.11 and the SPR data generated here. Their analysis also led to a global surface energy model for bound auxin, although this predates information about the TIR1/AFB receptor family and is unable to differentiate selective binding functions.
The popular sterically and spatially constrained aromatic platform binding model of Katekar10 is only partially supported, in that halogen substitutions increase the number of electrons but deactivate the aromaticity of the ring system. Again, this model predates knowledge of receptor proteins. There are interesting parallels between our findings and the conformational change binding site model.9 Our analysis suggests Efficiency factors drive ligand approach. Specificity factors are independent of Efficiency but will contribute to docking. Kaethner’s model9 allowed the approach of molecules in the “recognition conformation”. Only those with carboxylate side groups able to couple change to the “modulation conformation” with receptor movement are active. We can associate part of Kaethner’s receptor movement to co-receptor assembly,1,2 and the crystal structure of TIR1 showed that IAA is bound in the modulation conformation,5 equivalent to Tilted.11 There are no formalistic links between our Efficiency and Specificity functions and the two phases of binding implied by the Kaethner model, but all indicators point toward more than a single stage binding process.
The definition of auxin activity to which our data matches most closely is that of Veldstra,26 who recognized that the nonpolar ring system needed a high interface activity and the carboxy group (polar part) needed to be in a very definite and peripheral spatial position when bound. To his verbal definition we can now add some chemometrics based on quantum chemical calculations and similarity and self-similarity indices, and we have started to specify important distinctions in ligand preferences between TIR1/AFB family members.
Methods
Materials
All chemicals were of the highest purity available. All auxins and test compounds were dissolved to give 10 mM stock solutions in 50% ethanol or 100 mM in DMSO. Generally the final concentration of solvent was 0.1% or lower and never higher than 0.5%.
Baculovirus Expression and Protein Purification
Expression constructs for both TIR1 and AFB5 were engineered to give fusion proteins His-MBP-FLAG-TIR1 and GST-AFB5. These were cloned into baculovirus vectors that included His-ASK1 to give dual expression (Supplementary Figure S1). Generation of recombinant virus, selection, expression screening, and generation of high-titer viral stock was done by Oxford Expression Systems (Oxford, U.K.). Trichoplusia ni (T. ni High Five) was used throughout as the host cell line. Protein was harvested approx 92 h after infection.
Cells were harvested by centrifugation followed by lysis in Cytobust (Invitrogen) according to the manufacturer’s instructions. A protease inhibitor cocktail (Roche) and MG132 (final 10 μg mL–1, Sigma Aldrich) were included. Cells from 30 mL were lysed (600 μL Cytobust) and clarified by centrifugation. TIR1 lysate was loaded onto anti-FLAG Sepharose. After washing with Biacore buffer HBSEP (10 mM Hepes, 150 mM NaCl, 3 mM EDTA, pH 7.4, 0.005% P20), TIR1 was eluted with FLAG peptide. For AFB5, the lysate was loaded onto an FPLC Superose 12 (30 × 10) column equilibrated in HBSEP. Fractions were screened for activity using the Biacore assay.
Peptides and Compounds
Peptides were synthesized with biotin at the N-terminus to give biot-AKAQVVGWPPVRNYRKN. Key residues in this degron sequence are -GWPPVR-, and as an internal control we used a mutated version of this sequence, biot-AKAQVVEWSSGRNYRKN (IAAm7). Peptides (Thermo Fisher Scientific) were greater than 80% purity, although data sheets generally showed greater than 90% primary product. Peptide stocks (1 mg mL-1) were in deionized water and stored at −20 °C.
SPR
Biotinylated peptides (5 μg mL–1 in HBS EP buffer + 0.01% P20) were passed over streptavidin-coated chips (SA chips, GE Healthcare) to give, typically, around 700RU on the surface. In most cases, channel 1 was blocked with biocytin, channel 2 and channel 4 were coated with active Aux/IAA7 peptide, and channel 3 was coated with IAAm7. Both 2–1 and 4–3 data sets were recorded, although in practice little or no difference was found using either biocytin or IAAm7 as control channel. Sensorgrams were run in HBSEP + 0.01% P20 at 20 °C using a flow rate of 25 μL min–1. Purified TIR1 was mixed with appropriate auxin and incubated in the sample chamber before injection. Regeneration of the chip was with 50 mM NaOH, using injections of 30 s. Regeneration generally returned the sensorgram trace to zero, there was minimal baseline drift within experiments, and the chip could be used repeatedly over many weeks with little deterioration.
In general, association times were 180 s, and dissociation times were 600 s. Reference injections of receptor plus 500 μM IAA as well as receptor in the absence of auxin were included in each run at both start and end of a series, often with a further control midway. In bulk screening experiments all compounds were tested initially at 100 μM, association times were 60 s, and dissociation in buffer was 180 s, followed by regeneration.
Structure–Binding Relationship Analysis
Considering the molecules as functional units, we calculated the mean of binding activity per molecule for both TIR1 and AFB5 (BAmol = (BAmolTIRI + BAmol)/2). This allowed ordering of the molecules by increasing binding activity. A variance decomposition by factorial analysis quantified the different responses of both receptors to the population of molecules, and the equation ((xi – yi)2/(xi + yi)2) of the activity per receptor with respect to BAmol offers the points of the curve with change of activity.
The factorial analysis separates common and specific binding of both receptors. Variations of the curve resemble the changes of binding behaviors of the molecules. From this framework it was possible to focus on the ligand molecules using discriminant analysis, considering the curve of the common binding affinity values and the binomial function of binding specificity per receptor. Both functions are orthogonal.
The chemical structures of all compounds were optimized by quantum chemical calculations using the hybrid functional KMLYP, which gives accurate representations of geometry and electronic structure.33 The optimized geometries served as input for the calculations of the overlap (δ) and coulomb (J) matrixes of quantum chemical similarity measures (QMSM)12 and Hardness.27,33
Quantum chemical similarities of parent molecules were extended using the corresponding matrixes after substitutions of halogen atoms for hydrogen and nitrogen for carbon giving matrixes capturing the influence of halogens (three kind of matrixes for halogens Hal = F, Cl, and I) and N (for both N = Nring, NFunctional. Group) on the electronic structure of each molecule. The corresponding matrixes were factorized by principal component analysis in order to eliminate repetitive information. The following vectors of molecular quantum self-similarities and matrices of quantum chemical similarities in column-reduced form (k) are used in this work:
The similarity matrixes and self-similarity vectors were used as inputs for molecular information matrices to find the changes of electronic structure corresponding to binding mismatches between TIR1 and AFB5 (where they occurred) as well as the general competivity of binding processes using discriminant analysis34 and other statistical confirmatory techniques.35
Acknowledgments
We are grateful for support from the BBSRC (grant reference BB/F014651/1), The University Grants Commission, New Delhi, India and the Commonwealth Association, U.K. (ref INCF-2010-80) and to Dr. T. A. Walsh (Dow AgroSciences, Indianapolis, IN, USA) for access to compound DAS534.
Supporting Information Available
Figures showing the expression cassette and pH dependence and tables of binding and auxin classification data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Dharmasiri N.; Dharmasiri S.; Estelle M. (2005) The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. [DOI] [PubMed] [Google Scholar]
- Kepinski S.; Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451. [DOI] [PubMed] [Google Scholar]
- Gray W. M.; Kepinski S.; Rouse D.; Leyser O.; Estelle M. (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Woodward A. W.; Bartel B. (2005) A receptor for auxin. Plant Cell 17, 2425–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X.; Calderon-Villalobos L. I.; Sharon M.; Zheng C.; Robinson C. V.; Estelle M.; Zheng N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos L. I.; Lee S.; De Oliveira C.; Ivetac A.; Brandt W.; Armitage L.; Sheard L. B.; Tan X.; Parry G.; Mao H.; Zheng N.; Napier R.; Kepinski S.; Estelle M. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier R. M. (2001) Models of auxin binding. J. Plant Growth Regul. 20, 244–254. [Google Scholar]
- Ferro N.; Bredow T.; Jacobsen H.-J.; Reinard T. (2010) Route to novel auxin: Auxin chemical space toward biological correlation carriers. Chem. Rev. 110, 4690–4708. [DOI] [PubMed] [Google Scholar]
- Kaethner T. M. (1977) Conformational change theory for auxin structure-activity relationships. Nature 267, 19–23. [Google Scholar]
- Katekar G. F. (1979) Auxins: on the nature of the receptor site and molecular requirements for auxin activity. Phytochemistry 18, 223–233. [Google Scholar]
- Tomić S.; Gabdoulline R. R.; Kojić-Prodić B.; Wade R. C. (1998) Classification of auxin plant hormones by interaction property similarity indices. J. Comput.-Aided Mol. Des. 12, 63–79. [DOI] [PubMed] [Google Scholar]
- Ferro N.; Gallegos A.; Bultinck P.; Jacobsen H. J.; Carbó-Dorca R.; Reinard T. (2006) Coulomb and overlap self-similarities: a comparative selectivity analysis of structure-function relationships for auxin-like molecules. J Chem. Inf. Model. 46, 1751–1762. [DOI] [PubMed] [Google Scholar]
- Armstrong J. I.; Yuan S.; Dale J. M.; Tanner V. N.; Theologis A. (2004) Identification of inhibitors of auxin transcriptional activation by means of chemical genetics in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 14978–14983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S.; Baiga T. J.; Pojer F.; Dabi T.; Butterfield C.; Parry G.; Santner A.; Dharmasiri N.; Tao Y.; Estelle M.; Noel J. P.; Chory J. (2008) New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proc. Natl. Acad. Sci. U.S.A. 105, 15190–15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B.; Audenaert D.; Beeckman T.; Kepinski S. (2009) The past, present, and future of chemical biology in auxin research. ACS Chem. Biol. 4, 987–998. [DOI] [PubMed] [Google Scholar]
- Walsh T. A.; Neal R.; Merlo A. O.; Honma M.; Hicks G. R.; Wolff K.; Matsumura W.; Davies J. P. (2006) Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol. 142, 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N.; Dharmasiri S.; Jones A. M.; Estelle M. (2003) Auxin action in a cell-free system. Curr. Biol. 13, 1418–1422. [DOI] [PubMed] [Google Scholar]
- Huber W. (2005) A new strategy for improved secondary screening and lead optimization using high-resolution SPR characterization of compound-target interactions. J. Mol. Recognit. 18, 273–281. [DOI] [PubMed] [Google Scholar]
- Nordin H.; Jungnelius M.; Karlsson R.; Karlsson O. P. (2005) Kinetic studies of small molecule interactions with protein kinases using biosensor technology. Anal. Biochem. 340, 359–368. [DOI] [PubMed] [Google Scholar]
- Ramos J. A.; Zenser N.; Leyser O.; Callis J. (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of Domain II and is proteasome dependent. Plant Cell 13, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L. Z.; Cheung A. Y.; Nibau C.; Wu H.-M. (2005) RAC GTPases in tobacco and Arabidopsis mediate auxin-induced formation of proteolytically active protein bodies that contain Aux/IAA proteins. Plant Cell 17, 2369–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shena J.; Zenga Y.; Zhuanga X.; Sunb L.; Yaob X.; Pimpl P.; Jianga L. (2013) Organelle pH in the Arabidopsis endomembrane system. Mol. Plant 6, 1419–1437. [DOI] [PubMed] [Google Scholar]
- Dreher K. A.; Brown J.; Saw R. E.; Callis J. (2006) The Arabidopsis Aux/IAA family has diversified in degradation and auxin responsiveness. Plant Cell 18, 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D.; Mackay P.; Stirnberg P.; Estelle M.; Leyser H. M. O. (1998) Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ouellet F.; Overvoorde P. J.; Theologis A. (2001) IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13, 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldstra H. (1953) The relation of chemical structure to biological activity in growth substances. Ann. Rev. Plant Phys. 4, 151–198. [Google Scholar]
- Ferro N.; Bultinck P.; Gallegos A.; Jacobsen H. J.; Carbo-Dorca R.; Reinard T. (2007) Unrevealed structural requirements for auxin-like molecules by theoretical and experimental evidences. Phytochemistry 68, 237–250. [DOI] [PubMed] [Google Scholar]
- Ponec R.; Amat L.; Carbó-Dorca R. (1999) Molecular basis of quantitative structure-properties relationships (QSPR): a quantum similarity approach. J. Comput.-Aided Mol. Des. 13, 259–270. [DOI] [PubMed] [Google Scholar]
- Simon S.; Kubeš M.; Baster P.; Robert S.; Dobrev P. I.; Friml J.; Petrášek J.; Zažímalová E. (2013) Defining the selectivity of processes along the auxin response chain: a study using auxin analogues. New Phytol. 200, 1034–1048. [DOI] [PubMed] [Google Scholar]
- Hayashi K.; Tan X.; Zheng N.; Hatate T.; Kimura Y.; Kepinski S.; Nozaki H. (2008) Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 5632–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K.-I.; Neve J.; Hirose M.; Kuboki A.; Shimada Y.; Kepinski S.; Nozaki H. (2012) Rational design of an auxin antagonist of the SCF TIR1 auxin receptor complex. ACS Chem. Biol. 7, 590–598. [DOI] [PubMed] [Google Scholar]
- Strader L. C.; Wheeler D. L.; Christensen S. E.; Berens J. C.; Cohen J. D.; Rampey R. A.; Bartel B. (2011) Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23, 984–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro N.; Bredow T. (2010) Assessment of quantum-chemical methods for electronic properties and geometry of signaling biomolecules. J. Comput. Chem. 31, 1063–1079. [DOI] [PubMed] [Google Scholar]
- Mardia K. V., Kent J. T., and Bibby J. M. (1980) Multivariate Analysis (Probability and Mathematical Statistics), Academic Press, London. [Google Scholar]
- Rohlf F. J. and Sokal R. R. (2011) Biometry, W. H. Freeman, New York. . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.