Abstract
The broad host range plasmid RK2 replicates and regulates its copy number in a wide range of Gram-negative bacteria. The plasmid-encoded trans-acting replication protein TrfA and the origin of replication oriV are sufficient for controlled replication of the plasmid in all Gram-negative bacteria tested. The TrfA protein binds specifically to direct repeat sequences (iterons) at the origin of replication. A replication control model, designated handcuffing or coupling, has been proposed whereby the formation of coupled TrfA-oriV complexes between plasmid molecules results in hindrance of origin activity and, consequently, a shut-down of plasmid replication under conditions of higher than normal copy number. Therefore, according to this model, the coupling activity of an initiation protein is essential for copy number control and a copy-up initiation protein mutant should have reduced ability to form coupled complexes. To test this model for plasmid RK2, two previously characterized copy-up TrfA mutations, trfA-254D and trfA-267L, were combined and the resulting copy-up double mutant TFrfA protein TrfA-254D/267L was characterized. Despite initiating runaway (uncontrolled) replication in vivo, the copy-up double-mutant TrfA protein exhibited replication kinetics similar to the wild-type protein in vitro. Purified TrfA-254D, TrfA-267L, and TrfA-254D/267L proteins were then examined for binding to the iterons and for coupling activity using an in vitro ligase-catalyzed multimerization assay. It was found that both single and double TrfA mutant proteins exhibited substantially reduced (single mutants) or barely detectable (double mutant) levels of coupling activity while not being diminished in their capacity to bind to the origin of replication. These observations provide direct evidence in support of the coupling model of replication control.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Durland R. H., Helinski D. R. Replication of the broad-host-range plasmid RK2: direct measurement of intracellular concentrations of the essential TrfA replication proteins and their effect on plasmid copy number. J Bacteriol. 1990 Jul;172(7):3849–3858. doi: 10.1128/jb.172.7.3849-3858.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durland R. H., Helinski D. R. The sequence encoding the 43-kilodalton trfA protein is required for efficient replication or maintenance of minimal RK2 replicons in Pseudomonas aeruginosa. Plasmid. 1987 Sep;18(2):164–169. doi: 10.1016/0147-619x(87)90044-8. [DOI] [PubMed] [Google Scholar]
- Durland R. H., Toukdarian A., Fang F., Helinski D. R. Mutations in the trfA replication gene of the broad-host-range plasmid RK2 result in elevated plasmid copy numbers. J Bacteriol. 1990 Jul;172(7):3859–3867. doi: 10.1128/jb.172.7.3859-3867.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F. C., Helinski D. R. Broad-host-range properties of plasmid RK2: importance of overlapping genes encoding the plasmid replication initiation protein TrfA. J Bacteriol. 1991 Sep;173(18):5861–5868. doi: 10.1128/jb.173.18.5861-5868.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D. H., Meyer R. J., Helinski D. R. Suppression of Co1E1 replication properties by the Inc P-1 plasmid RK2 in hybrid plasmids constructed in vitro. J Mol Biol. 1979 Sep 25;133(3):295–318. doi: 10.1016/0022-2836(79)90395-4. [DOI] [PubMed] [Google Scholar]
- Filutowicz M., Uhlenhopp E., Helinski D. R. Binding of purified wild-type and mutant pi initiation proteins to a replication origin region of plasmid R6K. J Mol Biol. 1986 Jan 20;187(2):225–239. doi: 10.1016/0022-2836(86)90230-5. [DOI] [PubMed] [Google Scholar]
- Haugan K., Karunakaran P., Tøndervik A., Valla S. The host range of RK2 minimal replicon copy-up mutants is limited by species-specific differences in the maximum tolerable copy number. Plasmid. 1995 Jan;33(1):27–39. doi: 10.1006/plas.1995.1004. [DOI] [PubMed] [Google Scholar]
- Helsberg M., Ebbers J., Eichenlaub R. Mutations affecting replication and copy number control in plasmid mini-F both reside in the gene for the 29-kDa protein. Plasmid. 1985 Jul;14(1):53–63. doi: 10.1016/0147-619x(85)90032-0. [DOI] [PubMed] [Google Scholar]
- Ishiai M., Wada C., Kawasaki Y., Yura T. Replication initiator protein RepE of mini-F plasmid: functional differentiation between monomers (initiator) and dimers (autogenous repressor). Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3839–3843. doi: 10.1073/pnas.91.9.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
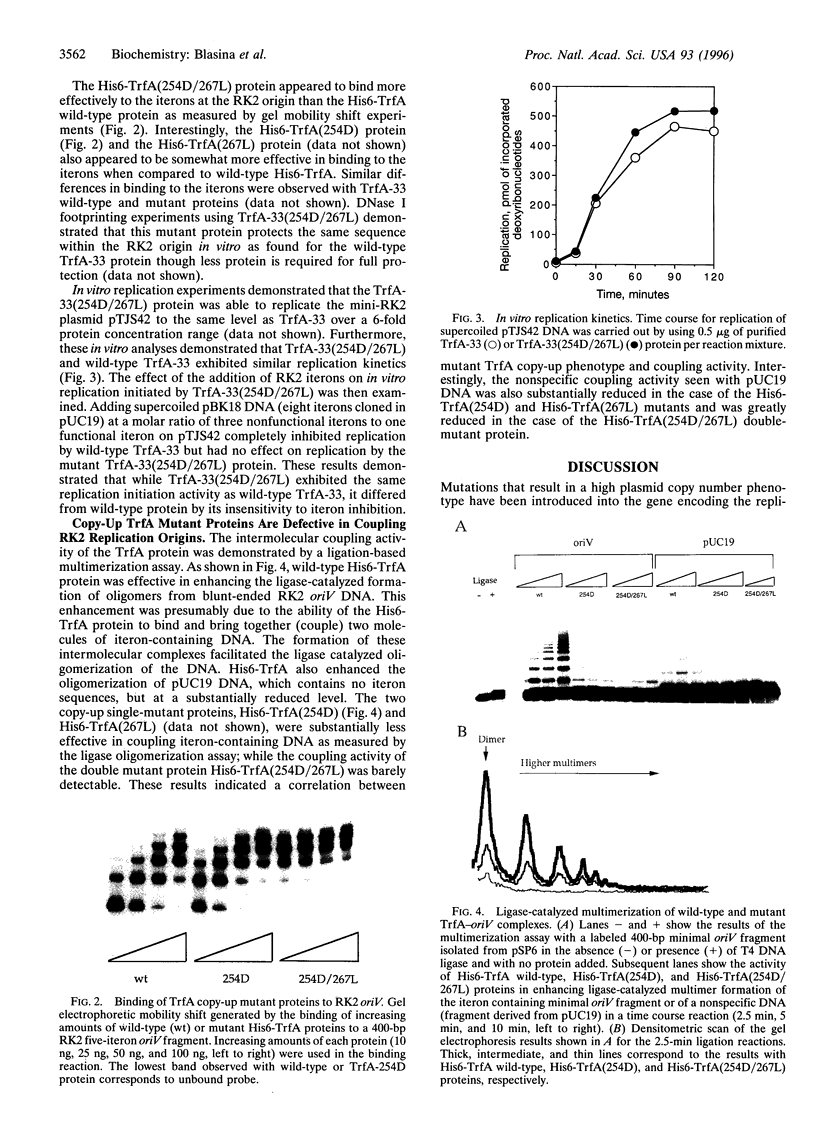
- Kittell B. L., Helinski D. R. Iteron inhibition of plasmid RK2 replication in vitro: evidence for intermolecular coupling of replication origins as a mechanism for RK2 replication control. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1389–1393. doi: 10.1073/pnas.88.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornacki J. A., West A. H., Firshein W. Proteins encoded by the trans-acting replication and maintenance regions of broad host range plasmid RK2. Plasmid. 1984 Jan;11(1):48–57. doi: 10.1016/0147-619x(84)90006-4. [DOI] [PubMed] [Google Scholar]
- Manen D., Upegui-Gonzalez L. C., Caro L. Monomers and dimers of the RepA protein in plasmid pSC101 replication: domains in RepA. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8923–8927. doi: 10.1073/pnas.89.19.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M. J., Bott M. A., Tooker P. A., Helinski D. R. Negative control of plasmid R6K replication: possible role of intermolecular coupling of replication origins. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7942–7946. doi: 10.1073/pnas.86.20.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron A., Mukherjee S., Bastia D. Activation of distant replication origins in vivo by DNA looping as revealed by a novel mutant form of an initiator protein defective in cooperativity at a distance. EMBO J. 1992 Mar;11(3):1205–1216. doi: 10.1002/j.1460-2075.1992.tb05161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron A., Patel I., Bastia D. Multiple pathways of copy control of gamma replicon of R6K: mechanisms both dependent on and independent of cooperativity of interaction of tau protein with DNA affect the copy number. Proc Natl Acad Sci U S A. 1994 Jul 5;91(14):6438–6442. doi: 10.1073/pnas.91.14.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay G., Sozhamannan S., Chattoraj D. K. Relaxation of replication control in chaperone-independent initiator mutants of plasmid P1. EMBO J. 1994 May 1;13(9):2089–2096. doi: 10.1002/j.1460-2075.1994.tb06484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S. K., Chattoraj D. K. P1 plasmid replication: initiator sequestration is inadequate to explain control by initiator-binding sites. J Bacteriol. 1988 Aug;170(8):3554–3560. doi: 10.1128/jb.170.8.3554-3560.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
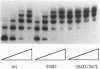
- Perri S., Helinski D. R. DNA sequence requirements for interaction of the RK2 replication initiation protein with plasmid origin repeats. J Biol Chem. 1993 Feb 15;268(5):3662–3669. [PubMed] [Google Scholar]
- Perri S., Helinski D. R., Toukdarian A. Interactions of plasmid-encoded replication initiation proteins with the origin of DNA replication in the broad host range plasmid RK2. J Biol Chem. 1991 Jul 5;266(19):12536–12543. [PubMed] [Google Scholar]
- Roberts R. C., Burioni R., Helinski D. R. Genetic characterization of the stabilizing functions of a region of broad-host-range plasmid RK2. J Bacteriol. 1990 Nov;172(11):6204–6216. doi: 10.1128/jb.172.11.6204-6216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokeach L. A., Søgaard-Andersen L., Molin S. Two functions of the E protein are key elements in the plasmid F replication control system. J Bacteriol. 1985 Dec;164(3):1262–1270. doi: 10.1128/jb.164.3.1262-1270.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser T. J., Filutowicz M., Helinski D. R. Replication of derivatives of the broad host range plasmid RK2 in two distantly related bacteria. Plasmid. 1983 May;9(3):325–330. doi: 10.1016/0147-619x(83)90010-0. [DOI] [PubMed] [Google Scholar]
- Schmidhauser T. J., Helinski D. R. Regions of broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of gram-negative bacteria. J Bacteriol. 1985 Oct;164(1):446–455. doi: 10.1128/jb.164.1.446-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingler V., Thomas C. M. Analysis of nonpolar insertion mutations in the trfA gene of IncP plasmid RK2 which affect its broad-host-range property. Biochim Biophys Acta. 1989 Apr 12;1007(3):301–308. doi: 10.1016/0167-4781(89)90152-8. [DOI] [PubMed] [Google Scholar]
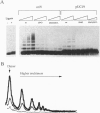
- Shingler V., Thomas C. M. Analysis of the trfA region of broad host-range plasmid RK2 by transposon mutagenesis and identification of polypeptide products. J Mol Biol. 1984 May 25;175(3):229–249. doi: 10.1016/0022-2836(84)90346-2. [DOI] [PubMed] [Google Scholar]
- Smith C. A., Thomas C. M. Nucleotide sequence of the trfA gene of broad host-range plasmid RK2. J Mol Biol. 1984 May 25;175(3):251–262. doi: 10.1016/0022-2836(84)90347-4. [DOI] [PubMed] [Google Scholar]
- Sozhamannan S., Chattoraj D. K. Heat shock proteins DnaJ, DnaK, and GrpE stimulate P1 plasmid replication by promoting initiator binding to the origin. J Bacteriol. 1993 Jun;175(11):3546–3555. doi: 10.1128/jb.175.11.3546-3555.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalker D. M., Filutowicz M., Helinski D. R. Release of initiation control by a mutational alteration in the R6K pi protein required for plasmid DNA replication. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5500–5504. doi: 10.1073/pnas.80.18.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalker D. M., Thomas C. M., Helinski D. R. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181(1):8–12. doi: 10.1007/BF00338997. [DOI] [PubMed] [Google Scholar]
- Stueber D., Bujard H. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1982;1(11):1399–1404. doi: 10.1002/j.1460-2075.1982.tb01329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Hussain A. A. The korB gene of broad host range plasmid RK2 is a major copy number control element which may act together with trfB by limiting trfA expression. EMBO J. 1984 Jul;3(7):1513–1519. doi: 10.1002/j.1460-2075.1984.tb02004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Meyer R., Helinski D. R. Regions of broad-host-range plasmid RK2 which are essential for replication and maintenance. J Bacteriol. 1980 Jan;141(1):213–222. doi: 10.1128/jb.141.1.213-222.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Stalker D. M., Helinski D. R. Replication and incompatibility properties of segments of the origin region of replication of the broad host range plasmid RK2. Mol Gen Genet. 1981;181(1):1–7. doi: 10.1007/BF00338996. [DOI] [PubMed] [Google Scholar]
- Toukdarian A. E., Helinski D. R., Perri S. The plasmid RK2 initiation protein binds to the origin of replication as a monomer. J Biol Chem. 1996 Mar 22;271(12):7072–7078. doi: 10.1074/jbc.271.12.7072. [DOI] [PubMed] [Google Scholar]
- Valla S., Haugan K., Durland R., Helinski D. R. Isolation and properties of temperature-sensitive mutants of the trfA gene of the broad host range plasmid RK2. Plasmid. 1991 Mar;25(2):131–136. doi: 10.1016/0147-619x(91)90025-r. [DOI] [PubMed] [Google Scholar]
- Wickner S., Hoskins J., McKenney K. Monomerization of RepA dimers by heat shock proteins activates binding to DNA replication origin. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]