Summary
The stability and activity of numerous signaling proteins in both normal and cancer cells depends on the dimeric molecular chaperone Heat Shock Protein 90 (Hsp90). Hsp90 function is coupled to ATP binding and hydrolysis, and requires a series of conformational changes that are regulated by co-chaperones and numerous posttranslational modifications (PTMs). SUMOylation is one of the least understood Hsp90 PTMs. Here we show that asymmetric SUMOylation of a conserved lysine residue in the N-domain of both yeast (K178) and human (K191) Hsp90 facilitates both recruitment of the ATPase activating co-chaperone Aha1 and, unexpectedly, the binding of Hsp90 inhibitors, suggesting that these drugs associate preferentially with Hsp90 proteins that are actively engaged in the chaperone cycle. Importantly, cellular transformation is accompanied by elevated steady-state N-domain SUMOylation and increased Hsp90 SUMOylation sensitizes yeast and mammalian cells to Hsp90 inhibitors, providing a mechanism to explain the sensitivity of cancer cells to these drugs.
Keywords: Heat Shock Protein 90, SUMOylation, Molecular Chaperones, Co-chaperones, Aha1
Introduction
The evolutionarily conserved molecular chaperone Heat Shock Protein 90 (Hsp90) is an essential component of the cellular homeostatic machinery in eukaryotes (Picard, 2002; Taipale et al., 2010). Cancer cells strongly depend on Hsp90 because of their need to cope with constitutive genetic instability and frequent environmental insults, including nutrient deprivation, hypoxia and proteotoxic stress (Trepel et al., 2010), and emerging clinical data identify Hsp90 inhibition as a promising therapeutic strategy to treat cancer (Neckers and Workman, 2012). Cancer cells appear to be particularly sensitive to Hsp90 inhibitors when compared to non-transformed cells (Bisht et al., 2003), and these drugs are retained by tumors in vivo far longer than in normal tissues (Trepel et al., 2010). However, the molecular basis for these phenomena remains undefined.
The Hsp90 molecule is a dimer, and each protomer can be divided into structurally and functionally distinct domains: i) an N-terminal domain that contains an ATP-binding pocket, which can also be occupied by small molecule inhibitors, as well as sites for co-chaperone and client binding, ii) a middle domain that provides additional co-chaperone and client interacting surfaces, and also completes the split ATPase motif of Hsp90, iii) a C-terminal domain providing sites for constitutive dimerization and for co-chaperone and client interaction (Ali et al., 2006; Shiau et al., 2006; Stebbins et al., 1997). Hsp90 chaperone function depends on an ordered sequence of dynamic conformational changes that are linked to binding and hydrolysis of ATP and are disrupted by drug occupancy of the ATP pocket (Hessling et al., 2009; Mayer, 2010; McClellan et al., 2007; Picard, 2002; Prodromou, 2012; Trepel et al., 2010). In eukaryotes, the Hsp90 conformational cycle is facilitated by a number of proteins, termed co-chaperones, that interact with distinct Hsp90 conformations and serve discrete functions (Cox and Johnson, 2011; Li et al., 2012). Growing evidence suggests that Hsp90 interacts asymmetrically with clients and some co-chaperones and that each Hsp90 protomer can hydrolyze ATP independently (Cunningham et al., 2008; Vaughan et al., 2006). A number of co-chaperones (e.g., p50/Cdc37, HOP/Sti1, p23, Aha1) modulate the rate of Hsp90-mediated ATP hydrolysis, thereby regulating the rate of the chaperone cycle and tailoring Hsp90 chaperone activity to the needs of specific clients (Panaretou et al., 2002; Retzlaff et al., 2010; Siligardi et al., 2004). Importantly, Aha1, by facilitating energy-intensive conformational changes necessary to establish Hsp90 ATPase competence, markedly stimulates the weak endogenous ATPase activity of Hsp90 and is thus considered to be a crucial component of active Hsp90 chaperone complexes (Li et al., 2013; Lotz et al., 2003; Panaretou et al., 1998).
In Saccharomyces cerevisiae, the abundance of Hsp90 far exceeds that of its co-chaperones (http://yeastgfp.yeastgenome.org/). Thus, mechanisms must exist to facilitate the controlled interaction of these proteins at sites of chaperone activity. Eukaryotic Hsp90 is subject to numerous posttranslational modifications (PTMs) that play significant roles in regulating association and dissociation of specific co-chaperones (Mollapour and Neckers, 2012; Muller et al., 2012; Soroka et al., 2012; Walton-Diaz et al., 2013; Xu et al., 2012). SUMOylation is a reversible PTM in which Small Ubiquitin-like Modifier (SUMO) proteins are covalently linked to lysine residues, in a process similar to ubiquitination, and in so doing affect the conformation, protein-protein interaction, or subcellular location of the modified protein (Hickey et al., 2012; Ulrich, 2009). Although SUMOylation of yeast Hsp82 (yHsp90) and human Hsp90α (hHsp90α) has been reported previously (Panse et al., 2004; Pountney et al., 2008; Zhou et al., 2004), identification of individual SUMOylated lysine residues and their impact on Hsp90 function remains unexplored. Here, we report that SUMOylation of an N-domain lysine conserved in both yeast and human Hsp90 initiates recruitment of Aha1 to the chaperone complex in cells. Unexpectedly, N-domain SUMOylation also facilitates binding of Hsp90 inhibitors and sensitizes yeast and mammalian cells to these drugs.
Results
Identification of a SUMOylated lysine in the Hsp90 N-domain
We identified a conserved lysine residue (K178) in the N-domain of yeast Hsp82 (yHsp90) that is within the SUMO recognition motif Ψ-K-x-D/E (Ψ is a hydrophobic amino acid and x is any amino acid) (Sampson et al., 2001) (Figure 1A). Lysine 178, in the N-terminal domain (open ATP lid), is situated in a solvent exposed loop and thus is potentially susceptible to post-translational modification (pdb: 1AMW), although the side-chain amine of lysine 178 makes a series of interactions with the main-chain carbonyl groups of isoleucine 20, phenylalanine 128 and leucine 129. In the closed state (pdb: 2CG9), however, lysine 178 becomes buried in the N-terminal dimerization interface and the side-chain amine of lysine 178 is now hydrogen bonded to one of the two carboxyl side-chain oxygens of aspartic acid 132 and to the main-chain carbonyl oxygens of alanine 131 and phenylalanine 128. Thus, posttranslational modification of lysine 178 is likely only possible prior to ATP lid closure and N-domain dimerization, but modification of this residue also appears to require additional conformational changes not visible in these crystal structures.
Figure 1. The N-domain of yeast and human Hsp90 is SUMOylated in vivo.

A) Schematic representation of yHsp90 and hHsp90α showing the putative N-domain SUMO consensus motif. Proposed SUMO modification sites are in red.
B) N-domain SUMOylation of wild-type (WT) and yHsp90-K178R (K178R) was detected by immunoblotting with anti-hexahistidine and anti-Smt3 antibodies after PreScission protease digestion.
C) Yeast cells expressing wild-type (WT) or yHsp90-K178R (K178R) and carrying SMT3-MYC under GAL1 promoter were grown on raffinose (–) or galactose (+). Hsp90 modification by Smt3-Myc was assessed by immunoblotting Hsp90 pulldowns.
D) SUMOylation of N-domain wild-type (WT) FLAG-hHsp90α and K191R mutant. Hsp90 proteins were immunoprecipitated, subjected to PreScission protease digestion, and immunoblotted with anti-FLAG and anti-SUMO-1 antibodies. Lysates from cells transfected with empty plasmid (C) were used as a control. See also Figure S1.
To ascertain whether this residue is SUMOylated in cells and to determine if it was the only SUMOylated residue in the yHsp90 N-domain, we mutated lysine 178 to non-SUMOylatable arginine. We also introduced a PreScission protease cleavage site between the N-domain and adjacent charged linker region of both wild-type yHsp90 and yHsp90-K178R, allowing us to separate the N-domain from the full-length Hsp90 protein (which is predicted to have additional SUMOylation sites in other domains) (Figure 1A). These yHsp90 proteins were hexahistidine epitope-tagged at their N-terminus and were expressed as the sole copy of yHsp90 in yeast. It is noteworthy that insertion of a PreScission protease site upstream of the N-domain did not interfere with the chaperone activity of wild-type yHsp90 (Figure S1A-D). The hexahistidine epitope-tag allowed us to isolate Hsp90 proteins from cell lysates using Ni-NTA agarose beads. We then treated the pulldowns with PreScission protease to isolate yHsp90 N-domains. Western blotting of the isolated N-domains with antibodies to both hexahistidine and Smt3 (the only SUMO protein in yeast) confirmed a slower migrating band to be SUMOylated yHsp90 (Figure 1B). These data also confirmed that lysine 178 is the only SUMOylated lysine in the yHsp90 N-domain (Figure 1B). Lysine can also be targeted by other PTMs, including ubiquitination and acetylation, however our data suggest that lysine 178 is only subject to SUMOylation (Figure S1E, F).
Smt3 is an essential protein in yeast. To query the impact of modulating cellular Smt3 expression on yHsp90 SUMOylation we over-expressed Myc-epitope-tagged Smt3 under galactose inducible promoter (GAL1) in yeast expressing either wild-type yHsp90 or yHsp90-K178R. After inducing SMT3-MYC expression by growing the cells on galactose media, we could detect Smt3-modified wild-type yHsp90 N-domain, but the N-domain of yHsp90-K178R remained unmodified (Figure 1C).
To determine whether the corresponding lysine residue (K191) in human Hsp90α (hHsp90α) (see Figure 1A) is also subject to SUMOylation, we introduced a PreScission site between the N-domain and charged linker of FLAG-tagged wild-type hHsp90α and hHsp90α -K191R. After verifying equivalent expression of both proteins in transiently transfected HEK293 cells (Figure 1D), we confirmed SUMOylation of lysine 191 in hHsp90α N-domain using anti-FLAG and anti-SUMO-1 antibodies (Figure 1D). Mammalian cells express three SUMO proteins: SUMO-1, SUMO-2 and SUMO-3. Re-probing the blot in Figure 1D with anti-SUMO-2/3 antibody did not yield a detectable signal, suggesting that the hHsp90α N-domain is modified only by SUMO-1 (data not shown). Unlike in yeast, lysine 191 also is subject to ubiquitination and acetylation (Figure S1G, H).
Hsp90 N-domain SUMOylation recruits the co-chaperone Aha1
PTMs can facilitate or inhibit interaction of co-chaperones with Hsp90 in cells (Mollapour and Neckers, 2012; Xu et al., 2012). Therefore, we examined the impact of N-domain SUMOylation on Hsp90 binding to selected co-chaperones. Hsp90 proteins from yeast cells expressing either hexahistidine epitope-tagged wild-type yHsp90 or the non-SUMOylatable K178R mutant were isolated with Ni-NTA agarose and associating co-chaperones were examined by Western blot. Interaction of yAha1 with the K178R mutant was abrogated (Figure 2A), as was interaction of non-SUMOylatable hHsp90α -K191R with human Aha1 (hAha1) (Figure 2B). In contrast, non-SUMOylatable yeast and human Hsp90 proteins readily associated with Cdc37p50, Sti1HOP and Sba1p23 (Figure S2A, S2B). Next, we examined whether overexpression of yeast SMT3-MYC or human SUMO-1-HA could ameliorate the observed reduction in Aha1 interaction. While overexpression of SMT3-MYC increased SUMOylation of wild-type yHsp90 (Figure 2C) and enhanced its interaction with yAha1 (Figure 2D, S2A), it did not restore yAha1 interaction with yHsp90-K178R (Figure 2D). We observed comparable results when SUMO-1-HA was co-expressed with either wild-type hHsp90α or hHsp90α -K191R in COS7 cells (Figure 2E, F, S2B).
Figure 2. Aha1 binds to N-domain SUMOylated Hsp90.
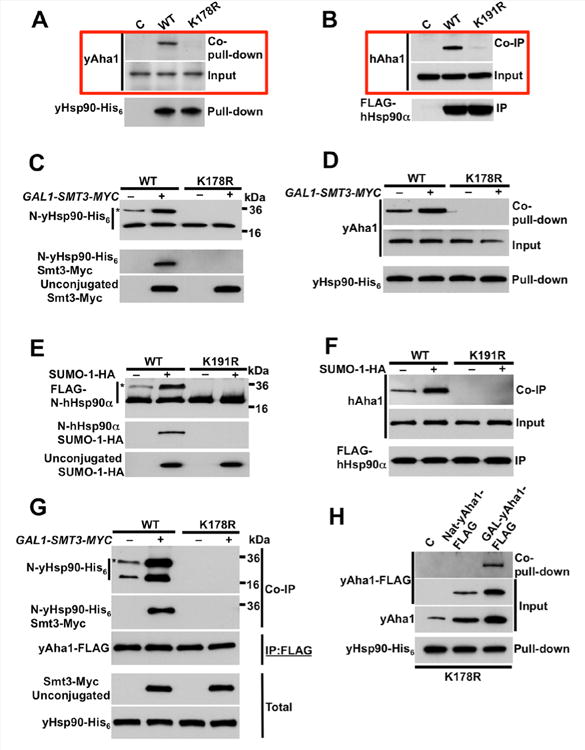
A) Protein extracts from control (C), wild-type yHsp90 (WT), and non-SUMOylatable yHsp90-K178R (K178R) expressing cells were precipitated using Ni-NTA agarose and yAha1 interaction was detected by immunoblotting.
B) COS7 cells were transfected with empty plasmid (C), FLAG-hHsp90α (WT), or non-SUMOylatable hHsp90α-K191R constructs. hAha1 interaction with these Hsp90 proteins was examined by immunoprecipitation (IP) and immunoblotting.
C) Yeast cells expressing wild-type yHsp90 (WT) or yHsp90-K178R (K178R), and carrying GAL1-SMT3-MYC plasmid, were grown on raffinose (–) or galactose (+). N-domain modification (*) by Smt3-Myc was assessed following precipitation with Ni-NTA agarose, PreScission protease digestion, and immunoblotting.
D) yAha1 interaction with wild-type yHsp90 (WT) or yHsp90-K178R (K178R) from cells over-expressing GAL1-SMT3-MYC and grown on raffinose (–) or galactose (+) was assessed following Ni-NTA agarose pulldown and immunoblotting.
E) COS7 cells transiently transfected with FLAG-hHsp90α (WT) or non-SUMOylatable hHsp90 α-K191R were co-transfected with SUMO-1-HA or empty plasmid. N-domain hHsp90α modification (*) by SUMO-1-HA was assessed following IP, PreScission protease digestion and immunoblotting.
F) hAha1 interaction with wild-type or non-SUMOylatable Hsp90 treated as in (E) was examined following IP and immunoblotting.
G) Yeast cells expressing yHsp90 (WT) or yHsp90-K178R (K178R) and co-expressing yAha1-FLAG and GAL1-SMT3-MYC plasmid were grown on raffinose (–) or galactose (+). yAha1-FLAG was immunoprecipitated with anti-FLAG-agarose and yAha1 interaction with SUMOylated (*) and non-SUMOylated yHsp90 was assessed by immunoblotting.
H) Non-SUMOylatable yHsp90-K178R-expressing cells were transformed with empty plasmid (C), or multicopy plasmid expressing yAha1-FLAG under native promoter (Nat) or GAL1 promoter (GAL). yHsp90-His6 was isolated by Ni-NTA agarose pulldown and yAha1-FLAG interaction was examined by immunoblotting. See also Figure S2.
Based on these data, we asked whether Aha1 preferentially interacts with N-domain SUMOylated yHsp90. Yeast Aha1-FLAG was expressed under its native promoter and was immunoprecipitated from cells expressing either wild-type yHsp90 or yHsp90-K178R. After PreScission protease cleavage, Western blotting for co-immunoprecipitated yHsp90 N-domains revealed a doublet of equal intensity corresponding to SUMOylated and non-SUMOylated N-domains (Figure 2G). Overexpression of SMT3-MYC enhanced the interaction of yAha1 with yHsp90 and proportionately enhanced the intensity of the yHsp90 N-domain doublet (Figure 2G). These data, together with the lack of detectable yAha1 binding to yHsp90-K178R, suggest a model in which yAha1 is recruited to asymmetrically SUMOylated yHsp90. Although it is not possible to detect yAha1 interaction with yHsp90-K178R under steady-state conditions (or even when yAha1 is over-expressed using its native promoter), association with yHsp90-K178R is detectable upon strong over-expression of yAha1 (using a GAL1 promoter on a yeast episomal plasmid, see Supplemental Information) (Figure 2H). This is consistent with binding data obtained using bacterially expressed (thus lacking posttranslational modifications) and purified yAha1 and yHsp90 proteins. In vitro, the affinities of both wild-type yHsp90 and yHsp90-K178R proteins for yAha1 were similar (wild-type yHsp90 Kd = 0.42 ± 0.08 µM; yHsp90-K178R Kd = 1.1 ± 0.1 µM); Figure S2C, D, respectively). These data are in agreement with previously published values (Xu et al., 2012). They confirm that lysine 178 mutation does not structurally compromise yHsp90 and they are consistent with the ability of yHsp90-K178R to support yeast viability (Figure S1A–D).
Detectable Aha1 binding to Hsp90 in cells requires SUMOylation of one protomer
Our data suggest that, in cells, yAha1 may interact with yHsp90 in which only one of the protomers is SUMOylated on lysine 178 (Figure 2G). To explore this possibility further, we co-expressed two copies of yHsp90 (with a PreScission cleavage site as in Figure 1A) in yeast, with one copy carrying a hexahistidine (His6) tag and the other copy carrying a GLU/GLU (GLU) epitope tag. The cells also overexpressed SMT3-MYC. First, we verified equal expression of both epitope-tagged yHsp90 proteins (Figure 3A, top two panels). Next, we isolated yHsp90-His6 using Ni-NTA agarose, followed by elution with imidazole and immunoprecipitation with antibody recognizing the GLU/GLU epitope tag (Figure 3A, middle panel). Immobilized yHsp90-His6/yHsp90-GLU (WTHis6/WTGLU) heterodimers were then treated with PreScission protease in order to isolate the yHsp90 N-domains (Figure 3A, cartoon of flow scheme). Similar experiments were conducted with yeast expressing combinations of wild-type and K178R yHsp90, or yHsp90-K178R only (e.g., WTHis6/K178RGLU; WTGLU/K178RHis6; K178RHis6/K178RGLU). After protease cleavage, Western blotting with anti-His6 or anti-GLU/GLU antibodies identified a doublet of equal intensity for WTHis6/WTGLU samples (Figure 3A, bottom two panels, first lane). In the WTHis6/K178RGLU and K178RHis6/WTGLU samples, the tag antibody recognizing the wild-type protomer also identified a doublet of equal intensity, while the tag antibody recognizing the mutated protomer identified only a single band whose molecular weight is consistent with lack of SUMOylation (Figure 3A, bottom two panels, lanes 2 & 3). Finally, when yHsp90 was comprised of two mutated protomers, antibodies recognizing either tag identified a single unmodified species (Figure 3A, bottom two panels, lane 4).
Figure 3. Aha1 binds to asymmetrically SUMOylated Hsp90.
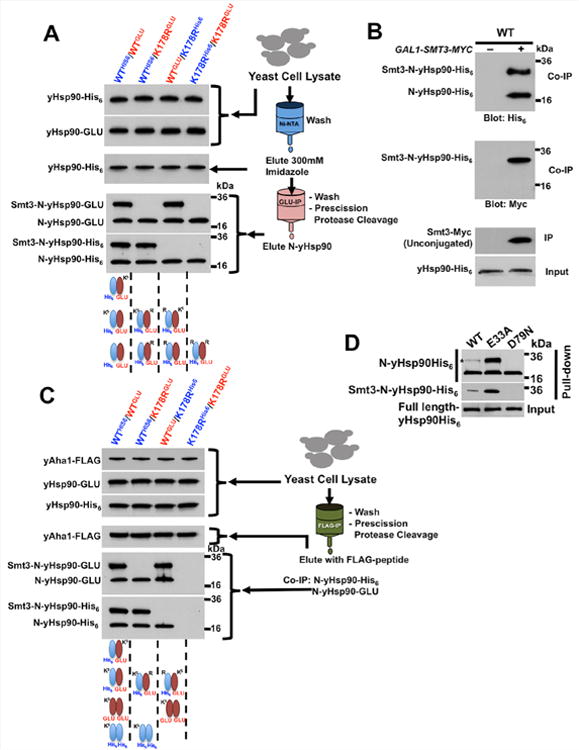
A) Lysate from yeast cells over-expressing GAL1-SMT3-MYC and co-expressing wild-type yHsp90 (WT) or yHsp90-K178R (K178R) with either a His6 or a GLU epitope tag (as indicated) were used to first precipitate His6-yHsp90 with Ni-NTA agarose. Eluates were subsequently used to immunoprecipitate GLU epitope-tagged yHsp90. After PreScission protease digestion, N-domain SUMOylation was assessed by immunoblotting.
B) Smt3-Myc was immunoprecipitated from wild-type yeast cells (WT) grown on either raffinose (–) or galactose (+). Immunoprecipitated Smt3-Myc and Smt3-Myc-modified proteins were treated with PreScission protease and N-domain SUMOylation of Smt3-Myc co-precipitated Hsp90 proteins was assessed by immunoblotting.
C) Yeast cells in (A) co-expressing yAha1-FLAG under native promoter were used to immunoprecipitate yAha1 with anti-FLAG agarose. N-domains from co-precipitated Hsp90 were then isolated by PreScission protease digestion and were analyzed by immunoblotting.
D) Wild-type yHsp90-His6 (WT), yHsp90-E33A-His6 (E33A) or yHsp90-D79N-His6 (D79N) constructs were expressed in yeast cells containing endogenous yHsp90. After Ni-NTA agarose pulldown and PreScission protease digestion, N-domain SUMOylation was assessed by immunoblotting.
These data suggest that a non-SUMOylated yHsp90 protomer readily dimerizes with either another non-SUMOylated protomer or with a SUMOylated protomer (Figure 3A, schematic diagram). To explore the possibility that two SUMOylated protomers might dimerize, we immunoprecipitated Smt3-Myc from yeast lysates expressing wild-type yHsp90His6. The immobilized yHsp90 proteins were treated with PreScission protease and isolated N-domains were visualized by Western blot. If two SUMOylated protomers dimerized, we would expect the abundance of the slower migrating (SUMOylated) N-domain to be greater than that of the faster migrating (non-SUMOylated) species. However, we always observed a doublet of nearly equal intensity upon probing with anti-His6 antibody (Figure 3B, upper panel). Probing the same blot with anti-Myc antibody confirmed that the slower migrating band reflected SUMOylated yHsp90 (Figure 3B, middle panel).
These findings prompted us to re-examine yAha1 binding to SUMOylated yHsp90. We immunoprecipitated FLAG-yAha1 from yeast cells over-expressing SMT3-MYC followed by PreScission protease treatment to isolate N-domains of the co-immunoprecipitated His6- or GLU-tagged yHsp90 proteins. We verified that yAha1 does not bind to yHsp90 comprised of two non-SUMOylated protomers (Figure 3C, lane 4), and we confirmed that the co-chaperone binds with equal efficiency to a yHsp90 molecule comprised of either one wild-type and one K178R protomer (Figure 3C, lanes 2 and 3) or of two wild-type protomers (Figure 3C, lane 1). These data provide further evidence that SUMOylation of one Hsp90 protomer is necessary and sufficient for optimal yAha1 recruitment in yeast cells.
To determine whether ATP-binding affects Hsp90 N-domain SUMOylation, we examined two well-characterized yHsp90 mutants: E33A, which efficiently binds but poorly hydrolyzes ATP (the equivalent hHsp90 mutant displays increased Aha1 association in cells (Xu et al., 2012)), and D79N, which does not bind ATP (Obermann et al., 1998; Panaretou et al., 1998). We expressed both hexahistidine-tagged mutants as well as wild-type yHsp90 in yeast cells and we examined their steady-state N-domain SUMOylation after affinity pull-down and PreScission cleavage. N-domain SUMOylation of yHsp90-E33A was markedly greater than that of wild-type yHsp90. In contrast, N-domain SUMOylation of yHsp90-D79N was not detectable (Figure 3D). These data suggest that ATP binding to Hsp90 is an important prerequisite for N-domain SUMOylation, consistent with the requirements for optimal Aha1 association. This model predicts that N-domain de-SUMOylation occurs prior to initiation of a new chaperone cycle.
Loss of Hsp90 N-domain SUMOylation minimally affects several Hsp90-dependent clients
Protein SUMOylation impacts numerous biological pathways, including nuclear-cytoplasmic transport, cell cycle control, and stress responses (Johnson, 2004; Ulrich, 2009). However, we found that neither α-factor-mediated G1 arrest nor heat shock caused an increase in steady-state yHsp90 N-domain SUMOylation (Figure S3A), and yeast expressing only yHsp90-K178R mounted a normal transcriptional response to heat shock (Figure 4A). Likewise, yeast cells expressing yHsp90-K178R displayed no growth or cell cycle defects (Figure S3B, C, D), nor did this mutation affect nuclear/cytoplasmic distribution of yHsp90-GFP (data not shown).
Figure 4. Lack of yHsp90 N-domain SUMOylation has little effect on most Hsp90 clients.
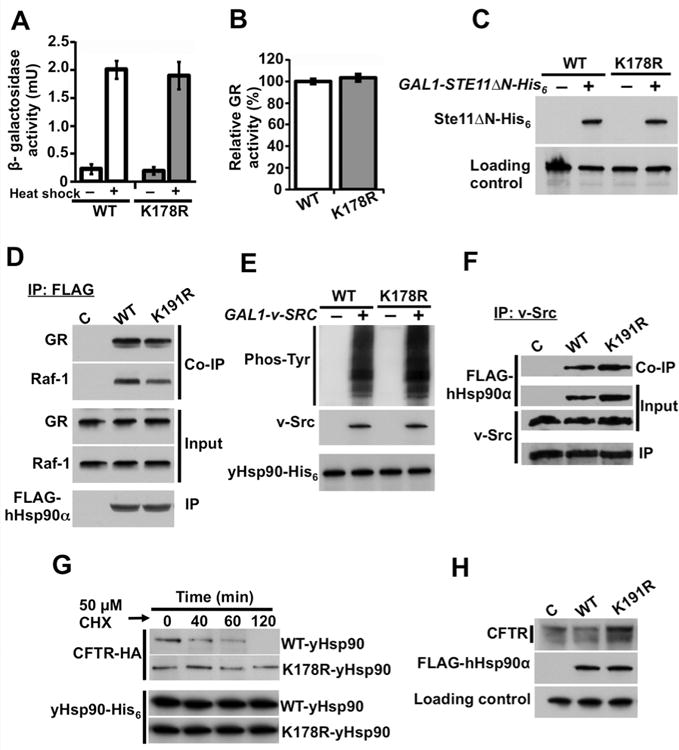
A) Wild-type (WT) or yHsp90-K178R (K178R) expressing yeast strains were heat shocked (40 min at 39°C). Heat shock response was measured in three independent experiments. All data represent mean ± S.D.
B) GR-lacZ activity was assessed in the same yeast strains as above. Data are expressed as a percentage of the activity observed in wild-type cells, and are depicted as the mean ± S.D. from values obtained in three independent experiments.
C) Wild-type (WT) and yHsp90-K178R (K178R) yeast strains co-expressing Ste11ΔN-His6 under GAL1 promoter were grown on raffinose (–) or galactose (+) media, and Ste11ΔN-His6 protein expression was examined by immunoblotting. a-tubulin was used as loading control.
D) COS7 cells were transfected with indicated FLAG-Hsp90a constructs. Hsp90 was immunoprecipitated with anti-FLAG agarose; co-precipitating (endogenous) GR and Raf-1 proteins were detected by immunoblotting.
E) Wild-type (WT) and yHsp90-K178R (K178R) yeast containing v-SRC under GAL1 promoter were grown on raffinose (–) or galactose (+) media. v-Src and total phosphotyrosine were detected by immunoblotting.
F) NIH3T3 cells stably expressing v-Src protein were transfected with empty plasmid (C), FLAG-hHsp90α (WT), or FLAG-hHsp90α-K191R (K191R) constructs. v-Src proteinwas immunoprecipitated and associated FLAG-Hsp90 proteins were detected by immunoblotting.
G) CFTR-HA was expressed in wild-type (WT) or yHsp90-K178R yeast; cells were treatedwith cycloheximide (CHX, 50 µM) and harvested at the indicated time points. CFTR wasanalyzed by immunoblotting.
H) HEK293 cells were transfected with wild-type CFTR and co-transfected with either wild-type FLAG-hHsp90α (WT) or FLAG-hHsp90α-K191R (K191R) constructs. After 24 h, CFTR and FLAG-hHsp90α were detected by immunoblotting. α-tubulin was used as loading control. See also Figure S3.
When we queried the ability of yHsp90-K178R or hHsp90α-K191R to interact with and chaperone several Hsp90 clients previously reported to require Aha1/Hsp90 association (Lotz et al., 2003; Panaretou et al., 2002), we found that this mutation failed to affect Hsp90-dependent chaperoning of the glucocorticoid receptor (GR), or the active form of Ste11 kinase (Ste11ΔN), the ortholog of mammalian Raf-1 (Flom et al., 2008; Louvion et al., 1998) (Figure 4B, C). Similarly, hHsp90α-K191R associated with GR and Raf-1 equivalently to wild-type hHsp90α (Figure 4D). Non-SUMOylatable yeast and human Hsp90 proteins also interacted with and chaperoned v-Src with equal efficiency to wild-type Hsp90 (Figure 4E, F). These data suggest that the attenuated interaction of yHsp90-K178R with yAha1 does not phenocopy yaha1 deletion (Lotz et al., 2003; Panaretou et al., 2002). This was further supported by comparing the growth of yeast expressing either yHsp90-K178R or wild-type yHsp90 but lacking yAha1 expression on glucose (YPDA) and respiratory (YPEG) growth media at elevated temperature. As previously reported (Lotz et al., 2003; Panaretou et al., 1998), yaha1Δ cells displayed a temperature sensitivity phenotype on YPDA and YPEG media (Figure S3E). In contrast, yeast expressing yHsp90-K178R displayed no growth defects on either media, consistent with the possibility that Aha1 may have Hsp90-independent activity (Sun et al., 2012).
Aha1 stimulates Hsp90-mediated ATP hydrolysis and thus reduces the dwell time of clients in the chaperone complex, which has deleterious consequences for hard-to-fold clients that require prolonged association with Hsp90 (such as the cystic fibrosis transmembrane conductance regulator protein, CFTR) (Loo et al., 1998; Youker et al., 2004). We examined the ability of non-SUMOylatable yHsp90 to promote CFTR stabilization and maturation. Yeast cells were co-transfected with HA-tagged CFTR (under the control of a constitutive promoter) and either wild-type yHsp90 or yHsp90-K178R, and we determined the rate of CFTR protein decay in the presence of cycloheximide. We found CFTR stability to be significantly enhanced in yeast expressing yHsp90-K178R compared to wild-type cells (Figure 4G). A similar result was obtained in HEK293 cells transiently transfected with wild-type CFTR and with either wild-type hHsp90α or hHsp90α-K191R. The greater abundance of slower migrating CFTR in cells over-expressing hHsp90α-K191R (Figure 4H) is consistent with previous observations that reduced Aha1 expression improves Hsp90-dependent CFTR maturation, presumably by allowing prolonged interaction of this-difficult-to-fold client with Hsp90 (Wang et al., 2006). Thus, the enhanced CFTR chaperone efficiency of N-domain lysine-mutated Hsp90 in both yeast and mammalian cell backgrounds likely reflects a physiological consequence of compromised Hsp90/Aha1 interaction.
Increased SUMOylation of the Hsp90 N-domain inhibits chaperone function
Over-expression of Smt3 and SUMO-1 increased N-domain SUMOylation of yHsp90 and hHsp90α, respectively (Figure 2C, E). In contrast to lack of SUMOylation, increased SUMOylation of the Hsp90 N-domain more generally affected Hsp90 chaperone activity. Over-expression of SMT3-MYC reduced CFTR stability in wild-type yeast but not in yeast expressing yHsp90-K178R (Figure 5A and S4). Over-expression of SUMO-1 in HEK293 cells co-transfected with CFTR and FLAG-tagged hHsp90α proteins had a similar effect (Figure 5B). Likewise, we found that SMT3-MYC overexpression in wild-type yeast resulted in reduced v-Src expression and v-Src mediated protein tyrosine phosphorylation (Figure 5C), as well as in a 70% reduction in GR activity (Figure 5D), but this was not seen in yeast expressing yHsp90-K178R.
Figure 5. Increased Hsp90 N-domain SUMOylation affects chaperone function.
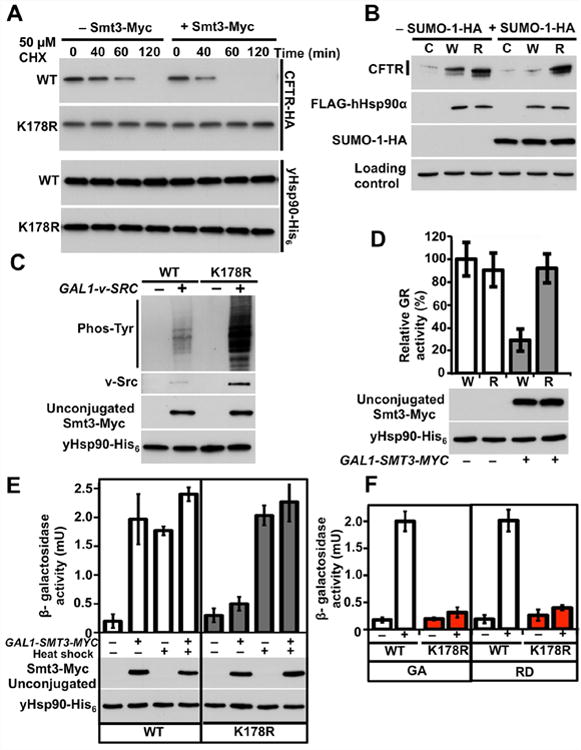
A) Wild-type (WT) or yHsp90-K178 (K178R) yeast cells co-expressing CFTR-HA and GAL1-SMT3-MYC were grown on raffinose (–) or galactose (+) media, treated with cycloheximide (CHX, 50 μM), and harvested at the indicated time points. CFTR was visualized by immunoblotting.
B) HEK293 cells were transfected with wild-type CFTR and co-transfected with empty plasmid (C), wild-type FLAG-hHsp90α (W), or FLAG-hHsp90α-K191R (R), co-transfection with (+) or without (–) SUMO-1-HA. After 24 h, CFTR, FLAG-hHsp90α and SUMO-1-HA were detected by immunobloting. α-tubulin was used as loading control.
C) Wild-type (WT) and yHsp90-K178R (K178R) expressing yeast containing GAL1-v-SRC and GAL1-SMT3-MYC were grown on raffinose (–) or galactose (+) media. v-Src, total phosphotyrosine, and Smt3-Myc were detected by immunoblotting.
D) GR-lacZ activity was assessed in wild-type (W) and yHsp90-K178R (R) yeast cells that also expressed GAL1-SMT3-MYC. Cells were grown on raffinose (–) or galactose (+) media. Data are expressed as a percentage of the activity observed in wild-type cells, and are depicted as the mean ± S.D. of three independent experiments.
E) Heat Shock Element (HSE)-lacZ reporter was used to monitor the heat shock response in yeast expressing wild-type yHsp90 (WT) or yHsp90-K178R (K178R); yeast also expressed GAL-SMT3-MYC. Cells were grown on raffinose (–) or galactose (+) media and heat shocked (40 min at 39°C). All data represent mean ± S.D. of three independent experiments.
F) Wild-type yHsp90 (WT) and yHsp90-K178R (K178R) cells were treated with GA (“+”, 50 µM, 1 h) or RD (“+”, 30 µM, 1 h). Heat shock response was measured from three independent experiments. All data represent mean ± S.D. See also Figure S4.
Finally, we studied the effect of SUMOylation on Hsf1 transcriptional activity. It is generally accepted that Hsp90 interaction suppresses Hsf1 activity (Zou et al., 1998), and compromised Hsp90 chaperone function, due to mutation or pharmacologic inhibition, leads to induction of Hsf1 activity in yeast even in the absence of heat shock (Hjorth-Sorensen et al., 2001). SMT3-MYC overexpression caused a heat shock response in wild-type yeast but not in yeast expressing yHsp90-K178R (Figure 5E). Similarly, we observed that the Hsp90 inhibitors geldanamycin (GA) and radicicol (RD) failed to induce a heat shock response in yHsp90-K178R-expressing yeast (Figure 5F). Importantly, mutation of lysine 178 did not affect the heat shock response induced by elevated temperature (Figure 4A). These data indicate that N-domain SUMOylation differentially affects the heat shock transcriptional response to elevated temperature and Hsp90 inhibitors, and they suggest possible similarities between deregulated Hsp90 SUMOylation and pharmacologic inhibition of the chaperone.
N-domain SUMOylation sensitizes cells to Hsp90 inhibition
To explore the possible impact of Hsp90 SUMOylation on drug sensitivity, we first examined effects of GA and RD on steady-state SUMOylation of lysine 178. Unlike the response to α-factor and heat shock (Figure S3A), N-domain SUMOylation increased following exposure to GA or RD (Figure 6A). Since these drugs did not affect endogenous Smt3 expression, we reasoned that they might preferentially interact with, and conformationally trap, N-domain SUMOylated Hsp90. We tested this possibility by overexpressing SMT3-MYC in wild-type yHsp90-expressing and yHsp90-K178R-expressing cells, and using biotinylated Hsp90 inhibitor ganetespib and streptavidin beads to affinity purify yHsp90 proteins from cell lysates. We found that SMT3 overexpression clearly enhanced ganetespib recognition of wild-type yHsp90, but had little effect on recognition of yHsp90-K178R (Figure 6B).
Figure 6. Increased Hsp90 N-domain SUMOylation sensitizes cells to Hsp90 inhibitors.
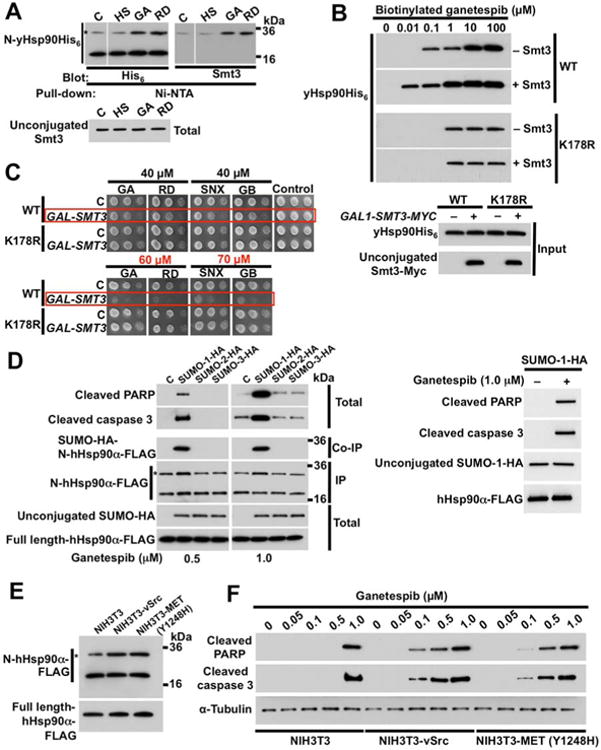
A) Effect of heat shock (HS) (40 min at 39°C), GA (50 µM, 1 h), or RD (30 µM, 1 h) on SUMOylation of yHsp90-K178 (*) was monitored by immunoblotting with anti-hexahistidine antibody. Unconjugated Smt3 was detected by anti-Smt3 antibody.
B) Yeast cells expressing wild-type yHsp90 (WT) or yHsp90-K178R (K178R), and harboring GAL1-SMT3-MYC, were grown on galactose. Hsp90 was isolated from yeast lysates by incubating with indicated amounts of biotinylated ganetespib followed by Streptavidin agarose beads, and detected by immunoblotting with anti-His6 antibody.
C) Wild-type or K178R-yHsp90-expressing yeast harboring GAL1-SMT3-MYC were grown on galactose media for 4 h. A 1:10 dilution series of 107cells/ml were spotted on YPDA agar containing indicated concentrations of the Hsp90 inhibitors GA, RD, SNX (SNX2112), or GB (ganetespib). Plates were incubated at 28°C for 4 days.
D) NIH3T3 cells were transfected with empty plasmid (C), SUMO-1-HA, SUMO-2-HA, or SUMO-3-HA. After 24 h, cells were treated with either 0.5 µM or 1.0 µM ganetespib for an additional 6 h. SUMO-1, 2 and 3, hHsp90α N-domain SUMOylation, cleaved PARP and cleaved caspase-3 were detected by immunoblotting.
E) Parental NIH3T3 cells and NIH3T3 stably expressing v-Src or constitutively active MET (Y1248H) mutant were transfected with wild-type FLAG-hHsp90α (containing a PreScission cleavage site). After 24 h, N-domain SUMOylation (*) of FLAG-hHsp90α was assessed by immunoblotting with anti-FLAG following PreScission protease digestion.
F) Parental NIH3T3 cells and NIH3T3 cells stably expressing v-Src or constitutively activated MET (Y1248H) mutant were treated with indicated amounts of ganetespib for 6h. Cleaved PARP and cleaved caspase-3 were detected by immunoblotting. α-tubulin was used as a loading control. See also Figure S5.
Based on these observations, we asked whether increased N-domain SUMOylation sensitized yeast cells to Hsp90 inhibition. Although we saw no difference in drug sensitivity between cells expressing wild-type yHsp90 or yHsp90-K178R under normal conditions (Figure 6C, ‘C’ rows), after SMT3 overexpression cells harboring wild-type yHsp90 displayed greater sensitivity to a panel of four N-domain inhibitors compared to yeast expressing yHsp90-K178R (Figure 6C, ‘GAL-SMT3’ rows). It is noteworthy that, at higher drug concentrations, lack of SUMOylation does not confer resistance to Hsp90 inhibitors (Figure S5A).
We carried out similar experiments in mammalian cells. Using immortalized NIH3T3 mouse fibroblasts transiently transfected with plasmids encoding HA-tagged SUMO-1, SUMO-2, or SUMO-3 proteins, we confirmed that only SUMO-1 overexpression sensitized the cells to ganetespib, as evidenced by induction of the pro-apoptotic markers cleaved caspase and cleaved PARP (Figure 6D). Likewise, Hsp90 N-domain SUMOylation was detectable only in SUMO-1 transfected cells. It is noteworthy that overexpression of SUMO-1 in the absence of Hsp90 inhibitor did not induce apoptotic markers in NIH3T3 cells (Figure 6D, right panel).
Cancer cells generally display greater sensitivity towards Hsp90 inhibitors than do their non-tumorigenic counterparts (Bisht et al., 2003). Therefore, we asked whether Hsp90 was SUMOylated to a greater degree in NIH3T3 cells stably transformed with either v-Src or mutated (constitutively active) MET (MET-Y1428H) compared to parental NIH3T3 cells, and whether steady-state Hsp90 N-domain SUMOylation intensity correlated with sensitivity to ganetespib. Indeed, both transformed cell lines displayed increased Hsp90 N-domain SUMOylation compared to non-transformed cells (Figure 6E), and oncogenic transformation rendered the cells more sensitive to ganetespib, as evidenced by a greater abundance of apoptotic markers (Figure 6F).
These findings suggest that Hsp90 inhibitors and Aha1 compete for binding to SUMOylated Hsp90. To test this possibility, we overexpressed SMT3-MYC in yeast cells expressing wild-type yHsp90 and then treated the cells with GA. Interaction of yHsp90 and yAha1 was examined by Hsp90 pull-down and Western blot. Although SMT3-MYC overexpression increased yHsp90 interaction with yAha1, association of the co-chaperone with Hsp90 was not detectable in cells treated with GA prior to lysis (Figure S5B). Finally, we showed earlier (Figure 3D) that yHsp90-E33A (which binds ATP but does not hydrolyze it) is markedly more SUMOylated than is wild-type yHsp90, and the equivalent mutation in hHsp90α-E47A promotes increased steady-state interaction with Aha1 in cells (Xu et al., 2012). However, GA-affinity bead pulldown of hHsp90α-E47A was less efficient than that of the wild-type protein and Aha1 was not present in either pulldown (Figure S5C), consistent with the hypothesis that Hsp90 inhibitors and Aha1 cannot interact concurrently with N-domain SUMOylated Hsp90.
Discussion
In this study, we identified SUMOylation of a conserved lysine residue in the N-domains of both yeast (lysine 178) and human (lysine 191) Hsp90. Mutation of this residue abrogated N-domain SUMOylation and prevented detectable intracellular interaction with the ATPase-stimulating co-chaperone Aha1. Based on AHA1 deletion studies in yeast, Aha1/Hsp90 interaction is considered to be important for chaperoning of Hsp90 clients, including those examined in this study (Holmes et al., 2008; Lotz et al., 2003; Panaretou et al., 2002). However, lack of detectable Aha1 interaction with yHsp90-K178R engendered observable effects only with the hard-to-fold Hsp90 client CFTR, consistent with published reports demonstrating improved Hsp90-dependent CFTR folding in cells with reduced Aha1 expression (Koulov et al., 2010; Wang et al., 2006). Our data suggest that reduced Aha1/Hsp90 interaction per se does not phenocopy yaha1Δ, implying that Aha1, like the co-chaperones Sba1p23 and Cdc37p50 (Echtenkamp et al., 2011; Kimura et al., 1997), may have additional activities in cells that are independent of (but perhaps function in parallel with) its role in modulating Hsp90 activity (Sun et al., 2012).
In yeast, Aha1 is expressed at a 10-fold lower concentration than is Hsp90 (Ghaemmaghami et al., 2003). Thus, although bacterially purified yAha1 binds to wild-type yHsp90 and yHsp90-K178R proteins with comparable affinity, it is reasonable to assume that in cells certain Hsp90 PTMs modulate Aha1 recruitment to active Hsp90 chaperone complexes. Indeed, phosphorylation and acetylation of specific sites on Hsp90 have been shown previously to affect interaction with Aha1 (Mollapour et al., 2010; Mollapour et al., 2011a; Retzlaff et al., 2010; Scroggins et al., 2007). Our current data suggest that N-domain SUMOylation is a PTM able to initiate recruitment of Aha1 to Hsp90 in both yeast and human cells. One molecule of Aha1 is sufficient to maximally stimulate Hsp90 ATPase activity (Panaretou et al., 2002; Soroka et al., 2012; Xu et al., 2012). The Aha1 N-domain is thought to interact first with the middle domain of one Hsp90 protomer, after which the Aha1 C-domain associates with the N-domain of the opposing protomer (or with a hydrophobic groove formed by transient, ATP-dependent dimerization of both Hsp90 N-domains) (Koulov et al., 2010; Meyer et al., 2003; Retzlaff et al., 2010). Although as yet we have not uncovered the mechanism by which Hsp90 N-domain SUMOylation facilitates Aha1 interaction, the asymmetry of this PTM (and its conservation in yeast and human cells) is consistent with a role in promoting the asymmetric interaction of Aha1 and Hsp90 in cells.
Increased SUMOylation of the Hsp90 N-domain, although predicted to accelerate the rate of chaperone cycling, functionally compromised several Hsp90 clients, including v-Src, GR, and CFTR. These data are consistent with previous studies examining the impact of other Hsp90 modifications that stimulate ATP hydrolysis. Thus, the yHsp90 mutant T22I demonstrates both enhanced ATPase activity and ATP-dependent N-domain dimerization (Prodromou et al., 2000), while possessing reduced ability to chaperone both GR and v-Src (Nathan and Lindquist, 1995). Similarly, although covalent N-terminal fusion of constitutively dimerized coiled-coil domains to yHsp90 enforces N-domain proximity and significantly increases basal ATPase activity and yAha1 binding affinity, coiled-coil yHsp90 chaperones GR and v-Src less efficiently than does wild-type yHsp90 (Pullen and Bolon, 2011).
We explored the temporal relationship between N-domain SUMOylation, ATP binding and ATP hydrolysis with the help of two yHsp90 point mutants. yHsp90-E33A efficiently binds but does not hydrolyze ATP and this mutant was markedly more SUMOylated at steady-state compared to wild-type yHsp90. In contrast, yHsp90-D79N does not bind ATP and was not detectably SUMOylated. The simplest model that is consistent with these data suggests that ATP binding is a prerequisite for N-domain SUMOylation. Although we cannot absolutely rule out the possibility that SUMOylation may also stabilize an ATP-bound Hsp90 conformation, the fact that SUMOylation of yHsp90-D79N was not observed reduces the likelihood of this hypothesis.
Surprisingly, ATP-competitive Hsp90 inhibitors increased steady-state N-domain SUMOylation and readily recognized wild-type, but not non-SUMOylatable, yHsp90 in protein lysates of yeast overexpressing SMT3. Since association of Aha1 and these inhibitors with Hsp90 is mutually exclusive (Figure S5B), our data suggest that inhibitors preferentially bind to SUMOylated Hsp90 by displacing previously bound ATP prior to lid closure and Aha1-facilitated Hsp90 commitment to N-domain dimerization and ATP hydrolysis, by which point conformational changes have rendered the ATP pocket inaccessible to these drugs. Drug binding to Hsp90 prevents N-domain dimerization and Aha1 recruitment, trapping SUMOylated Hsp90 in an ‘open’ conformation unable to proceed further in the chaperone cycle (Figure 7). This model is consistent with a previous report that Aha1 knockdown sensitizes cancer cells to Hsp90 inhibitors (Holmes et al., 2008), and with our finding that the hydrolysis-incompetent yHsp90-E33A, which is trapped in an ATP-bound, lid-closed conformation, is SUMOylated to a greater degree than is wild-type yHsp90 yet is poorly recognized by Hsp90 inhibitor (Figure S5C). Perhaps unexpectedly, this model predicts that Hsp90 inhibitors do not bind to all N-domain undimerized (open) Hsp90 conformations with equal affinity, but can distinguish ATP-bound Hsp90 (prior to lid closure) from the ATP-unbound chaperone.
Figure 7. Hsp90 SUMOylation facilitates Aha1 association but also enhances binding of ATP-competitive inhibitors.

The Hsp90 chaperone cycle is initiated by ATP binding to N-domain(s) (1). This promotes asymmetric N-domain SUMOylation (2). Hsp90 inhibitors preferentially bind to SUMOylated Hsp90 by replacing ATP in the nucleotide binding pocket (3). In the absence of N-domain inhibitors, asymmetric N-domain SUMOylation facilitates Aha1 association, accelerating the ATPase-driven Hsp90 chaperone cycle (4). SUMOylation is dynamic and Hsp90 de-SUMOylation is predicted to occur prior to initiation of a new chaperone cycle (5).
Finally, since Hsp90 inhibitors have much higher binding constants compared to ATP, this model predicts that drug trapping of SUMOylated Hsp90 preferentially removes a fraction of the actively cycling chaperone pool. Our observation that the intensity of Hsp90 N-domain SUMOylation increases with cellular transformation is consistent with this proposal and provides mechanistic insight that helps clarify an observation made some years ago by Kamal and colleagues (Kamal et al., 2003), experimentally supported by subsequent studies but still not satisfactorily explained, that Hsp90 isolated from tumor cells simultaneously demonstrates both greater ATPase activity and greater affinity for Hsp90 inhibitors compared to Hsp90 isolated from non-transformed cells. The tumor cell selectivity of these drugs thus may reflect a greater dependence of cancer cells (relative to non-transformed cells) on the constitutive engagement of a significant fraction of Hsp90 in the chaperone cycle.
Experimental Procedures
Yeast Strains, Plasmids and Growth Media
Yeast strain pp30 (MAT a, trp1-289, leu2-3,112, his3-200, ura3-52, ade2-101, lys2-801, hsc82KANMX4, hsp82KANMX4) was used in this study (Panaretou et al., 1998). Plasmids were constructed as described previously (Mollapour et al., 2011b). Detailed procedures, a list of primers (Table S1), and media conditions for both yeast and mammalian cells are provided in the Supplemental Data.
Protein Extraction, Immunoprecipitation, PreScission Protease Cleavage, and Immunoblotting
Total protein extracts were prepared and analyzed by Western blotting, as described previously (Mollapour et al., 2010), with the exception that N-ethylmaleimide (NEM, 20 mM) was included in the protein lysis buffer. Detailed methods for protein precipitations, cleavage by PreScission protease, and detection by Western blotting for both yeast and mammalian systems are presented in Supplemental Data.
Assays for Hsp90 Client Activity
v-Src induction and activation were analyzed as described previously (Mollapour et al., 2011b). Expressed v-Src protein was detected with EC10 mouse antibody (Millipore) and v-Src activity with 4G10 mouse anti-phosphotyrosine antibody (Millipore). GR assay was performed as described previously (Garabedian and Yamamoto, 1992), as was measurement of HSE-LacZ expression (Hjorth-Sorensen et al., 2001). Ste11ΔN induction was analyzed as described previously (Flom et al., 2008; Louvion et al., 1998). Ste11ΔN plasmid is a gift of Jill Johnson. Additional details are found in Supplemental Data.
Isothermal Titration Calorimetry (ITC) and Kd Determinations
ITC and Kd determinations were performed as described previously (Prodromou et al., 2000,Prodromou et al., 2000). Additional information can be found in Supplemental Data.
Supplementary Material
Highlights.
Yeast and human Hsp90 are subject to asymmetric N-domain SUMOylation
Hsp90 N-domain SUMOylation facilitates both Aha1 recruitment and drug binding
Cellular transformation is accompanied by increased Hsp90 N-domain SUMOylation
Increased Hsp90 N-domain SUMOylation sensitizes cells to Hsp90 inhibitors
Acknowledgments
We thank Dr. J. Johnson (University of Idaho) for STE11ΔN plasmid, Dr. J. L. Brodsky (University of Pittsburgh) for CFTR-HA plasmid, Dr. T. Haystead (Duke University) for the Hsp90 inhibitor SNX-2112, Dr. W. Ying (Synta Pharmaceuticals) for the Hsp90 inhibitor ganetespib, and Dr K. Robzyk (Memorial Sloan-Kettering Cancer Center) for SUMO-1, -2, and -3 HA-tagged plasmids. We are also grateful to Dr. A. Zuehlke (NCI) for helpful discussion. This work was supported with funds from the Intramural Research Program of the National Cancer Institute (L.N.) and from the SUNY Upstate Medical University (M.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht KS, Bradbury CM, Mattson D, Kaushal A, Sowers A, Markovina S, Ortiz KL, Sieck LK, Isaacs JS, Brechbiel MW, et al. Geldanamycin and 17-allylamino-17-demethoxygeldanamycin potentiate the in vitro and in vivo radiation response of cervical tumor cells via the heat shock protein 90-mediated intracellular signaling and cytotoxicity. Cancer Res. 2003;63:8984–8995. [PubMed] [Google Scholar]
- Cox MB, Johnson JL. The Role of p23, Hop, Immunophilins, and Other Co-chaperones in Regulating Hsp90 Function. Methods Mol Biol. 2011;787:45–66. doi: 10.1007/978-1-61779-295-3_4. [DOI] [PubMed] [Google Scholar]
- Cunningham CN, Krukenberg KA, Agard DA. Intra- and intermonomer interactions are required to synergistically facilitate ATP hydrolysis in Hsp90. J Biol Chem. 2008;283:21170–21178. doi: 10.1074/jbc.M800046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echtenkamp FJ, Zelin E, Oxelmark E, Woo JI, Andrews BJ, Garabedian M, Freeman BC. Global functional map of the p23 molecular chaperone reveals an extensive cellular network. Mol Cell. 2011;43:229–241. doi: 10.1016/j.molcel.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flom GA, Lemieszek M, Fortunato EA, Johnson JL. Farnesylation of Ydj1 is required for in vivo interaction with Hsp90 client proteins. Mol Biol Cell. 2008;19:5249–5258. doi: 10.1091/mbc.E08-04-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabedian MJ, Yamamoto KR. Genetic dissection of the signaling domain of a mammalian steroid receptor in yeast. Mol Biol Cell. 1992;3:1245–1257. doi: 10.1091/mbc.3.11.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hessling M, Richter K, Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2009;16:287–293. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol. 2012;13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjorth-Sorensen B, Hoffmann ER, Lissin NM, Sewell AK, Jakobsen BK. Activation of heat shock transcription factor in yeast is not influenced by the levels of expression of heat shock proteins. Mol Microbiol. 2001;39:914–923. doi: 10.1046/j.1365-2958.2001.02279.x. [DOI] [PubMed] [Google Scholar]
- Holmes JL, Sharp SY, Hobbs S, Workman P. Silencing of HSP90 cochaperone AHA1 expression decreases client protein activation and increases cellular sensitivity to the HSP90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2008;68:1188–1197. doi: 10.1158/0008-5472.CAN-07-3268. [DOI] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, Burrows FJ. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Rutherford SL, Miyata Y, Yahara I, Freeman BC, Yue L, Morimoto RI, Lindquist S. Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes Dev. 1997;11:1775–1785. doi: 10.1101/gad.11.14.1775. [DOI] [PubMed] [Google Scholar]
- Koulov AV, Lapointe P, Lu B, Razvi A, Coppinger J, Dong MQ, Matteson J, Laister R, Arrowsmith C, Yates JR, 3rd, et al. Biological and Structural Basis for Aha1 Regulation of Hsp90 ATPase Activity in Maintaining Proteostasis in the Human Disease Cystic Fibrosis. Mol Biol Cell. 2010;21:871–884. doi: 10.1091/mbc.E09-12-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Richter K, Reinstein J, Buchner J. Integration of the accelerator Aha1 in the Hsp90 co-chaperone cycle. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2502. [DOI] [PubMed] [Google Scholar]
- Li J, Soroka J, Buchner J. The Hsp90 chaperone machinery: Conformational dynamics and regulation by co-chaperones. Biochim Biophys Acta. 2012;1823:624–635. doi: 10.1016/j.bbamcr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Loo MA, Jensen TJ, Cui L, Hou Y, Chang XB, Riordan JR. Perturbation of Hsp90 interaction with nascent CFTR prevents its maturation and accelerates its degradation by the proteasome. EMBO J. 1998;17:6879–6887. doi: 10.1093/emboj/17.23.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz GP, Lin H, Harst A, Obermann WM. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J Biol Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- Louvion JF, Abbas-Terki T, Picard D. Hsp90 is required for pheromone signaling in yeast. Mol Biol Cell. 1998;9:3071–3083. doi: 10.1091/mbc.9.11.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP. Gymnastics of molecular chaperones. Mol Cell. 2010;39:321–331. doi: 10.1016/j.molcel.2010.07.012. [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Mollapour M, Neckers L. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim Biophys Acta. 2012;1823:648–655. doi: 10.1016/j.bbamcr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Tsutsumi S, Donnelly AC, Beebe K, Tokita MJ, Lee MJ, Lee S, Morra G, Bourboulia D, Scroggins BT, et al. Swe1Wee1-dependent tyrosine phosphorylation of Hsp90 regulates distinct facets of chaperone function. Mol Cell. 2010;37:333–343. doi: 10.1016/j.molcel.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Tsutsumi S, Kim YS, Trepel J, Neckers L. Casein kinase 2 phosphorylation of Hsp90 threonine 22 modulates chaperone function and drug sensitivity. Oncotarget. 2011a doi: 10.18632/oncotarget.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Tsutsumi S, Truman AW, Xu W, Vaughan CK, Beebe K, Konstantinova A, Vourganti S, Panaretou B, Piper PW, et al. Threonine 22 phosphorylation attenuates hsp90 interaction with cochaperones and affects its chaperone activity. Mol Cell. 2011b;41:672–681. doi: 10.1016/j.molcel.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Ruckova E, Halada P, Coates PJ, Hrstka R, Lane DP, Vojtesek B. C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene. 2012 doi: 10.1038/onc.2012.314. [DOI] [PubMed] [Google Scholar]
- Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B, Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, Singh S, Millson SH, Clarke PA, Naaby-Hansen S, Stein R, et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem. 2004;279:41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pountney DL, Raftery MJ, Chegini F, Blumbergs PC, Gai WP. NSF, Unc-18-1, dynamin-1 and HSP90 are inclusion body components in neuronal intranuclear inclusion disease identified by anti-SUMO-1-immunocapture. Acta Neuropathol. 2008;116:603–614. doi: 10.1007/s00401-008-0437-4. [DOI] [PubMed] [Google Scholar]
- Prodromou C. The ‘active life’ of Hsp90 complexes. Biochim Biophys Acta. 2012;1823:614–623. doi: 10.1016/j.bbamcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Panaretou B, Chohan S, Siligardi G, O'Brien R, Ladbury JE, Roe SM, Piper PW, Pearl LH. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen L, Bolon DN. Enforced N-domain proximity stimulates Hsp90 ATPase activity and is compatible with function in vivo. J Biol Chem. 2011;286:11091–11098. doi: 10.1074/jbc.M111.223131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzlaff M, Hagn F, Mitschke L, Hessling M, Gugel F, Kessler H, Richter K, Buchner J. Asymmetric activation of the hsp90 dimer by its cochaperone aha1. Mol Cell. 2010;37:344–354. doi: 10.1016/j.molcel.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, et al. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Siligardi G, Hu B, Panaretou B, Piper PW, Pearl LH, Prodromou C. Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J Biol Chem. 2004;279:51989–51998. doi: 10.1074/jbc.M410562200. [DOI] [PubMed] [Google Scholar]
- Soroka J, Wandinger SK, Mausbacher N, Schreiber T, Richter K, Daub H, Buchner J. Conformational switching of the molecular chaperone Hsp90 via regulated phosphorylation. Mol Cell. 2012;45:517–528. doi: 10.1016/j.molcel.2011.12.031. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- Sun L, Prince T, Manjarrez JR, Scroggins BT, Matts RL. Characterization of the interaction of Aha1 with components of the Hsp90 chaperone machine and client proteins. Biochim Biophys Acta. 2012;1823:1092–1101. doi: 10.1016/j.bbamcr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. The SUMO system: an overview. Methods Mol Biol. 2009;497:3–16. doi: 10.1007/978-1-59745-566-4_1. [DOI] [PubMed] [Google Scholar]
- Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MM, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton-Diaz A, Khan S, Bourboulia D, Trepel JB, Neckers L, Mollapour M. Contributions of co-chaperones and post-translational modifications towards Hsp90 drug sensitivity. Future medicinal chemistry. 2013;5:1059–1071. doi: 10.4155/fmc.13.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, Gurkan C, Kellner W, Matteson J, Plutner H, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- Xu W, Mollapour M, Prodromou C, Wang S, Scroggins BT, Palchick Z, Beebe K, Siderius M, Lee MJ, Couvillon A, et al. Dynamic Tyrosine Phosphorylation Modulates Cycling of the HSP90-P50(CDC37)-AHA1 Chaperone Machine. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol Biol Cell. 2004;15:4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ryan JJ, Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem. 2004;279:32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


