Summary
Virtual ligand screening (VLS) is a powerful technique to identify potential hits of targets and to increase hit rates. Here, we describe how we used this technique combined with NMR 15N HSQC experiments to obtain small molecules that bind to the PDZ domain of Dvl targeting the Wnt signaling pathway.
Keywords: Virtual ligand screening, database search, docking, NMR, 15N HSQC, SAR
1. Introduction
Wnt signaling pathways play key developmental and growth regulatory roles that significantly affect many biological processes; abnormal Wnt activity has been implicated in cancer and other human diseases. Dishevelled (Dvl) relays the signal from the membrane-bound Wnt receptors of the Frizzled (Fz) family to downstream substrates via its PDZ domain (1). The special role of the Dvl PDZ domain in the Wnt signaling pathway (1) makes it a potential pharmaceutical target (2). Small organic inhibitors of the PDZ domain in Dvl (3, 4) might be useful in dissecting molecular mechanisms and formulating pharmaceutical agents that target cancers or other diseases in which Wnt signaling is involved.
1.1 Structure-based Virtual Ligand Screening for Inhibitor of the Dvl PDZ Domain
Like many other PDZ domains (5), the structure of the Dvl PDZ domain is known (6). This has permitted us to use structure-based virtual ligand screening (VLS) to computationally access potential ligands. By using a UNITY search for compounds with the potential to bind to the PDZ domain, FlexX docking of candidates into the binding site, Cscore ranking of binding modes, and chemical shift perturbation NMR experiments, we identified a small organic molecule (NSC668036) from the National Cancer Institute (NCI) small-molecule library that can bind to the mDvl1 PDZ domain. Further NMR experiments confirmed that the compound binds to the peptide-binding site on the surface of the PDZ domain. In addition, we carried out molecular dynamics (MD) simulations of the interaction between this compound and the PDZ domain as well as that between the C-terminal region of a known PDZ domain inhibitor (Dapper) and the PDZ domain, and we compared the binding free energies of these interactions calculated via the molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) method (7-9).
1.2 Optimizing Dvl PDZ Domain Inhibitor by Virtually Exploring Chemical Space
With the advancement of technology in the field of drug discovery, hits of a potential therapeutic reagent can be identified in a comparatively straightforward fashion by using high-throughput screening (10, 11). However the follow-up hit-to-lead process and lead optimization still remain as challenging problems in the drug discovery process (12, 13). One of the most frequently taken approaches in the hit-to-lead process is hit evolution (13). During hit evolution, analogues of the most promising hits are synthesized for the development of structure–activity relationship (SAR) data. The SAR is then used to guide the synthesis and optimization of lead compounds to improve their potencies and physico-chemical properties, and to reduce off-target activities. The synthesis processes are usually long and labor-intensive. Virtual screening of databases consisting of physically available compounds may help us to take advantage of the chemistry that has already been done and speed up projects, especially with the ever growing list of existing compounds. Indeed, the Zinc database has 13 million compounds (14) and the iResearch™ Library (ChemNavigator, San Diego, CA) has more than 50 million unique chemicals. Although the databases of available compounds are still under-sampled (15), the chemical space represented by those millions of compounds should never be neglected. We believed that the large chemical space of available compounds offers us with an opportunity to explore SAR of known hits; and as a proof of principle test, we searched the ChemDiv database for the Dvl PDZ domain inhibitors based on an inhibitor identified above (16). In our studies, we first developed a pharmacophore model based on NSC668036; based on the model, we then screened the ChemDiv database by using an algorithm that combines similarity search and docking procedures; finally, we selected potent inhibitors based on docking analysis and examined them by using NMR spectroscopy. NMR experiments showed that all the 15 compounds we chose bound to the PDZ domain tighter than NSC668036.
2. Materials
2.1 Virtual Screening
The structure of the Dvl PDZ domain was obtained from the protein data bank (PDB entry 1L6O (6)).
The three-dimensional (3D) NCI database of small-molecule is available from NCI at no cost, and it includes the coordinates of more than 250,000 druglike chemical compounds. The Tripos format of NCI database comes with SYBYL® (St Louis, MO) and is ready for UNITY search. For ChemDiv database, UNITY in SYBYL® was used to convert the SLN files obtained from ChemDiv Inc. (San Diego, CA).
2.2 NMR Experiments
The 15N-labeled mouse Dvl1 PDZ domain (residues 247–341 of mouse Dvl1) was prepared as described previously (1, 17) by the protein production facility at St. Jude Children's Research Hospital. For NMR, the 0.3 mM 15N-labeled Dvl PDZ domain was in 100 mM potassium phosphate buffer (pH 7.5), 10% D2O, and 0.5 mM EDTA.
Compounds from the NCI library were obtained from Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, National Cancer Institute (http://129.43.27.140/ncidb2/). The 15 ChemDiv compounds were purchased from ChemDiv Inc. (San Diego, CA). Compounds were dissolved in the 100 mM potassium phosphate buffer (pH 7.5) with 10% D2O, and 0.5 mM EDTA. DMSO were added for compounds not soluble in the above buffer.
3. Methods
3.1 Structure-based Ligand Screening of Small Compounds Binding to the PDZ Domain
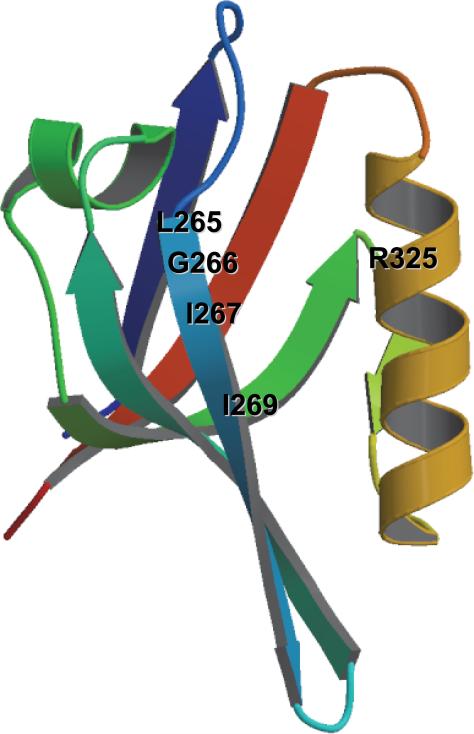
The UNITY module of the SYBYL software package (Tripos, Inc.) was used to screen the NCI small-molecule three-dimensional database for chemical compounds that could fit into the peptide-binding groove of the Dvl PDZ domain. Based on the complex structure of PDZ domain and Dapper peptide (6), a search query was designed; it consists of two hydrogen bond donors (backbone amide nitrogens of Gly266 and Ile269) and two hydrogen bond acceptors (carbonyl oxygens of Ile267 and Ile269) on the PDZ domain, with 0.3 Å tolerances for spatial constraints (Figure 1).
The Flex search module of UNITY was used to explore the 3D small-molecule NCI database to identify compounds that met the requirements of the query. The Flex search option of UNITY considers the flexibility of compounds, and it uses the Directed Tweak algorithm to conduct a rapid and conformationally flexible 3D search (18). The search yielded 108 organic compounds as the initial hits.
These 108 hits then were “docked” into the binding site of the PDZ domain by using the FlexX program of SYBYL. FlexX is energy minimization modeling software that varies the conformation of the ligand to fit it into the protein-binding site (19). The receptor's binding site was defined by residues Gly266, Ile269, and Arg325 with a selection radius of 5.9 Å, and a core subpocket was defined by Gly266 with a selection radius of 5.9 Å (Figure 1). The docking condition was validated by docking the dapper peptide to the PDZ domain as in Note 4.2.
The results of the docking procedure were evaluated manually and those compounds that were not docked into the binding pocket of the PDZ domain were removed. The Cscore program of SYBYL was then used to rank the remaining compounds on the basis of their predicted ability to bind to the binding pocket. Cscore generates a relative, consensus score, based on the individual scoring functions of the protein-ligand complex (20). One of the scoring functions in Cscore, the F_score, is particularly useful. F_score considers polar and nonpolar interactions in calculating the binding free energy of ligand and protein. Based on this notion, nine compounds whose F_scores were better than that of the control Dapper-PDZ interaction were then further characterized; and subsequently obtained from the Developmental Therapeutics Program of the NCI.
Figure 1. The structure of the Dvl PDZ domain.
The ligand site is located at α-helix B (yellow) and β-sheet B (blue) regions.
3.2 Testing VLS Hits with NMR Spectroscopy
The abilities of the nine compounds obtained from DTP to bind to the PDZ domain were tested by NMR spectroscopy, mainly the chemical shift perturbation experiment (21). NMR 15N HSQC experiments were performed by using a Varian Inova 600 MHz NMR spectrometer at 25 °C.
HSQC of the 15N-labeled Dvl PDZ domain (residues 247–341 of mouse Dvl1) was recorded as a reference.
Compound was added to the solution PDZ domain and NMR spectrum of the mixture was recorded. If the compound generated perturbations to the PDZ domain, different amounts of compound were added to the PDZ domain and series of NMR spectra were recorded for calculations of compound's dissociation constant (KD) to the PDZ domain.
NMR spectra were processed with NMRpipe (22).
NMR spectra were analyzed by using Sparky (23). We monitored whether added compounds generated chemical shift perturbations to the resonances of the Dvl PDZ domain and compared if the chemical shift perturbations were similar to those caused by binding of the Dapper peptide and Fz7 peptide, which was derived from a Fz membrane receptor (1). Similar perturbations would suggest that compound binds to the same binding site as native PDZ domain-binding partners such as Dapper and Fz. Among nine tested compounds, NSC668036 perturbed the PDZ domain similarly as Dapper and Fz suggesting it binds to the same binding site as these two native peptides.
- Calculating KD by using HSQC titration spectra. The binding affinities (KD) of PDZ ligands were calculated using HSQC spectra by following the method described by Worrall et al. (24). The mean chemical-shift perturbation changes caused by the binding of ligands were calculated using Eq. 1. KD was then calculated using Eqs. 2 and 3 by applying a one-site binding model with corrections for dilutions, where R was the ligand to protein molar ratio, P was the protein concentration before titration, C was the ligand stock concentration, and KD was the dissociation constant. Two-parameter nonlinear least-squares fitting was performed with program Prism (GraphPad Software, La Jolla, CA).
(1) (2) (3)
3.3 Molecular Dynamics Simulationof Verified PDZ Ligand
To further investigate the interaction between the PDZ domain and NSC668036, the AMBER software (25) suite was used to conduct a MD simulation study of the NSC668036-PDZ domain complex.
The starting structures of ligand-protein complexes were prepared using the output from the FlexX docking studies. To sample sufficient possible binding modes during the MD simulation, the entire output of the initial FlexX docking results were re-examined. Based on structure alignment, the 30 docked NSC668036 conformers can be clustered into three clusters. Manual inspection of these docking conformers led us to select 10 conformers from the three clusters representing the 30 conformers as starting points for the MD simulations.
After neutralization these complexes with Na+ or Cl-, they were dissolved in a periodic rectangular TIP3P water box, with each side 10 Å from the edge of the complex. AM1-BCC charges and parameters from GAFF force field were assigned to NSC668036 by using the Antechamber module (26) in AMBER 8. For protein, ions and water, parm99 force field (25, 27) was used.
Systems were minimized by a 1000-step steepest descent minimization followed by a 9000-step conjugated gradient minimization (25, 27).
MD simulation was performed by using the sander program in AMBER 8 with a time step of 2 fs and the nonbonded cutoff set to 9.0 Å. Constant volume (NVT) and constant pressure (NPT) ensemble simulations were carried out to equilibrate the system. In detail, a 50 ps NVT simulation was used to increase the temperature from 100 to 300 K; then the 50 ps NPT ensemble was used to adjust the solvent density, and another 100 ps NPT ensemble was used to gradually reduce the harmonic restraints from 5.0 kcal mol-1 Å-2 to none (25, 27).
MD simulations were performed in explicit water for 5 ns after equilibration with the particle mesh Ewald (PME) method (28, 29) using the NPT ensemble. During the production run, snapshots were saved every 5 ps. Other simulation parameters were set to values similar to those described in the work by Gohlke et al. (30).
3.4 Binding Free Energy Calculation
- The MM-PBSA algorithm (7-9) was then used to calculate the binding free energy of the interaction between the PDZ domain and NSC668036 using Eq. 4 with the mm_pbsa.pl script in AMBER 8, which employs an MM-PBSA approach.
Where(4) (5) (6)
where the gas phase energy, Hgas, is the sum of internal (bond, angle, and torsion), van der Waals, and electrostatic energies in the molecular mechanical force field with no cutoff, as calculated by molecular mechanics (31). Htrans/rot is 3RT (R being the gas constant) because of six translational and rotational degrees of freedom. The solvation free energy, Gsolvation, was calculated by using the PB model (8, 30, 31). In PB calculations, the polar solvation energy, , was obtained by solving the PB equation with Delphi using parse radius, parm94 charges (for the PDZ domain and the Dapper peptide), and AM1-BCC charges (for the compound). The nonpolar contribution was calculated by Eq. 7. In this equation, A is the solvent accessible area calculated by the Molsurf module in Amber 8 and γ (surface tension) and b (a constant) were 0.00542 kcal mol-1 Å-2 and 0.92 kcal mol-1, respectively.(7) All of the energy terms given above were averaged from 150 snapshots extracted every 20 ps, and the entropy TS was estimated by normal-mode analysis using 15 snapshots extracted every 200 ps during the last 3 ns production run.
During the 10 MD simulation runs, the simulation that started with conformer 22 had the lowest and most stable binding free energy, suggesting that this conformer represents the true PDZ domain-bound conformation of NSC668036 in solution.
3.5 Deduction of Pharmacophore Based on a PDZ-Inhibitor Complex Structure and Two Non-binders
A pharmacophore is composed of functional groups essential and necessary to receptor-ligand binding; without those groups, ligands will no longer bind to receptors and lose their activities (32). Pharmacophore-based approaches have been widely used and shown successes in the field of computer-aided drug design (33, 34). In order to identify more PDZ domain inhibitors, we took such an approach, derived the pharmacophore of PDZ ligands and used it to screen for PDZ domain inhibitors in the ChemDiv database.
The complex structure of NSC668036 and the PDZ domain generated by docking and extensive molecular dynamics simulations was used to build a pharmacophore by using LigandScout (Inte:Ligand, Austria). LigandScout extracts 3-D pharmacophores based on complex structures (35).
The LigandScout pharmacophore model of NSC668036 is consistent with the structural analysis. The carboxyl group contributes three hydrogen-bond acceptors and the isopropyl group contributes a hydrophobic interaction to the pharmacophore.
The NSC668036 compound was compared with other compounds that did not bind to the PDZ domain as verified by NMR experiments. By aligning two non-binders, NSC344681 (((((2-amino-3-hydroxybutanoyl) amino)acetyl)amino)acetic acid) and NSC119132 (methyl 3-((2-amino-3-(aminooxy)propanoyl)amino)-2-((aminooxy) methyl)-3-oxopropanoate) against NSC668036, the differences among these structures that might render NSC344681 and NSC119132 inactive were identified. NSC334681 does not have a hydrophobic group at the 2-position; and NSC119132 is an ester instead of a free acid. This finding strongly suggested that both the carboxyl group and the hydrophobic group next to it were important to the binding and might compose the pharmacophore of PDZ ligands since compounds without them did not bind to the protein.
3.6 Pharmacophore-based Virtual Screening
Based on the pharmacophore proposed above, a 2-D search to retrieve all the compounds with 2-(3-methylbutanoic acid) groups in the ChemDiv database by using UNITY in SYBYL® (Tripos, Inc.) was carried out and the search returned 116 hits.
To reduce the number of compounds needed to test experimentally and to select ligands with higher potentials to bind to the PDZ domain, the hits were further filtered by using FlexX docking and “Cscore” ranking following the procedure described in Section 3.1. After manually inspecting top-ranked compounds, 15 compounds were chosen for further examinations based on the following four criteria: i) the compounds were docked into the designated binding site, ii) their docking conformations were complementary to that of the PDZ domain, and iii) the docked compounds formed hydrogen-bonds with the βA–βB loop as well as iv) additional hydrogenbonds with the PDZ domain.
3.7 Build SAR Models of PDZ Domain Inhibitors
The interactions between the 15 compounds and the Dvl PDZ domain were examined experimentally by using NMR chemical shift perturbation experiments as described in Section 3.2. All the 15 compounds bound to the PDZ domain at the same binding site as the Fz7 peptide.
The above verified compounds were then ranked by their KD to the PDZ domain, which were calculated by using chemical-shift perturbation changes as described in Section 3.2.
The complex structures of these compounds with the PDZ domain were modeled. The FlexX docking models of the 15 PDZ ligands were refined by using Glide (Schrödinger Inc.). During Glide docking, compounds’ amide bonds were kept rigid; hydrogen-bond pharmacophores were designed on the protein to induce ligands to form hydrogen-bonds with the βA–βB loop and the βB strand of the PDZ domain. Other than fixed amide bonds and H-bond pharmacophores, default docking parameters were used. All ligand binding poses generated by Glide have reasonable Glide scores, suggesting that they are likely very close to the true binding modes. For example, the Glide score of compound ChemDiv 5435-0027 is -7.57. According to Schrödinger Inc., low-micromolar inhibitors should have scores around -7. Glide 2.5 predicted binding affinities of a set of 125 crystallized complexes with an RMSD of 2.2 kcal/mol against experimental data (36). Based on the facts that compound ChemDiv 5435-0027 binds to the PDZ domain with a moderate binding affinity and its glide score fits with the experimental data, it is likely that this docking conformation is close to the true binding mode. This conformation was then used to generate complex structures for compounds ChemDiv 2509-0036 and 2509-0040 which had different docking poses than those of the rest of 13 compounds. Complex structures of compounds ChemDiv 2509-0036 and 2509-0040 were modeled by superimposing them onto docked compound ChemDiv 5435-0027 followed by ligand minimization in the ligand binding pocket of the PDZ domain with LigandScout.
Structure-based pharmacophore models of these compounds were built using LigandScout as in Section 3.5.
These 15 compounds are similar in structure but have considerable differences in binding affinities, and thus provide an opportunity to study their SAR to gain insights into the molecular determinants of PDZ-ligand binding which might facilitate further ligand optimization. These 15 compounds have similar scaffolds except that some of them have an extra double bond in their scaffold. According to their scaffolds, these compounds can be grouped into two classes and by comparing their structures and binding strengths in the context of docking complex structures and complex-structure-based pharmacophore model, the SAR models of these 15 compounds can be built. In turn, the SAR models can be used to guide furthering optimization of PDZ ligands.
4. Notes
4.1 Using the Structure of a Homologous Protein in VLS and Docking
To build the search query for the virtual screening stage, we used the crystal structure of the PDZ domain of Xenopus Dvl bound with the Dapper peptide (7) instead of the NMR solution structure of the apo-PDZ domain of mouse Dvl (1). The two PDZ domains share a high degree of homology, especially around the peptide-binding sites; near the binding sites, there is only a single amino acid difference between the two PDZ domains (Glu323 in the PDZ domain of mDvl1 vs. Asp326 in the PDZ domain of Xenopus Dvl), and the side chain of this residue points away from the peptide-binding cleft. The peptide-binding cavity of the domain is smaller in the apo form of the solution structure than in the crystal structure of the Dapper-bound PDZ domain of Xenopus Dvl. This difference is consistent with the classic “induce-and-fit” mechanism, in which, upon the binding of a peptide or a small organic molecule, the binding sites in the PDZ domain undergo conformational change to accommodate the bound ligand. However, this flexibility cannot be fully explored through a UNITY search and the FlexX docking protocols. Therefore, although the PDZ domain of mouse Dvl was used in the experimental studies, the crystal structure of the PDZ domain of Xenopus Dvl provides a better template for the virtual screening steps. Indeed, the binding free energies calculated from MD simulation of the PDZ domain-NSC668036 and PDZ domain-Dapper peptide complexes fit well with the experimental binding data.
4.2 Docking Validation
As a control, we also docked the Dapper peptide into the PDZ domain using FlexX. Under this condition, the docked Dapper peptide had a conformation similar to that found in the crystal structure of the complex with a backbone RMSD of 2.04 Å. In particular, the backbone RMSD for the last six C-terminal amino acids is 1.22 Å, indicating that the docking procedure we used was able to dock ligand into the binding site of the PDZ domain with reasonable accuracy.
4.3 Compound Handling
To test compounds using NMR experiments, they should be dissolved in the same buffer as in which the protein was dissolved. pH should be adjusted back to 7.5 if changed to ensure that perturbations on the PDZ domain would only come from the compounds but not from changes in the condition. For compounds with small solubility in the protein buffer, small amount of DMSO was added so that there would be no more than 5% of DMSO in the final mixture of the PDZ domain with the compound. NMR experiments showed that < 5% DMSO does not change the spectra of the PDZ domain. Compound solutions were stored in the freezer at -20 °C to avoid hydrolysis and/or other reactions that might change compound identities.
4.4 Test Selectivity of Identified Ligands
We tested two other PDZ domains: the first PDZ domain of PSD-95, PSD95a (37) (PDB entries 1IU0 and 1IU2), which belongs to the class I PDZ domains, and the PDZ7 domain of the glutamate receptor-interacting protein (38) (PDB entry 1M5Z), a member of the class II PDZ domains. NSC668036 binds to both of these PDZ domains extremely weakly.
References
- 1.Wong H-C, et al. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the c-terminal region of Frizzled. Molecular Cell. 2003;12:1251–1260. doi: 10.1016/s1097-2765(03)00427-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 3.Grandy D, et al. Discovery and characterization of a small molecule inhibitor of the PDZ domain of dishevelled. J Biol Chem. 2009;284:16256–16263. doi: 10.1074/jbc.M109.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee HJ, et al. Sulindac inhibits canonical Wnt signaling by blocking the PDZ domain of the protein Dishevelled. Angew Chem Int Ed Engl. 2009;48:6448–6452. doi: 10.1002/anie.200902981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HJ, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheyette BNR, et al. Dapper, a Dishevelled-associated antagonist of b-Catenin and JNK signaling, is required for Notochord formation. Developmental Cell. 2002;2:449–461. doi: 10.1016/s1534-5807(02)00140-5. [DOI] [PubMed] [Google Scholar]
- 7.Gohlke H, Case DA. Converging free energy estimates: MM-PB(GB)SA studies on the protein–protein complex Ras–Raf. Journal of Computational Chemistry. 2004;25:238–250. doi: 10.1002/jcc.10379. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, et al. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. Journal of the American Chemical Society. 2001;123:5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, et al. BIOMOLECULAR SIMULATIONS: Recent developments in force fields, simulations of enzyme catalysis, protein-ligand, protein-protein, and protein-nucleic acid noncovalent interactions. Annual Review of Biophysics and Biomolecular Structure. 2001;30:211–243. doi: 10.1146/annurev.biophys.30.1.211. [DOI] [PubMed] [Google Scholar]
- 10.Burbaum JJ, Sigal NH. New technologies for high-throughput screening. Current Opinion in Chemical Biology. 1997;1:72–78. doi: 10.1016/s1367-5931(97)80111-1. [DOI] [PubMed] [Google Scholar]
- 11.Liu B, et al. Technological advances in high-throughput screening. Am J Pharmacogenomics. 2004;4:263–276. doi: 10.2165/00129785-200404040-00006. [DOI] [PubMed] [Google Scholar]
- 12.Bleicher KH, et al. Hit and lead generation: beyond high-throughput screening. Nat Rev Drug Discov. 2003;2:369–378. doi: 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- 13.Keseru GM, Makara GM. Hit discovery and hit-to-lead approaches. Drug Discovery Today. 2006;11:741–748. doi: 10.1016/j.drudis.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Irwin JJ, Shoichet BK. ZINC -- A free database of commercially available compounds for virtual screening. Journal of Chemical Information and Modeling. 2004;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hann MM, Oprea TI. Pursuing the leadlikeness concept in pharmaceutical research. Current Opinion in Chemical Biology. 2004;8:255–263. doi: 10.1016/j.cbpa.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Shan J, et al. Identification of a specific inhibitor of the Dishevelled PDZ domain. Biochemistry. 2005;44:15495–15503. doi: 10.1021/bi0512602. [DOI] [PubMed] [Google Scholar]
- 17.London TBC, et al. Interaction between the internal motif KTXXXI of Idax and mDvl PDZ domain. Biochemical and Biophysical Research Communications. 2004;322:326–332. doi: 10.1016/j.bbrc.2004.07.113. [DOI] [PubMed] [Google Scholar]
- 18.Hurst T. Flexible 3D searching: The directed tweak technique. Journal of Chemical Information and Computer Sciences. 1994;34:190–196. [Google Scholar]
- 19.Rarey M, et al. A fast flexible docking method using an incremental construction algorithm. Journal of Molecular Biology. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 20.Clark RD, et al. Consensus scoring for ligand/protein interactions. Journal of Molecular Graphics and Modelling. 2002;20:281–295. doi: 10.1016/s1093-3263(01)00125-5. [DOI] [PubMed] [Google Scholar]
- 21.Zheng J, et al. Identification of the binding site for acidic phospholipids on the PH domain of dynamin: Implications for stimulation of GTPase activity. Journal of Molecular Biology. 1996;255:14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]
- 22.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. Journal of Biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Goddard TD, Kenller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- 24.Worrall JAR, et al. Transient protein interactions studied by NMR spectroscopy: The case of cytochrome c and adrenodoxin. Biochemistry. 2003;42:7068–7076. doi: 10.1021/bi0342968. [DOI] [PubMed] [Google Scholar]
- 25.Case DA, et al. AMBER 8. Scripps Research Institute; La Jolla, CA: 2004. [Google Scholar]
- 26.Wang J, et al. Development and testing of a general amber force field. Journal of Computational Chemistry. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 27.Cornell WD, et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. Journal of the American Chemical Society. 1995;117:5179–5197. [Google Scholar]
- 28.Cerutti DS, et al. Staggered Mesh Ewald: An extension of the Smooth Particle-Mesh Ewald method adding great versatility. J Chem Theory Comput. 2009;5:2322. doi: 10.1021/ct9001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmerling C, et al. Combined locally enhanced sampling and Particle Mesh Ewald as a strategy to locate the experimental structure of a nonhelical nucleic acid. Journal of the American Chemical Society. 1998;120:7149–7155. [Google Scholar]
- 30.Gohlke H, et al. Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. Journal of Molecular Biology. 2003;330:891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 31.Kollman PA, et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Accounts of Chemical Research. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 32.Gund P. Three-dimensional pharmacophoric pattern searching. Progress in molecular and subcellular biology. 1977;5:17. [Google Scholar]
- 33.Guner OF. Pharmacophore perception, development, and use in drug design. International University Line; 2000. [Google Scholar]
- 34.Langer T, Hoffmann RD. Pharmacophores and pharmacophore searches. Wiley-VCH; 2006. [Google Scholar]
- 35.Wolber G, Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. Journal of Chemical Information and Modeling. 2004;45:160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 36.Friesner RA, et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 37.Long J-F, et al. Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95. Journal of Molecular Biology. 2003;327:203–214. doi: 10.1016/s0022-2836(03)00113-x. [DOI] [PubMed] [Google Scholar]
- 38.Feng W, et al. PDZ7 of glutamate receptor interacting protein binds to its target via a novel hydrophobic surface area. J Biol Chem. 2002;277:41140–41146. doi: 10.1074/jbc.M207206200. [DOI] [PubMed] [Google Scholar]