Abstract
Many bacteria glide smoothly on surfaces, but with no discernable propulsive organelles on their surface. Recent experiments with Myxococcus xanthus and Flavobacterium johnsoniae show that both distantly related bacterial species glide utilizing proteins that move in helical tracks, albeit with significantly different motility mechanisms. Both species utilize proton motive force for movement. However, the motors that power gliding in M. xanthus have been identified, while the F. johnsoniae motors remain to be discovered.
“Physics can tell us what cannot happen, and it can tell us what could happen. But only experiments tell us what does happen.”—A. Katchalsky
Introduction
The most common propulsive machine for bacteria is the flagellum, propelling cells by rotary motors powered by trans membrane ionmotive potential [1]. Since bacteria are very small, they live in a ‘low Reynolds number’ world where inertial forces are negligible [2]. Thus they cannot swim using ‘reciprocal’ motions; that is, a motions that when filmed look the same when the film is run backwards. To solve this problem, bacteria like Escherichia coli swim using stiff rotating helical flagella [2] [3]. Most spirochetes have helically-shaped internal flagella that are located in the periplasmic space between the inner membrane and the cell wall. In these bacteria, the rotation of the periplasmic flagella allows them to swim by generating backward-moving waves [4].
Flagellar motility is effective for bacteria that swim in water, and even allows some to 'swarm' over very moist surfaces, but what about microorganisms that move on surfaces that are covered with only a thin aqueous film? For these bacteria, two radically different modes of locomotion have evolved: ‘twitching motility’, which involves intermittent, ‘jerky’ cell movements, and ‘gliding motility,’ where the cell motion is smooth. Of course, these terms are strictly descriptive, and give no clue as to the underlying physical mechanisms. Twitching motility is driven by the extension, adhesion, and retraction of fibrous cellular protrusions called Type IV pili [5–7]. In Myxococcus xanthus this is called Social, or S-motility, since the extended pili stick not only to the substrate but also to other cells, and so are important for coordinated group movements of the bacteria.
Gliding motility, by contrast, is not well understood. In the myxobacteria it is called Adventurous, or A-motility because it can drive the movement of isolated bacteria, even when pili are not present. These A-motile cells glide slowly at about one body length (~ 5 µm) per minute, and reverse direction periodically every 8–14 minutes, suggesting that there is some internal ‘clock’ regulating reversals [8]. A-motility appears to require the secretion of slime; in myxobacteria these include a viscous polysaccharide gel [9]. An early model for myxobacterial gliding suggested that the cell was driven by the hydration and extrusion of slime from protein ‘nozzles’ that cluster mostly at the cell poles [9]. However, recent experimental data suggest that the motion of internal proteins rather than the extrusion of polysaccharides drives cell movement. [10–13].
In this review we describe recent progress in understanding the different ways that bacteria employ helical tracks to glide over surfaces.
Helical tracks and protein motors
Using high-resolution fluorescence microscopy of moving M. xanthus cells, Nan et al. [12] demonstrated that AgmU, a critical A-motility protein labeled with a fluorescent tag (mCherry) decorated a helical ribbon that spanned the length of the cells in a closed loop (see Figure 1). Astoundingly, these helices appeared to rotate within the cell cytoplasm as they moved forward, and when the cells reversed their gliding direction the helices rotated in the opposite direction. These results recalled previously published images that showed M. Xanthus cell bodies helically twisted, as though the cell membrane had been shrink-wrapped around a helical cytoskeletal structure [14, 15]. Based on these findings, a model for gliding motility was proposed in which helical waves sweep over the cell surface as the helical rotor inside the cell rotates. Could this be the elusive A-motor, ‘pushing’ on the substrate to move the cell forward? Such a mechanism would be similar to that used by snails [16]. The surface waves in snails, however, arise from the neuro-musculature of the snail’s mantle, while the waves in gliding bacteria appear to arise from the rotation of an internal helix.
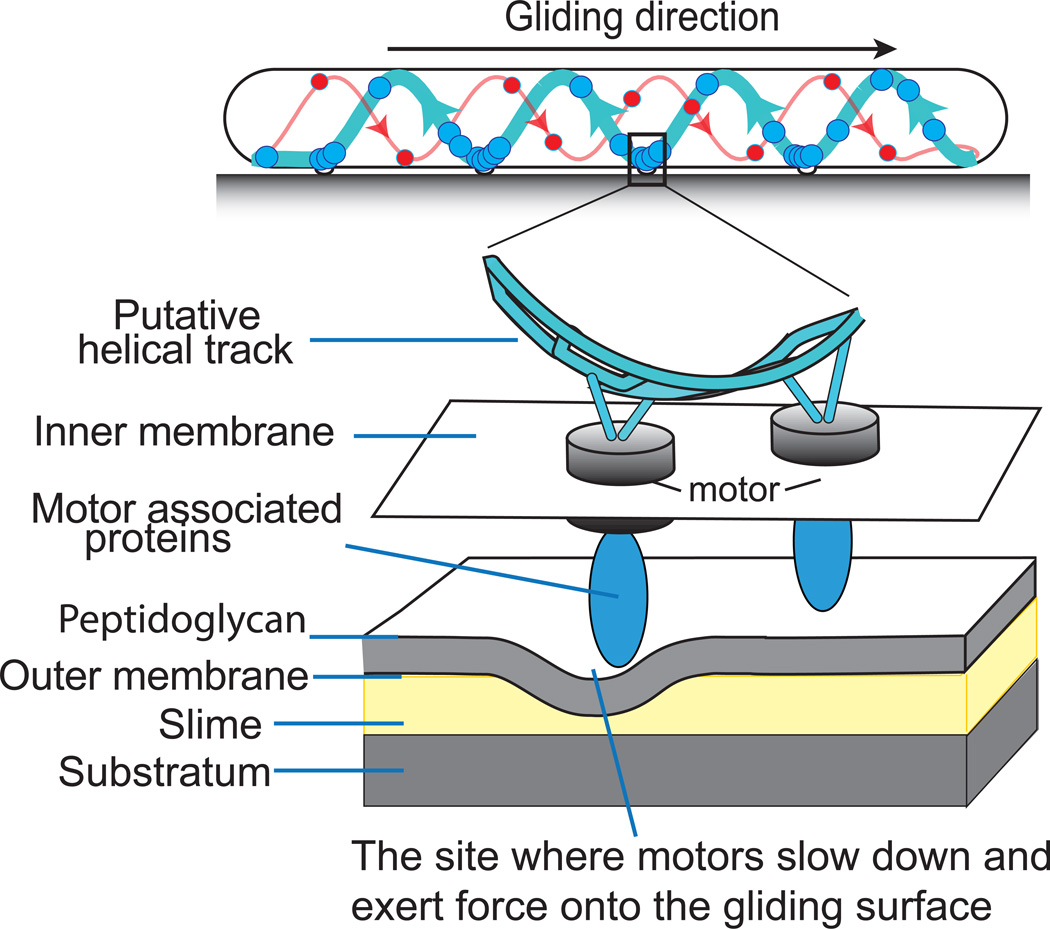
Figure 1.
The helical rotor mechanism in M. xanthus. A schematic of the endless helical protein track on which the motors (large circles) walk. The thick lines show the leftward motion of the motors that drive the rightward direction of gliding. The thin red lines show the fewer number of rightward moving motors on the opposite strand. The model shows that the motors slow down in the ventral ‘traffic jam’ where they encounter the high drag region. The higher resolution inset shows the motors walking on the cytoplasmic track carrying large ‘cargo’ of motor associated proteins that deform the cell wall. The deformation pushes on the external slime providing the thrust that drives the cell gliding.
The mechanism of myxobacterial gliding has other similarities to snail movement. The rearward-propagating waves in a crawling snail do not push directly on the substrate, but on the slime that the snail secretes. This slime sticks more tightly to the substrate than to the snail’s foot, and so the propulsive force of the mantle waves that push on the slime is transmitted to the substrate. The same seems to be true for M. Xanthus since the slime that the bacteria secrete appears necessary for cell locomotion and is present in all the gliding myxobacteria. Moreover the slime does indeed adhere more strongly to the surface than to the cell [17], allowing the helical waves to transmit the propulsive force to the substrate via the slime. But what makes the internal helix rotate to generate the surface waves?
A careful examination of single motors labeled with photo-activatablem Cherry revealed that they move around a helical track. Motor movement is powered by the proton gradient across the cytoplasmic membrane, also referred to as the proton motive force (PMF) [11]. The motors are comprised of the proteins AglR and either AglQ or AglS. AglR is related to the well-studied bacterial flagellar motor protein MotA, and AglQ and AglS are similar to MotA's partner, MotB. MotA and MotB form a complex that harvests the PMF and drives rotation of the flagellar filament [18, 19]. The MotAB proteins of the bacterial flagellar motor are anchored to the peptidogly can cell wall, and function as ‘stators’ since they ‘walk in place’ to drive rotation of the flagellum [20]. The myxobacterial motors, however, are not anchored to the cell wall, but are free to move within the cytoplasmic membrane, walking along an apparently helical cytoskeletal track [11]. Thus the gliding motion of M. Xanthus operates differently from E. coli flagellar swimming.
Here Newton’s laws of motion come into play. As the motors walk on the helix, their propulsive force exerts an equal and opposite reaction force on the helix. This reaction force can be resolved into two components: one parallel to the cell axis and one tangential to the cell radius. The longitudinal force pushes the helix in the direction of gliding (towards the ‘leading’ pole), while the tangential force tends to rotate the helix about the cell axis. By Newton’s Third Law, the tangential force that rotates the helix creates a reaction force that tends to rotate the cell in the opposite direction, but because the myxobacterial slime is so viscous, there is but a small counter-rotation of the cell. Thus the motors moving on the helical track rotate the helix. In principle, this could generate a wave of helical deformations on the cell surface to propel the cell, similar to a snail. It turns out, however, that things are not so simple, and before the mechanical picture is complete we need to address how the rotary force on the helix is transmitted to the cell surface.
Motor complexes and traffic-jams
The images of helically-shaped cells mentioned above were prepared by dehydrating the cells, in effect shrink-wrapping the cell body about the helical cytoskeleton. In a live myxobacterium, the helical shape is not prominent. Therefore, another mechanism must transmit the force exerted by the motors on the rotating helical track to the external surface. Currently, the nature of the helical track is not known although it may be MreB or MreB-associated proteins, since both gliding motility and the movement of motors are sensitive to the MreB inhibitor A22. MreB is an actin homolog involved in many functions such as the determination of cell shape, cell wall biosynthesis and chromosome segregation [21–24]. In E. coli, MreB forms short ‘patchy’ filaments; the previously reported helical conformation of MreBmay bean artifact created by a fluorescent tag fused at its N-terminus [25]. However, the M. Xanthus MreB filaments appear helical when stained with antibody-conjugated fluorescent dyes, where the fluorescent tag inducing the helical artifact was not used [26]. This implies that the conformation and function of M. xanthus MreB differs from its homolog in E. coli.
Importantly, Nan et al. observed that moving motors in M. Xanthus always slowed down dramatically when passing through the ventral turn of the track closest to the slime-covered surface [11] and formed accumulations. This is because the surface distortion created by the motor-cargo complex (see Box 1) has to push its way through the ventral slime layer where the drag is highest because the slime sticks more to the substrate than to the cell. [1]
Box 1: The peptidogly can problem.
Myxobacteria are unusually flexible since single cells are often observed to bend sharply and turn. Despite their flexibility, cells require a peptidogly can matrix under their outer membrane that enables them to maintain their cylindrical shape despite the interior osmotic pressure that would otherwise inflate the cell into a sphere, or worse yet, burst the cell. Thus the integrity of the peptidogly can is essential. But how then are the motile motors to transmit their propulsive force through the peptidogly can layer without destroying it?
A plausible solution has been provided by recent experiments by Nan, et al. (unpublished data). These experiments show that MglA, a GTPase in the Ras family that is localized largely at the cell’s leading pole, is responsible for assembling AglR motors along with associated ‘cargo’ proteins (usually large proteins such as AgmU) onto the helical tracks. These assembled motor complexes can be large enough to deform the peptidogly can without tearing it. We propose that the rearward motion of the cargo proteins towards the trailing pole will create surface deformations that will slow down when they enter the high drag environment of the ventral traffic jams. It is this viscous coupling of the surface deformation to the substratum via the slime that enables the motors to drive cells forward.
Thus motors entering into the ventral region are retarded by ‘traffic-jams’ from which they eventually escape to continue on their way towards the trailing cell pole. The traffic jams were visualized by fluorescently labeled motility proteins and they appear as nearly stationary clusters to an observer looking at the cell surface. These ventral accumulations were seen earlier by Mignot et al. [27] who viewed them as ‘focal adhesions’, analogous to similar adhesion plaques in crawling eukaryotic cells. A motor complex temporarily trapped in one of these traffic-jams slows down because it is exerting propulsive thrust on the slime, which transmits the force to the substrate. Since there are about 4–8 turns in the helix track of M. xanthus cells (about one turn per µm of cell length), there are the same number of ventral traffic jams. Accordingly, the thrust developed by the motors in these aggregates act as 4–8 force generators to propel the cell forward. In time-lapse movies it appears as though the motors ‘crawl through’ each traffic jam. When a jam reaches the trailing pole it dissipates as the motors escape, and a new jam is generated at the leading cell pole as motors encounter the increased drag from ventral slime (Figure 1) [12, 27]. In moving cells, force generation by traffic jams is invisible since each traffic jam remains almost stationary with respect to the substratum. Sun et al. visualized the propulsion of the traffic jam sites by attaching polystyrene beads to the surface of cells. In their report, instead of using moving cells, the authors immobilized cells onto a coated glass surface. In this case, beads were propelled from the leading to the lagging cell pole; significantly the beads co-localized with the traffic jam sites [19].
But if the helical rotor is a closed loop, what about the cargo proteins that pass through the trailing pole and commence moving towards the leading pole? Won’t they generate an equal and opposite force and cancel out the propulsive thrust? This would indeed be the case if equal numbers of cargo proteins of similar size moved in opposite directions. But if there were a consistent difference in the number and/or size of cargo proteins migrating in the opposite direction, then the thrust would not cancel and the cell would move alternately forwards and backwards depending on the relative amounts of cargo moving in each direction at any time, as is observed. Nan, et al. tagged and followed motors cycling on the two strands and found that there was a persistent difference in the number of motors moving towards the trailing and leading poles, with the majority consistently moving opposite to the direction of gliding (Nan et al, unpublished data). If this asymmetric distribution of motors persists, then that would explain the periodic directional reversals. What remains to be explained is the behavior of mutants that glide without reversing. This may occur by the repeated assembly of motor complexes at only one cell pole with recycling occurring by free diffusion. This could also explain the ability of wild type cells to move in a directional manner in response to a stimulus.
Although the chemistry is still unclear, it appears that the reversals are slave to a biochemical oscillator at the cell poles that couple the motors and their cargo to the helical looped track. The nature of the mechanical coupling remains to be investigated. Cells may gain some control of their movements by breaking free of this rigid oscillation. A model that might explain how this could occur exploits the similarity between the chemotactic signal transduction (Che) system that controls the direction of rotation of the bacterial flagellar motor and the Frz signaling system in myxobacteria that controls cell reversal frequency [28].
Do other gliding bacteria move in the same way?
Many bacteria belonging to different phyla exhibit gliding motility, and for most of these the mechanisms of movement have not been studied [29]. Some of these bacteria may utilize the helical rotor mechanism described for M. xanthus. However others clearly do not, suggesting that bacterial 'gliding motility' is not a single phenomenon. For example, the wall-less bacterium Mycoplasma mobile glides by a mechanism that is powered by ATP hydrolysis rather than by PMF [30]. Mycoplasma gliding involves large cell surface proteins that appear to function as 'legs' [31, 32]. ATP driven movements of these legs are thought to allow Mycoplasma cells to walk centipede-like over surfaces. Filamentous cyanobacteria also glide, although the mechanism is not well-understood. Recent evidence has implicated type IV pili [33] and polysaccharide extrusion [9, 34], either or both of which could propel these cells.
Flavobacterium johnsoniae (previously called 'Cytophagajohnsonae') is a gliding bacterium for which extensive molecular analyses of motility have been performed. F. johnsoniae, a member of the bacterial phylum Bacteroidetes, is not closely related to M. xanthus. Many members of the phylum Bacteroidetes exhibit rapid gliding motility and F. johnsoniae is a model organism for studies of this form of movement. There are many parallels between M. xanthus and F. johnsoniae gliding suggesting that they might use a common mechanism, but a deeper look also reveals differences. At first glance F. johnsoniae appears to be a supercharged version of M. xanthus that glides about 25 times faster. Like M. xanthus, cell movement is powered by PMF [35, 36]. Cells are long and somewhat flexible, and they move in the direction of either pole, occasionally reversing direction. Interestingly, motility of both bacteria seems to utilize a helical track [12, 35].
Decades of research on F. johnsoniae and on M. xanthus have revealed many proteins required for gliding in each organism [37–39]. Surprisingly, however, there are few similarities between these components at the molecular level, suggesting that these systems may have evolved independently. For example, for F. johnsoniae we know a great deal about the cell surface and cell envelope components, but the motors that drive movement and links to cytoplasmic components are unclear [40–42]. As discussed above, however, the motors associated with motilityin M. xanthus have been identified, as have components in the cytoplasm that may interact with and control these motors. By contrast, the components on the cell surface of M. xanthus that transmit the force to the substratum are less well understood. Further study of both systems may thus reveal common elements.
Some motility behaviors of F. johnsoniae and related bacteria differ from those of M. xanthus, and are difficult to explain by the helical rotor model. First, cells floating in liquid or attached to and gliding on a surface, bind to and rapidly propel small particles such as latex microspheres [36, 43, 44]. When asymmetrical particles are examined they often appear to be rigidly attached to the cell surface and maintain the same orientation at their point of attachment as they travel the length of the cell, around the pole, and back toward the opposite pole. Second, cells gliding on a glass surface often flip end-over-end [35, 43]. These observations suggest that outer membrane adhesins are propelled along the cell surface, which may drive Flavobacterium gliding.
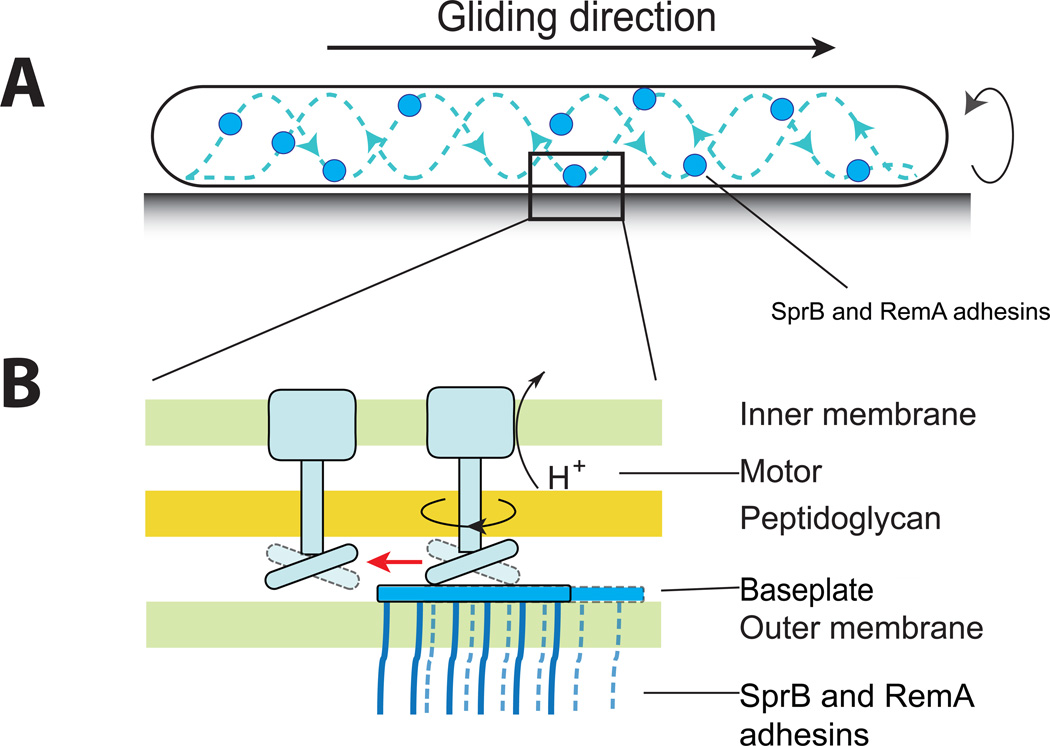
Genetic experiments identified a candidate adhesin: the huge 669 kDa protein, SprB [40]. SprB proteins decorated with fluorescently labeled antibodies are propelled rapidly (2 µm per second) along the cell surface on what appears to be a closed helical loop [35]. SprB proteins traverse the length of the cell, loop around the pole, and migrate toward the opposite pole within 5 to 10 seconds. As a cell moves forward, some SprB protein son the cell surface remain attached to the substratum, suggesting that the action of a motor pushing against these adhesins propels the cell. Dissipation of the PMF with chemical un couplers rapidly and reversibly inhibits cell gliding and SprB movements [35]. F. johnsoniae has other motility adhesins, such as the carbohydrate-binding lectin RemA, that are propelled along the cell surface [41]. Secreted polysaccharides that interact with RemA have also been implicated in gliding [41]. These polysaccharides may coat the substratum, and form a 'road' that interacts with specific 'tires', such as RemA. The ability to use different adhesins, and perhaps different polysaccharides, may explain how F. johnsoniae cells can crawl over diverse surfaces.
The motor involved in F. johnsoniae gliding is not known, but we expect it to span the cytoplasmic membrane and be anchored to a large structure such as the peptidogly can layer. Such a motor complex could have components that reach through the peptidogly can to propel the outer membrane adhesins. One model envisions large ‘base plates’ adjacent to the inner face of the outer membrane to which multiple adhesins are attached (Figure 2). The large repetitive protein SprD, which supports SprB function, might form such a structure. There may be multiple positions at which the motor complex could engage the base plate and propel it a short distance. The same motor could then repeat this process multiple times to move the base plate and attached adhesins over a longer distance, at which point it could be engaged by another motor complex.
Figure 2.
Speculative model for movement of F. johnsoniae cell-surface adhesins. A) The surface adhesins move along a looped helical track. B) A portion of the cell envelope is shown. The motors, anchored to the peptidogly can, harvest the proton gradient across the inner membrane. A portion of the motor complex extends through the peptidogly can layer and interacts with an outer membrane associated protein (baseplate) that carries the cell surface adhesins. Repeated movements of this portion of the motor propels the base plate and attached adhesins along the cell surface until they are engaged by the next motor.
This model leaves unaddressed how the motor is controlled to result in directed movement. In M. xanthus a chemotactic signal transduction system similar to those found in flagellated bacteria is thought to perform this function, but the core components of such systems (methyl-accepting chemotaxis proteins, CheA, CheW) are lacking in F. johnsonaie [38]. The closed helical loop track presents the same problem for F. johnsoniae that it does for M. xanthus. Since different adhesins move simultaneously toward the front and back of the cell on different regions of the helical track, how does the cell make any net progress? Are only some adhesins actively engaged in binding to the substratum, and if so, how is this controlled? Identification of the actual motor complex and its interactions with other proteins will help to reveal the mechanism of F. johnsoniae cell movement and directional control. It may also help to determine whether there are underlying similarities between the myxobacterial and flavobacterial solutions to movement over a surface.
In summary, bacterial gliding remains largely mysterious, although experiments are beginning to shed light on some mechanisms. Experiments on myxobacteria have provided sufficient evidence to propose a working model. The mechano chemical mechanism driving flavobacterial gliding, however, remains mysterious despite extensive knowledge of the proteins involved. Nevertheless, it is intriguing that both motility mechanisms are powered by transmembrane ionmotive potentials and involve the ubiquitous presence of helical structures.
Acknowledgements
GO and JC were supported by NIH Grant R01-GM104979. BN and DRZ were supported by NIH grant RO1-GM020509. MM was supported by NSF grant MCB1021721.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Purcell EM. The efficiency of propulsion by a rotating flagellum. Proc Natl Acad Sci U S A. 1997;94:11307–11311. doi: 10.1073/pnas.94.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lauga E. Life around the scallop theorem. Soft Matter. 2011;7 [Google Scholar]
- 4.Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb MA, Wolgemuth CW. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol. 2012;66:349–370. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maier B, Potter L, So M, Long CD, Seifert HS, Sheetz MP. Single pilus motor forces exceed 100 pN. Proc Natl Acad Sci U S A. 2002;99:16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 8.Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 9.Wolgemuth C, Hoiczyk E, Kaiser D, Oster G. How myxobacteria glide. Curr Biol. 2002;12:369–377. doi: 10.1016/s0960-9822(02)00716-9. [DOI] [PubMed] [Google Scholar]
- 10.Ehlers K, Oster G. On the mysterious propulsion of Synechococcus. PLoS One. 2012;7:e36081. doi: 10.1371/journal.pone.0036081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nan B, Bandaria JN, Moghtaderi A, Sun IH, Yildiz A, Zusman DR. Flagella stator homologs function as motors for myxobacterial gliding motility by moving in helical trajectories. Proc Natl Acad Sci U S A. 2013;110:E1508–E1513. doi: 10.1073/pnas.1219982110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nan B, Chen J, Neu JC, Berry RM, Oster G, Zusman DR. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci U S A. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nan B, Zusman DR. Uncovering the mystery of gliding motility in the myxobacteria. Annu Rev Genet. 2011;45:21–39. doi: 10.1146/annurev-genet-110410-132547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lunsdorf H, Schairer HU. Frozen motion of gliding bacteria outlines inherent features of the motility apparatus. Microbiology. 2001;147:939–947. doi: 10.1099/00221287-147-4-939. [DOI] [PubMed] [Google Scholar]
- 15.Pelling AE, Li Y, Shi W, Gimzewski JK. Nanoscale visualization and characterization of Myxococcus xanthus cells with atomic force microscopy. Proc Natl Acad Sci U S A. 2005;102:6484–6489. doi: 10.1073/pnas.0501207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan B, Balmforth NJ, Hosoi AE. Building a better snail: lubrication and adhesive locomotion. Phys Fluids. 2005;17:113101–113110. [Google Scholar]
- 17.Ducret A, Valignat MP, Mouhamar F, Mignot T, Theodoly O. Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc Natl Acad Sci U S A. 2012;109:10036–10041. doi: 10.1073/pnas.1120979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luciano J, Agrebi L, A LG, M W, Fiegna F, Ducret A, Brochier-Armanet C, Mignot T. Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet. 2011;7:e1002268. doi: 10.1371/journal.pgen.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci U S A. 2011;108:7559–7564. doi: 10.1073/pnas.1101101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kojima S, Blair DF. The bacterial flagellar motor: structure and function of a complex molecular machine. Int Rev Cytol. 2004;233:93–134. doi: 10.1016/S0074-7696(04)33003-2. [DOI] [PubMed] [Google Scholar]
- 21.Divakaruni AV, Baida C, White CL, Gober JW. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol Microbiol. 2007;66:174–188. doi: 10.1111/j.1365-2958.2007.05910.x. [DOI] [PubMed] [Google Scholar]
- 22.Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci U S A. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 25.Swulius MT, Jensen GJ. The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-Terminal yellow fluorescent protein tag. J Bacteriol. 2012;194:6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauriello EM, Mouhamar F, Nan B, Ducret A, Dai D, Zusman DR, Mignot T. Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. Embo J. 2010;29:315–326. doi: 10.1038/emboj.2009.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. Evidence that focal adhesion complexes power bacterial gliding motility. Science. 2007;315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckhert E, 1, Davis A, Oster G, Berleman J. Dual Biochemical Oscillators controls Cellular Reversals in Myxococcus xanthus. 2013 doi: 10.1016/j.bpj.2014.09.046. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride MJ. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu Rev Microbiol. 2001;55:49–75. doi: 10.1146/annurev.micro.55.1.49. [DOI] [PubMed] [Google Scholar]
- 30.Uenoyama A, Miyata M. Gliding ghosts of Mycoplasma mobile. Proc Natl Acad Sci U S A. 2005;102:12754–12758. doi: 10.1073/pnas.0506114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyata M. Unique centipede mechanism of Mycoplasma gliding. Annu Rev Microbiol. 2010;64:519–537. doi: 10.1146/annurev.micro.112408.134116. [DOI] [PubMed] [Google Scholar]
- 32.Chen J, Neu J, Miyata M, Oster G. A mechanochemical model for mycoplasma motility. Biophys. J. 2009;97:2930–2938. doi: 10.1016/j.bpj.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duggan PS, Gottardello P, Adams DG. Molecular analysis of genes in Nostoc punctiforme involved in pilus biogenesis and plant infection. J Bacteriol. 2007;189:4547–4551. doi: 10.1128/JB.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Risser DD, Meeks JC. Comparative transcriptomics with a motility-deficient mutant leads to identification of a novel polysaccharide secretion system in Nostoc punctiforme. Mol Microbiol. 2013;87:884–893. doi: 10.1111/mmi.12138. [DOI] [PubMed] [Google Scholar]
- 35.Nakane D, Sato K, Wada H, McBride MJ, Nakayama K. Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1219753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pate JL, Chang L-YE. Evidence that gliding motility in prokaryotic cells is driven by rotary assemblies in the cell envelopes. Curr. Microbiol. 1979;2:59–64. [Google Scholar]
- 37.Spormann AM. Gliding motility in bacteria: insights from studies of Myxococcus xanthus. Microbiol Mol Biol Rev. 1999;63:621–641. doi: 10.1128/mmbr.63.3.621-641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBride MJ, Xie G, Martens EC, Lapidus A, Henrissat B, Rhodes RG, Goltsman E, Wang W, Xu J, Hunnicutt DW, et al. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl Environ Microbiol. 2009;75:6864–6875. doi: 10.1128/AEM.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luciano J, Agrebi R, Le Gall AV, Wartel M, Fiegna F, Ducret A, Brochier-Armanet C, Mignot T. Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet. 2011;7:e1002268. doi: 10.1371/journal.pgen.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson SS, Bollampalli S, McBride MJ. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol. 2008;190:2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shrivastava A, Rhodes RG, Pochiraju S, Nakane D, McBride MJ. Flavobacterium johnsoniae RemA is a mobile cell surface lectin involved in gliding. J Bacteriol. 2012;194:3678–3688. doi: 10.1128/JB.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shrivastava A, Johnston JJ, van Baaren JM, McBride MJ. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell surface gliding motility adhesins SprB and RemA. J Bacteriol. 2013;195:3201–3212. doi: 10.1128/JB.00333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lapidus IR, Berg HC. Gliding motility of Cytophaga sp. strain U67. J Bacteriol. 1982;151:384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beatson PJ, Marshall KC. A proposed helical mechanism for gliding motility in three gliding bacteria (order Cytophagales) CanJMicrobiol. 1994;40:173–183. [Google Scholar]