Maturation of rRNA precursors requires an ordered series of endonucleolytic cleavages. One such cleavage site, designated A′, upstream of mature 18S rRNA has been observed in metazoa. The studies in this paper show that cleavage at A′ is not a prerequisite for downstream processing events but may be a quality control step for pre-rRNA transcripts.
Keywords: nuclease, nucleolus, rRNA processing, ribosome, snoRNA
Abstract
During eukaryotic ribosome biogenesis, three of the mature ribosomal (r)RNAs are released from a single precursor transcript (pre-rRNA) by an ordered series of endonucleolytic cleavages and exonucleolytic processing steps. Production of the 18S rRNA requires the removal of the 5′ external transcribed spacer (5′ETS) by endonucleolytic cleavages at sites A0 and A1/site 1. In metazoans, an additional cleavage in the 5′ETS, at site A′, upstream of A0, has also been reported. Here, we have investigated how A′ processing is coordinated with assembly of the early preribosomal complex. We find that only the tUTP (UTP-A) complex is critical for A′ cleavage, while components of the bUTP (UTP-B) and U3 snoRNP are important, but not essential, for efficient processing at this site. All other factors involved in the early stages of 18S rRNA processing that were tested here function downstream from this processing step. Interestingly, we show that the RNA surveillance factors XRN2 and MTR4 are also involved in A′ cleavage in humans. A′ cleavage is largely bypassed when XRN2 is depleted, and we also discover that A′ cleavage is not always the initial processing event in all cell types. Together, our data suggest that A′ cleavage is not a prerequisite for downstream pre-rRNA processing steps and may, in fact, represent a quality control step for initial pre-rRNA transcripts. Furthermore, we show that components of the RNA surveillance machinery, including the exosome and TRAMP complexes, also play key roles in the recycling of excised spacer fragments and degradation of aberrant pre-rRNAs in human cells.
INTRODUCTION
The production of the eukaryotic ribosome, which contains four ribosomal (r)RNAs and about 80 proteins, is a highly complicated process involving over 300 trans-acting factors (Henras et al. 2008). Three of the four mature rRNAs, 18S, 5.8S, and 28S (25S in yeast), are transcribed by RNA polymerase I as a single precursor, which also contains long internal (ITS) and external (ETS) transcribed spacers (Fig. 1A). The mature rRNAs are released by an ordered series of endonucleolytic cleavages and exonucleolytic processing. Most of the early insights into pre-rRNA processing and ribosome assembly were derived from work performed in the yeast, Saccharomyces cerevisiae (Venema and Tollervey 1999; Henras et al. 2008). Recently, however, the functions of several novel ribosome assembly factors, not present in yeast, have been described in human cells, and it has emerged that the pre-rRNA processing pathway and the nucleolytic activities involved are also notably different in metazoans (Mullineux and Lafontaine 2012; Preti et al. 2013; Sloan et al. 2013b). These findings have been highly relevant for understanding the basis of genetic diseases that arise as a result of impaired ribosome biogenesis (Narla and Ebert 2010; McCann and Baserga 2013); novel ribosomopathies, such as North American Indian childhood cirrhosis and Bowen-Conradi syndrome, caused by mutations in UTP4 and EMG1, respectively, have been identified (Chagnon et al. 2002; Prieto and McStay 2007; Armistead et al. 2009), while the classification of others as ribosomopathies has been questioned (Sloan et al. 2013b). Furthermore, a comprehensive understanding of ribosome production in human cells is very important, as ribosome biogenesis is coupled to the regulation of the tumor suppressor p53 via MDM2 and p14ARF and directly linked to the regulation of cellular proliferation (Stumpf and Ruggero 2011; Sloan et al. 2013a).
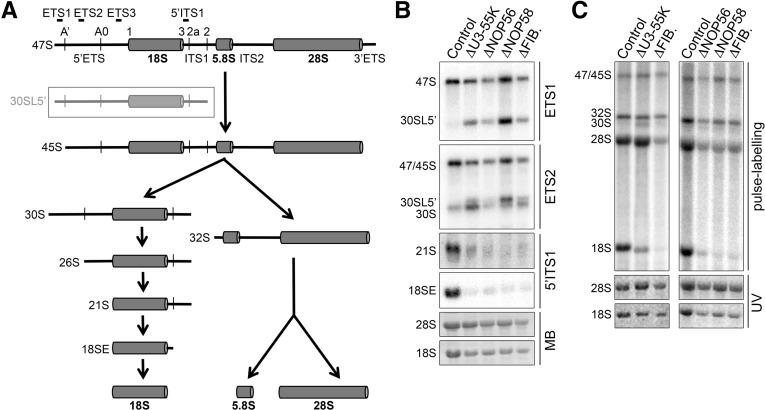
FIGURE 1.
Box C/D snoRNP proteins are important for efficient A′ processing but essential for downstream removal of the 5′ETS. (A) Schematic outline of the pre-rRNA processing pathway in human cells. Cleavages important for 18S rRNA processing are indicated. Probes used in this study are indicated above the schematic of the 47S rRNA. (ETS) external transcribed spacer, (ITS) internal transcribed spacer. An aberrant pre-rRNA, 30SL5′, which accumulates when A′ processing is inhibited, is shown in gray within a box. (B) RNA from control cells, or those depleted of box C/D snoRNP proteins by RNAi, was separated by agarose-glyoxal gel electrophoresis and transferred to nitrocellulose membrane. Northern blotting, using probes hybridizing in the 5′ETS or ITS1, was performed and pre-rRNAs detected using a phosphorimager. Mature rRNAs were visualized using methylene blue staining (MB). (C) HeLa cells were transfected with either control siRNAs or those targeting box C/D snoRNP proteins, and 48 h later, cells were pulse-labeled using 32P orthophosphate. RNA was extracted, separated by agarose-glyoxal gel electrophoresis, and visualized using a phosphorimager. Total rRNA (28S/18S) was visualized by ethidium bromide staining (UV).
In yeast, assembly of the small ribosomal subunit (SSU) is mediated by the SSU processome (Phipps et al. 2011). This complex assembles cotranscriptionally onto the nascent pre-rRNA and can be visualized as the “terminal knobs” in Miller spreads (Mougey et al. 1993). The SSU processome is composed of several subcomplexes, including the U3 snoRNP, the tUTP, bUTP, cUTP, Mpp10, and Bms1/Rcl1 complexes, and several individual proteins, such as Rrp5 and nucleolin (Phipps et al. 2011). A hierarchical assembly process has been described for the yeast SSU processome with association of the transcription (t)UTP (also known as UTP-A) complex, components of which are linked to RNA polymerase I and required for pre-rRNA transcription, being a prerequisite for recruitment of all other components (Perez-Fernandez et al. 2007, 2011). The U3 snoRNP contains the U3 snoRNA, the U3-specific protein Rrp9 (U3-55K in humans), and the core box C/D proteins, Snu13 (15.5K), Nop56, Nop58, and Nop1 (fibrillarin) (Watkins and Bohnsack 2012). The U3 snoRNA base-pairs with regions of the 5′ETS and the 18S rRNA in order to both orchestrate pre-rRNA processing and to regulate rRNA folding (Phipps et al. 2011). The RNA-binding proteins Imp3 and Imp4, which function together with Mpp10, are reported to help mediate the U3 snoRNA-pre-rRNA base-pairing interactions (Granneman et al. 2003; Gerczei and Correll 2004). Although many SSU components are conserved in higher eukaryotes, much less is known about their functional importance in ribosome production. Several proteins have been shown to be associated with the human pre-rRNA (Turner et al. 2009, 2012), but differences in the composition of yeast and human SSU subcomplexes have also been observed. For example, NOL11 has been identified as a novel component of the human tUTP complex, while UTP8 and UTP9 are missing in mammals (Prieto and McStay 2007; Freed et al. 2012). However, as in yeast, the human tUTP proteins have been linked to rRNA transcription and processing (Prieto and McStay 2007) and are required for the recruitment of the U3 snoRNP to the SSU processome (Turner et al. 2009).
Compared to yeast, an additional precursor 18S precursor, 18SE, was identified in humans (Fig. 1A). We and others have recently demonstrated that this is generated by exonucleolytic processing of ITS1 (Preti et al. 2013; Sloan et al. 2013b; Tafforeau et al. 2013). Removal of the 5′ETS in yeast involves two consecutive endonucleolytic cleavages at sites A0 and A1 (Henras et al. 2008). Surprisingly, the nucleases responsible for these cleavages remain elusive. In metazoans, an additional cleavage at site A′ in the 5′ETS has been identified (Fig. 1A), but a detailed characterization of this step and an understanding of how A′ cleavage is coordinated with assembly of the early preribosomal particle is lacking (Mullineux and Lafontaine 2012). In humans, A′ processing is the only cleavage event proposed to occur cotranscriptionally (Lazdins et al. 1997). In vitro data suggested that the major nucleolar protein, nucleolin, is required for A′ cleavage, but in vivo, nucleolin was shown to be important for RNA polymerase I transcription but not for pre-rRNA processing events (Ginisty et al. 1998, 2000). In vitro, A′ cleavage was also shown to be dependent on the U3, U14, U17, and E3 snoRNPs (Enright et al. 1996). A role for the U3 snoRNP in A′ processing is supported by RNAi knockdown data for the U3-specific protein hU3-55K (Prieto and McStay 2007), but beyond this, the general role of box C/D snoRNPs in this cleavage event is not clear. Furthermore, the tUTP proteins UTP10 and UTP4 have been demonstrated to be important for this cleavage event in vivo (Prieto and McStay 2007). Interestingly, A′ processing defects have also been observed upon depletion of the 5′-3′ exonuclease, XRN2, but the significance of this observation is not yet fully understood (Zakrzewska-Placzek et al. 2010; Wang and Pestov 2011; Sloan et al. 2013b). We recently observed that reducing the levels of XRN2 affects the balance of alternative ITS1 processing pathways, and we hypothesize that XRN2 may play a more general role in coordinating the optimal order of multiple pre-rRNA cleavages (Sloan et al. 2013b).
The transcription rate of pre-rRNA is very high, and more than half of each transcript is released and subsequently degraded as the mature rRNAs are formed. The turnover of aberrant pre-rRNAs and excised spacer regions is, therefore, important for maintaining nucleotide levels in the cell. In yeast, where both the quality control of pre-rRNA transcripts and the turnover of spacer fragments have been best studied, Rat1 (Xrn2) and the 3′-5′ exonuclease complex, the exosome, as well as its cofactors, including the TRAMP complex, are the key activities involved in performing these functions (Henras et al. 2008). The combined endo- and exonucleolytic activities of Rrp44 (Dis3) are responsible for degradation of the fragment between the A0 site and at the 5′ end of the pre-rRNA (Lebreton et al. 2008; Schaeffer et al. 2009; Schneider et al. 2009), while a fragment generated by cleavages at A0 and the 5′ end of the mature 18S rRNA accumulates when Rat1 is deleted (Petfalski et al. 1998). Similar roles for the mouse homologs of these enzymes have been suggested (Kent et al. 2009; Wang and Pestov 2011), but 5′ETS fragment turnover and aberrant pre-rRNA degradation have not been described in human cells.
Removal of the 5′ETS in metazoans involves an additional cleavage step (A′) compared to yeast, and currently little is known about how this processing event is coordinated with assembly of the early pre-ribosomal complex and what factors are necessary. Here, we show that, in human cells, only a subset of SSU processome factors is required for efficient A′ cleavage, while interestingly, we reveal a novel role for the RNA surveillance factor, MTR4, in this step. Furthermore, we show that A′ cleavage can be bypassed, and, taken together, our data suggest that A′ cleavage represents a quality control step in pre-rRNA processing.
RESULTS
The core box C/D snoRNP proteins are required for 18S rRNA production in human cells
In yeast, the box C/D snoRNPs U3 and U14 are essential for cleavage steps in the 5′ETS (A0 and A1) and ITS1 (A2) that are required for 18S rRNA production (Henras et al. 2008). In metazoans, an additional cleavage site (A′) upstream of A0 and site 1 (A1 in yeast) (Fig. 1A), ∼420 nt from the 5′ end of the pre-rRNA transcript, has been described (Mullineux and Lafontaine 2012). The U3 snoRNA base-pairs with the pre-rRNA immediately downstream from the A′ site (Watkins and Bohnsack 2012), and the U3 box C/D snoRNP-specific protein, hU3-55K, has been shown to be important for A′ processing (Prieto and McStay 2007). However, the importance of hU3-55K, and the other box C/D snoRNP proteins for downstream processing events and the overall production of the mature 18S rRNA has yet to be elucidated. To address this, RNAi was used to deplete the core box C/D snoRNP proteins, fibrillarin, NOP56, and NOP58, as well as the U3-specific protein hU3-55K, as previously reported (Watkins et al. 2004; Knox et al. 2011; Sloan et al. 2013b), and the production of ribosomal RNA was then analyzed by Northern blotting and pulse-chase labeling.
Northern blot analysis of RNA from cells depleted of hU3-55K, fibrillarin, NOP56, or NOP58, using a probe hybridizing upstream of the A′ cleavage site (ETS1) (Fig. 1A) revealed accumulation of the 30SL5′ pre-rRNA, demonstrating that A′ processing is inefficient in the absence of these proteins (Fig. 1B). However, a significant accumulation of the normal 30S pre-rRNA was detected using a probe hybridizing downstream from the A′ site (ETS2), indicating that the majority of pre-rRNA transcripts eventually undergo A′ processing and that the box C/D snoRNP proteins are important but not critical for this cleavage. Interestingly, 21S and 18SE pre-rRNA accumulation was severely reduced by knockdown of any of these proteins (Fig. 1B), signifying that they are essential for cleavage at sites A0 and 1, after A′ cleavage. Consistent with this, pulse-chase labeling experiments demonstrated that depletion of any of these proteins resulted in a severe inhibition of 18S production (Fig. 1C). Consistent with previous data (Sloan et al. 2013b), depletion of fibrillarin, NOP56, or NOP58, but not hU3-55K, also reduced the production of the 28S rRNA. This is likely to reflect the reported role for the U8 box C/D snoRNP in ITS2 processing (Srivastava et al. 2010). Together, these data indicate that the U3 snoRNP is important, but not essential, for A′ processing. The U3 snoRNP, and potentially other box C/D snoRNPs, are, however, critical for the downstream 5′ETS processing steps required for 18S rRNA production.
A subset of SSU processome components is required for A′ cleavage
The observation that the U3 snoRNP is important but not essential for A′ cleavage, combined with the reported differences in the human SSU processome composition compared to yeast, prompted us to determine the roles of other SSU factors in the early steps of ribosome biogenesis. We, therefore, used RNAi to deplete representative components of each of the major SSU subcomplexes: UTP10 (tUTP complex), UTP12 (bUTP complex), MPP10, IMP3, IMP4 (MPP10 complex), and BMS1 and RCL1 (Bms1/Rcl1 complex), as well as the putative endonucleases UTP23 and UTP24 (Fig. 2A; Sloan et al. 2013b).
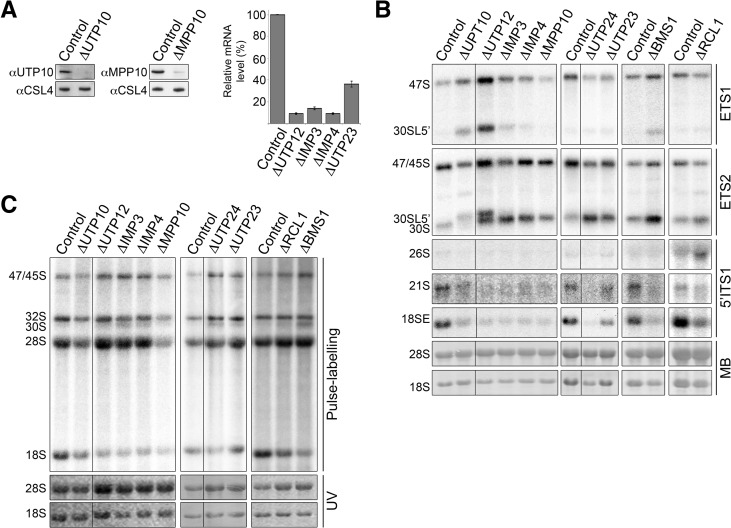
FIGURE 2.
A subset of SSU components is required for A′ cleavage. (A) HeLa cells were transfected with control siRNAs or those targeting SSU processome components. For UTP12, IMP3, IMP4, and UTP23, mRNA levels (normalized to GAPDH) were determined by qPCR and are given graphically. For UTP10 and MPP10, proteins were extracted and analyzed by Western blotting. We have previously reported the effectiveness of the other knockdowns (Sloan et al. 2013b). (B) RNA was extracted from RNAi-treated cells, separated by agarose-glyoxal gel electrophoresis, and analyzed by Northern blotting using probes hybridizing in the 5′ETS or ITS1 as indicated. Mature rRNAs were visualized by methylene blue staining (MB). (C) HeLa cells depleted of various SSU processome components were pulse-labeled, and RNA was analyzed by agarose-glyoxal gel electrophoresis and visualized using a phosphorimager.
Northern blot analysis of RNA derived from the knockdown cells, using probes specific to the 5′ETS, revealed that depletion of UTP10 or UTP12 resulted in accumulation of the unprocessed 47S pre-rRNA and the aberrant precursor 30SL5′, which is detected when A′ processing is impaired (Fig. 2B). RNAi-mediated depletion of BMS1, IMP3, or IMP4 caused a milder accumulation of these precursors. In contrast, depletion of RCL1, UTP23, UTP24, or MPP10 had little or no effect on the levels of 47S pre-rRNA and did not cause accumulation of 30SL5′ (Fig. 2B). Interestingly, Northern blotting using the ETS2 probe revealed that, with the exception of UTP10 and RCL1, the level of the 30S precursor was significantly increased by knockdown of each of these proteins. Depletion of RCL1 also results in the accumulation of 26S, indicating that A0 cleavage occurs as normal but that cleavage at site 1 is inhibited. Our data, therefore, imply that A′ cleavage is only blocked when the levels of the tUTP component, UTP10, are reduced (Fig. 2B). We conclude that UTP12, IMP3, IMP4, MPP10, BMS1, RCL1, UTP23, and UTP24 all have essential functions downstream from 30S pre-rRNA production, while UTP10 is required earlier in the pathway, before A′ cleavage. The mild effects observed following RCL1 and UTP23 knockdown are consistent with the relatively inefficient depletion of the protein (Fig. 2A; Sloan et al. 2013b).
Pulse-chase labeling of cells depleted of these proteins by RNAi revealed that UTP12, IMP3, IMP4, BMS1, RCL1, UTP23, and UTP24 are needed for 18S rRNA accumulation (Fig. 2C). Interestingly, depletion of UTP10 or MPP10 caused a reduction in both 18S and, to a lesser extent, 28S rRNA accumulation. These knockdowns also resulted in decreased levels of newly synthesized pre-rRNAs, including 32S, the precursor of the 28S rRNA and the 47/45S initial transcripts. These effects could be due either to impaired pre-rRNA transcription, as was previously suggested for UTP10 (Prieto and McStay 2007), or alternatively, could imply that the initial pre-rRNA transcript is unstable when these proteins are depleted (see Discussion).
Together, our data indicate that, of the proteins tested, only the tUTP component, UTP10, is essential for A′ cleavage, while the bUTP complex protein UTP12 and the U3 snoRNP are important for efficient processing. A′ cleavage was unaffected, or only mildly impaired, by depletion of representative components of the other major subcomplexes of the SSU processome. However, depletion of any of these factors blocked cleavages at sites A0 and site 1. This indicates that, surprisingly, only a subset of SSU processome components, which do not include the putative nucleases UTP23, UTP24, and RCL1, are important for A′ cleavage and that this processing event is uncoupled from the rest of the 18S rRNA processing steps.
XRN2 and MTR4 are required for A′ cleavage
As well as core SSU components, exonucleases are also predicted to have important roles in removal of the 5′ETS. The major nuclear 5′ to 3′ exonuclease XRN2 is important for A′ cleavage in several different organisms (Zakrzewska-Placzek et al. 2010; Wang and Pestov 2011; Sloan et al. 2013b) and has also been shown to be important for turnover of the 5′ETS fragment generated by this cleavage in mouse cells (ETS1) (Fig. 3A). It has recently been proposed that the involvement of exonucleases in various steps of pre-rRNA processing represents a way of monitoring and degrading aberrant pre-rRNAs (Granneman et al. 2011), so we hypothesized that A′ cleavage is an early quality control step in ribosome biogenesis. In addition to XRN2, the exosome and its cofactors, the TRAMP and NEXT complexes, are central components of the nuclear RNA surveillance machinery (Schneider and Tollervey 2013). We, therefore, investigated whether key activities of these complexes are important for 5′ETS processing and degradation steps in human cells. Cells were transfected with siRNAs targeting XRN2, the exosome catalytic subunit RRP6, or the TRAMP/NEXT complex component, the RNA helicase MTR4 (Sloan et al. 2013b), and the effect on rRNA processing was analyzed by Northern blotting. In addition to confirming the role of XRN2 in A′ processing, our Northern blot analysis revealed that depletion of MTR4 also resulted in the accumulation of 30SL5′, indicating a novel role for this protein in A′ processing (Fig. 3B). Knockdown of XRN2 also resulted in the accumulation of the 47S, rather than 45S, pre-rRNA. These defects in A′ processing were confirmed by S1 mapping analysis of the pre-rRNA using a probe spanning the A′ cleavage site (note that the increase in A′ cleaved products following MTR4 depletion is likely to represent accumulation of an excised ETS fragment [see below]) (Fig. 3C). Interestingly, a defect in A′ processing was not observed after knockdown of RRP6, which blocks later stages of 18S rRNA processing (Sloan et al. 2013b).
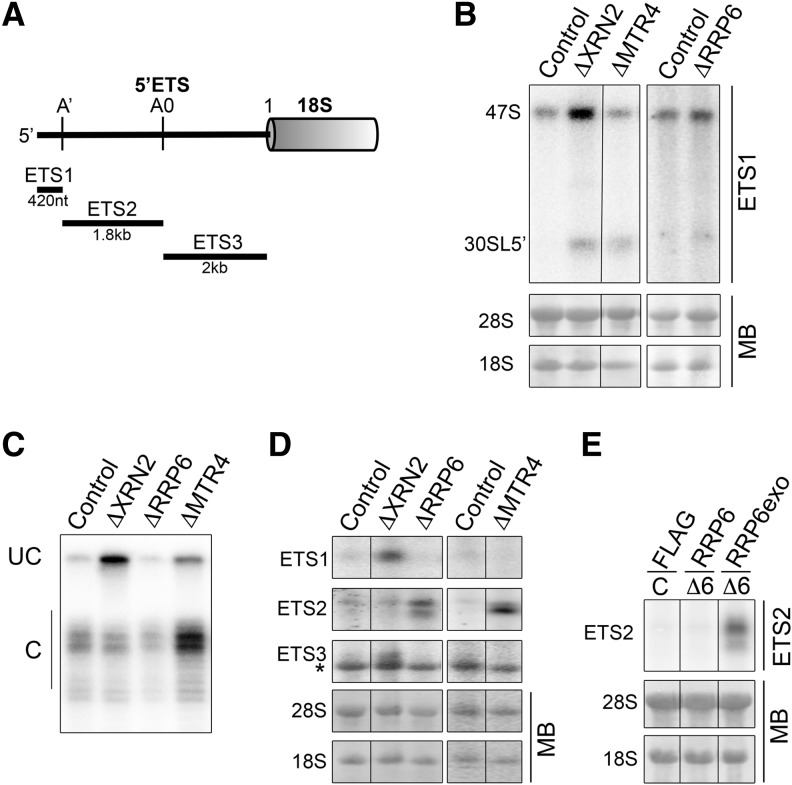
FIGURE 3.
XRN2, RRP6, and MTR4 function in A′ cleavage and turnover of 5′ETS fragments. (A) Schematic outline of the metazoan 5′ETS showing the cleavage sites and fragments generated. (B) RNA from HeLa cells depleted of XRN2, RRP6, or MTR4 was separated by agarose-glyoxal gel electrophoresis and analyzed by Northern blotting using an ETS1 probe. Mature rRNAs were visualized using methylene blue staining. (C) RNA from siRNA-treated cells was analyzed by S1 nuclease mapping using a probe hybridizing across the A′ cleavage site. The resulting DNA fragments ([UC] uncleaved, [C] cleaved) were separated by denaturing polyacrylamide gel electrophoresis and visualized using a phosphorimager. Note that, relative to the control, the ▵RRP6 lane is underloaded. (D) HeLa cells were transfected with control siRNAs or those targeting XRN2, MTR4, or RRP6, and 60 h later, RNA was harvested. This was analyzed by agarose-glyoxal gel electrophoresis followed by Northern blotting using probes hybridizing between the 5′-A′ (ETS1), A′-A0 (ETS2), and A0-site 1 (ETS3) cleavage sites. Note that the ETS3 probe cross-reacts with 18S—indicated by an asterisk (*). (E) HEK293 cells stably transfected with plasmids enabling expression of the FLAG-tag alone, FLAG-RRP6, or an exonucleolytically inactive form of RRP6 (RRP6exo) were transfected with control siRNAs (C) or those against RRP6 (Δ6). RNA was analyzed by Northern blotting using an ETS2 probe.
Knockdown of XRN2 in HeLa cells resulted in the accumulation of the 5′-A′ fragment (ETS1) and, to a lesser extent, the A0-site 1 fragment (ETS3) generated by endonucleolytic cleavages in the 5′ETS (Fig. 3D), as was previously observed in mouse cells (Wang and Pestov 2011). Knockdown of RRP6 or MTR4 resulted in the accumulation of the A′-A0 fragment (ETS2). This fragment accumulated as a doublet, suggesting that degradation of this fragment may be a multistep event. Using an RNAi-rescue system (Sloan et al. 2013b), we were also able to demonstrate that the accumulation of the ETS2 fragment occurred in cells expressing the catalytically inactive form of RRP6 (Fig. 3E), indicating that the exonuclease activity of RRP6 is required for the turnover of this fragment.
Therefore, in addition to XRN2, our data reveal a novel function for the TRAMP complex helicase MTR4 in A′ cleavage. Furthermore, our data demonstrate that, in human cells, different exonucleolytic activities are primarily responsible for the degradation of the cleaved 5′ETS fragments; while ETS1 and ETS3 are turned over from their 5′ ends by XRN2, RRP6 degrades the ETS2 fragment from the 3′ end.
The exosome is important for 5′ETS fragment turnover and degradation of aberrant pre-rRNAs
MTR4 functions as part of both the TRAMP and NEXT complexes in human cells and has also been shown to be associated with the exosome complex (Lubas et al. 2011). Our identification of a novel role for MTR4 in A′ processing and turnover of the ETS2 fragment raised the question of whether MTR4 performs these functions in the context of the TRAMP complex. In addition, RRP6 normally functions as one of the catalytic subunits of the multiprotein complex, the exosome (Schneider and Tollervey 2013), but has also been shown to function independently. Other components of the exosome complex, in particular, the other active subunit, DIS3, might therefore also participate in turnover of the ETS2 fragment. To test this, we used RNAi to deplete the exosome components RRP46 and DIS3 and the exosome cofactors C1D, MPP6, and the MTR4-associated polyA polymerases PAPD5 and POLS (Fig. 4A; Sloan et al. 2013b). Knockdown of the core exosome component, RRP46, but not DIS3, resulted in the accumulation of the ETS2 fragment (Fig. 4B). This observation is consistent with earlier results demonstrating that DIS3 is not present in the nucleolus and not required for nucleolar pre-rRNA processing (Tomecki et al. 2010; Sloan et al. 2013b). Furthermore, ETS2 fragment accumulation was detected after knockdown of the exosome cofactor MPP6, but not C1D. Interestingly, depletion of either of the MTR4-associated polyA polymerases, PAPD5 or POLS, did not cause ETS2 accumulation, suggesting that MTR4 is functioning independently of the TRAMP complex. Our data indicate that a defined subset of the RNA turnover machinery is involved in the degradation of the released 5′ETS fragments. With the exception of MTR4, depletion of any of the RNA surveillance factors tested did not perturb A′ cleavage, suggesting that MTR4 functions independently of both the exosome and TRAMP complexes in this cleavage event.
FIGURE 4.
Cofactors of the exosome and TRAMP complexes play different roles in 5′ETS fragment and aberrant pre-rRNA degradation. (A) HeLa cells were depleted of various subunits and cofactors of the exosome and TRAMP complexes. For PAPD5, mRNA levels (normalized to GAPDH) were measured using qPCR. Proteins extracted from control cells and those depleted of POLS were separated by SDS-PAGE and analyzed by Western blotting. (B) RNAs from HeLa cells described in A were analyzed by agarose-glyoxal gel electrophoresis, followed by Northern blotting using a probe hybridizing between the A′ and A0 cleavage sites (ETS2). Note that the increased signal seen for ETS2 in the DIS3 lane is due to the fact that this lane is slightly overloaded relative to the control. (C) RNA from control cells or those depleted of RRP6 by RNAi was analyzed by Northern blotting using probes hybridizing in the 5′ETS (ETS1, ETS2) and at the 5′ end of 28S (28S1302, 28S1727). (D) Schematic representations of the full-length pre-rRNA transcript and the aberrant 37S* pre-rRNA. The positions of additional probes used in Northern blotting are indicated above the 47S pre-rRNA.
Interestingly, knockdown of RRP46, RRP6, C1D, MPP6, MTR4, and PAPD5, but not POLS or DIS3, resulted in the accumulation of a novel pre-rRNA species we termed 37S* (Fig. 4B). Using a series of Northern probes, we determined that this rRNA extends between the A′ cleavage site and 1302–1727 nt downstream from the 5′ end of the 28S rRNA (Fig. 4C). The 37S* RNA, which was not detectable with the ETS1 probe and has, therefore, already undergone A′ processing, is likely to be derived from either a premature termination product in 28S rRNA or a partially degraded aberrant transcript. Interestingly, our data indicate that a different subset of the 3′-5′ degradation machinery is involved in the turnover of this aberrant 37S* transcript than the degradation of excised 5′ETS fragments.
A′ cleavage can be bypassed when XRN2 is depleted
While we observed accumulation of excised 5′ETS fragments following knockdown of surveillance/turnover proteins, it is unlikely that this accumulation represents a significant fraction of the 5′ETS fragments produced. This suggests that multiple activities are involved in the turnover of each of the 5′ETS fragments. We, therefore, asked whether XRN2 and the exosome function together in this recycling, and we compared 5′ETS fragment accumulation after knockdown of XRN2 alone or together with RRP6 or MTR4. Northern blotting revealed that codepletion of these factors resulted in an increase in the accumulation of the ETS2 fragment (Fig. 5A,B; note increase consistently more significant with RRP6 knockdown). However, to our surprise, codepletion of RRP6 or MTR4 with XRN2 resulted in a major reduction in the levels of the ETS1 fragment compared to knockdown of only XRN2 and the accumulation of a fragment that spanned from the 5′ end of the pre-rRNA to the site A0, i.e., an RNA fragment encompassing both ETS1 and ETS2 fragments, indicating that the A′ cleavage had been skipped (5′-A0) (Fig. 5A). Importantly, no change in the levels of ETS3 was observed upon codepletion of either RRP6 or MTR4 with XRN2 (data not shown). Together, our data indicate that, despite the fact that only specific 5′ETS fragments are detected after depletion of individual exonucleases, in fact, these activities work cooperatively to degrade excised fragments of the 5′ETS. A weak accumulation of the fragment spanning the 5′ end to site A0 was observed when only XRN2, but not MTR4, was depleted on its own (Fig. 5B). This indicates that, in the absence of XRN2, but not MTR4, the A′ cleavage is bypassed and A0 cleavage is the first processing event in the 5′ETS.
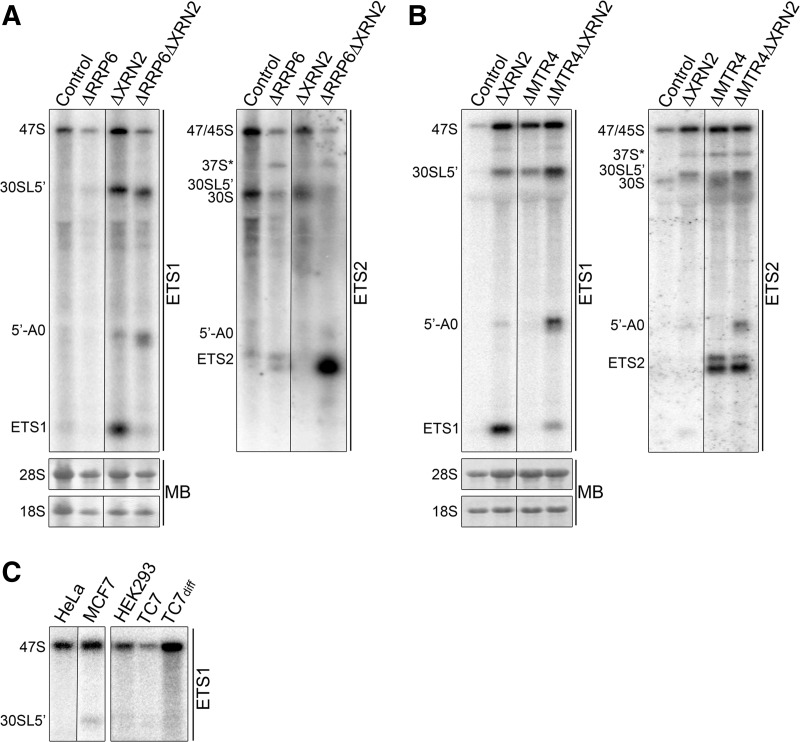
FIGURE 5.
A′ cleavage can be bypassed. (A) HeLa cells were transfected with control siRNAs or those targeting XRN2 or RRP6, either individually or together. Sixty hours later, RNA was harvested and analyzed by Northern blotting using probes hybridizing between the 5′ end of the pre-rRNA transcript and A′ (ETS1) or A′-A0 (ETS2). Mature rRNAs were visualized by methylene blue staining. (B) The experiment was repeated using siRNAs targeting MTR4 instead of RRP6. Note that the 37S* band seen with the XRN2 knockdown was not consistently observed. (C) RNA was extracted from various different cell lines and after differentiation for TC7 cells. This was analyzed by glyoxal gel electrophoresis, followed by Northern blotting using a probe hybridizing upstream of the A′ cleavage site (ETS1).
ITS1 processing can occur naturally before A′ cleavage in MCF7 cells
The observation that cleavage at A′ can be bypassed when XRN2 is depleted, without impeding subsequent 5′ETS processing steps, raised the possibility that this pathway could occur naturally in some cells or represent an alternative pathway used in different developmental stages. We, therefore, compared the levels of pre-rRNA species in different cell lines. RNA from HeLa (fibroblasts), HEK293 (human embryonic kidney), MCF7 (breast cancer), and both undifferentiated and differentiated TC7 (colon carcinoma) cells was analyzed by Northern blotting using a probe hybridizing upstream of the A′ cleavage site to detect the 47S and 30SL5′ pre-rRNAs. Interestingly, 30SL5′ was detectable in MCF7 cells but not in HeLa, HEK293, or TC7 cells (Fig. 5C). With the exception of the large amount of 47S seen in the differentiated TC7 cells, no other significant differences in pre-rRNA levels were seen between the RNAs extracted from the different cell lines (data not shown). This indicates that, in MCF7 cells, the normally aberrant 30SL5′ pre-rRNA represents a regular processing intermediate, and a significant proportion of pre-rRNAs are cleaved in ITS1 prior to removal of the 5′ETS, thus indicating that A′ is not obligatorily the primary pre-rRNA processing event in this cell line.
DISCUSSION
Here, we have investigated the removal of the 5′ETS in human cells, including a metazoan-specific pre-rRNA processing event, A′ cleavage. Furthermore, we have characterized factors that are involved in the degradation of the 5′ETS fragments after excision from the human pre-rRNA, and we have identified components of the early preribosome that are important for A′ cleavage. Interestingly, our data indicate that this processing event is not essential, can be bypassed, and may form part of a quality control mechanism. In addition, we have also determined that the surveillance factors XRN2 and MTR4 are required for both A′ processing and the turnover of the excised 5′ETS fragments.
The SSU processome and human 18S rRNA processing
While the importance of most SSU processome components in 18S rRNA production in yeast is now well established, studies into the roles of their human counterparts are considerably less advanced. This is of particular interest regarding their functions in two additional pre-rRNA processing steps that are specific to higher eukaryotes, 18SE production and A′ cleavage (Mullineux and Lafontaine 2012). We recently characterized the role of several SSU processome components in ITS1 processing and 18SE production (Sloan et al. 2013b), and here, we focus on A′ cleavage. Our data reveal that, as in yeast, the SSU processome components analyzed, which include components of the tUTP, bUTP, U3 snoRNP, BMS1/RCL1, and MPP10 complexes and the two putative nucleases UTP23 and UTP24, are important for 18S rRNA production. This is consistent with our earlier observations that many of these proteins localize to the nucleolus and are associated with pre-rRNA (Turner et al. 2009, 2012). However, not all of these components are essential, or even important, for A′ cleavage. Indeed, our data indicate that just the tUTP, bUTP, and U3 snoRNP proteins are needed for A′ processing to occur efficiently.
The processing of the 5′ETS at site A′ is proposed to occur cotranscriptionally, and interestingly, we found that the tUTP protein UTP10, which is required for pre-rRNA transcription (Prieto and McStay 2007), was the only protein tested that was essential for this processing event. The tUTP proteins, which are required for U3 snoRNP recruitment to the pre-rRNA, have been shown to bind the human pre-rRNA early in the processing pathway and are released either during or shortly after A′ cleavage (Turner et al. 2009). It is worth noting, however, that not all components of the tUTP complex show defects in RNA pol I transcription when depleted (Prieto and McStay 2007). Intriguingly, depletion of MPP10 also resulted in an overall reduction in the synthesis of the rRNAs from both subunits. This suggests that either MPP10, like UTP10, is important for pre-rRNA transcription or alternatively, MPP10 is important for the stability of the primary pre-rRNA transcript. A key difference between these two proteins is that, unlike UTP10, MPP10 is not important for A′ cleavage, perhaps favoring the latter alternative. Interestingly, the overall reduction in pre-rRNA levels was not seen after depletion of IMP3 and IMP4, two proteins shown to associate with MPP10 in both yeast and humans (Lee and Baserga 1999; Granneman et al. 2003).
It is surprising that depletion of either U3 snoRNP proteins or bUTP complex components only partially impaired A′ cleavage, with 18S rRNA processing fully inhibited downstream from the 30S precursor. In particular, the U3 snoRNA base-pairs to a region just downstream from the A′ cleavage site and was, based on in vitro data, thought to be essential for this processing step (Enright et al. 1996). Furthermore, depletion of the RNA chaperones, IMP3 and IMP4, which are predicted to be important for U3 snoRNA base-pairing to the 5′ETS (Gerczei and Correll 2004), hardly affected A′ cleavage. This, therefore, indicates that either these proteins are not important for the base-pairing event or that the U3-5′ETS base-pairing itself is not required for A′ cleavage. Several of the other SSU processome proteins tested, including MPP10, RCL1, BMS1, UTP23, and UTP24, as well as RRP5 (Sloan et al. 2013b) are not important for A′ cleavage. All of these proteins were, however, needed for later processing steps. Taken together, our data suggest a pathway in which A′ cleavage is largely independent of, and not essential for, the remaining 18S rRNA processing steps. The A′ cleavage is clearly facilitated by only a subset of the SSU processome.
The role of RNA surveillance factors in A′ cleavage
The extent of the A′ processing defect seen when XRN2 was depleted is significant, and we observe that there is very little 45S pre-rRNA present in these cells. Indeed, our data suggest that A′ cleavage is predominantly not the primary/initial cleavage when the levels of XRN2 are decreased. We found evidence that this bypass of A′ occurs naturally in cells, as what is considered an aberrant precursor, 30SL5′, is naturally present in MCF7 cells. Furthermore, the fact that a fragment that spans from the 5′ end of the initial transcript to the A0 site accumulates when turnover of the excised 5′ETS fragments is blocked indicates that this cleavage is not only delayed but that it can also be skipped without significantly impacting 18S rRNA production. Determining the level at which the A′ cleavage is skipped will only be resolved when XRN2′s roles in both A′ cleavage and the turnover of the ETS1 fragment can be uncoupled.
XRN2 is linked to transcription termination by RNA polymerase I and also to the surveillance of the pre-rRNA transcripts and turnover of premature transcription termination products (Shcherbik et al. 2010). The A′ cleavage is predicted to occur cotranscriptionally, and it is possible that, in the absence of XRN2, the link between the processing and transcription machineries is uncoupled. Indeed, it is interesting to speculate that, if the transcription and processing machineries are uncoupled, this would result in a change in the order of pre-rRNA cleavages, including promoting endonucleolytic cleavage at site 2a in ITS1 (Wang and Pestov 2011; Sloan et al. 2013b), perhaps explaining the results seen after depletion of XRN2. Interestingly, A′ cleavage was also affected by the depletion of another surveillance factor, MTR4. However, the 5′-A0 fragment did not accumulate after MTR4 knockdown, suggesting that A′ cleavage is not bypassed in the absence of this protein. Furthermore, MTR4 knockdown does not affect other endonucleolytic cleavages. The other major nucleolar surveillance factors analyzed, such as the exosome and PAPD5, were not required for any endonucleolytic cleavages, indicating that MTR4 acts on its own with respect to A′ cleavage.
The involvement of XRN2 in the processing event likely reflects the tight coupling of pre-rRNA processing to surveillance. As has been suggested for yeast 5.8S rRNA processing, the involvement of XRN2 in the processing reaction could reflect the fact that it is poised to degrade the pre-rRNA if something goes wrong in the process. It is possible that the A′ cleavage functions as a quality control step in the early stages of ribosome biogenesis and that, once A′ cleavage has occurred, XRN2 is no longer involved in the surveillance process. Indeed, XRN2 does not appear to play an important role in degradation of the aberrant pre-rRNA 37S*, which is either a partially degraded pre-rRNA or a premature transcription termination product, as accumulation of this pre-rRNA was not consistently observed upon depletion of XRN2 (Fig. 5, cf. panels A and B). Instead, the exosome, along with its cofactors, MTR4, PAPD5, C1D, and MPP6, represents the main activity required for turnover of 37S* pre-rRNA. The role of MTR4 in A′ cleavage is not so clear. It could be directly needed for the processing reaction itself, with its helicase activity perhaps being important for efficient unwinding/remodeling of the A′ cleavage site. Alternatively, MTR4 could provide an additional contact between the polymerase/processing complexes and the surveillance machinery.
Different activities are involved in the turnover of excised pre-rRNA spacer fragments and aberrant pre-rRNAs
Our data indicate that XRN2 plays a role in the turnover of all three 5′ETS fragments. XRN2 is essential for the degradation of ETS1 and ETS3 and contributes to the turnover of ETS2. In contrast, the exosome, the TRAMP complex, and MPP6 are essential for the degradation of ETS2. In yeast, both endo- and exonucleolytic activities of Rrp44 are involved in turnover of the cleaved 5′ETS. However, degradation of the human ETS2 fragment does not involve the exosome component DIS3. This is not entirely unexpected, as this protein is excluded from the human nucleolus (Lubas et al. 2011) and is not required for ITS1 processing (Sloan et al. 2013b), and provides further evidence of the diversification of active subunit function between human and yeast exosomes.
It is not entirely clear why different activities are required to degrade the different 5′ETS fragments. This could reflect the binding of factors to conserved elements within the ETS fragments dictating which end is more accessible for digestion. For example, the ETS2 fragment contains a conserved element at its 5′ end, which includes the U3 snoRNA base-pairing element. The presence of proteins bound at this end may block degradation, prompting turnover from the 3′ end. Conversely, the different degradation machineries may be specifically recruited at different stages of the processing pathway or to different regions of the pre-rRNA. Interestingly, the TRAMP complex (PAPD5, MTR4, and ZCCHC7) and RRP6 are all associated with bUTP proteins (Lubas et al. 2011). It is believed that the bUTP proteins, along with many of the SSU processome components, leave the preribosomes together with the cleaved excised 5′ETS (Hoang et al. 2005). Furthermore, the bUTP proteins are associated with pre-rRNA transcripts after treatment with actinomycin D (Turner et al. 2009), when the tUTP proteins and the U3 snoRNP are not stably bound, suggesting that they could interact with the pre-rRNA and function to tether the exosome/TRAMP complexes to the preribosomes. This could be to enhance 3′ processing of the 18S rRNA (Sloan et al. 2013b) or to couple surveillance to pre-rRNA processing.
From our data, it is interesting to see how different exosome cofactors are required for the degradation/processing of different pre-rRNAs linked to 18S rRNA processing. The exosome/RRP6 and MTR4 are important for 3′ processing of 21S pre-rRNA (Sloan et al. 2013b), the turnover of ETS2, and the 37S* aberrant transcript. Knockdown of MPP6 had a minor effect on 21S pre-rRNA processing (Sloan et al. 2013b), but MPP6 is required for ETS2 and 37S* degradation. PAPD5 and C1D are just required for 37S* degradation, and their depletion has no impact on ETS2 levels or 21S processing (Fig. 4C; data not shown). Our data, therefore, suggest that TRAMP and C1D are only involved in the degradation of aberrant transcripts. This is consistent with earlier work in mouse cells that showed that PAPD5 polyadenylated premature transcription termination products (Shcherbik et al. 2010). Therefore, different activities assist the exosome based on whether the complex is processing, degrading normal transcripts, or degrading aberrant transcripts. Human RRP6 is a more processive exonuclease than its yeast counterpart (Januszyk et al. 2011). It is, however, still not a very processive enzyme on its own and, especially given the high GC-content of the human pre-rRNA transcribed spacers, it will be interesting to investigate how this enzyme degrades long stretches of RNA.
Taken together, our results provide another step forward in understanding key aspects of the human pre-rRNA processing pathway that are different from yeast. Analyzing key components of the SSU processome and nuclear RNA surveillance components in parallel highlights the interplay between these two import machineries in the processing of the 5′ETS and the degradation of the released fragments.
MATERIALS AND METHODS
Cell culture and RNAi
HeLa, HEK293, MCF7, and TC7 cells were cultured according to standard protocols. HEK293 Flp-In cell lines stably transfected with plasmids for expression of the FLAG-tag, FLAG-RRP6, and FLAG-RRP6exo have been described previously (Sloan et al. 2013b). Differentiation of TC7 cells was achieved by incubation on polycarbonate membranes for 22 d (Knox et al. 2011).
RNAi depletion of factors of interest was performed using Lipofectamine RNAiMAX reagent (Invitrogen). The siRNAs used to deplete fibrillarin, NOP56, NOP56, hU3-55K, RCL1, BMS1, UTP24, XRN2, RRP6, MTR4, RRP46, C1D, MPP6, and DIS3 have been described previously, and the effect of these knockdowns on the levels of the target protein has been documented (Watkins et al. 2004; Knox et al. 2011; Sloan et al. 2013b). The following additional siRNAs (listed 5′ to 3′) were used here: UTP10 (Prieto and McStay 2007) (UAAAGAAGCUUGAAAGUGUTT), UTP12 (UAUGGGAUGUGAUCAAUGATT; CAAGGUGACAGUUACUUUATT; GCAACUGGCUCCGCUGAUATT; ACACUUAGCUGUUGGGUAUTT), IMP3 (CGAGCAGAAGCUGCUGAAGTT; GGAUCUCCUCCCUUGUUUATT; GAACUUGGACUACUGAUUATT), IMP4 (GGAUCCCAAGGUUAUGAUCTT; CCAGGACGACUACAUAUCATT; UGAACCGAGGUCGACAUGATT), MPP10 (UGAGCAGAUUUGGCAACAATT; UGAGGAGGAGGAAGAUAUUTT; UGAAGAUGAUGACCUUCAATT), UTP23 (UACAUGAUCCUGACAAGAA; CCACGAGGGCUAUGUAUGU; ACGCAAAGAUUACCUUAAG; GCUAAGAACUGCACUGCUU), PAPD5 (CAUCAAUGCUUUAUAUCGATT; GGACGACACUUCAAUUAUUTT; GAUAAAGGAUGGUGGUUCATT; GAAUAGACCUGAGCCUUCATT), and POLS (GGAGUGACGUUGAUUCAGATT; CGGAGUUCAUCAAGAAUUATT; AAACAGAGACGCCGAAAGUTT; GCGAAUAGCCACAUGCAAUTT). Cells were harvested 60 h after siRNA transfection and RNA or proteins extracted and analyzed by Northern and Western blotting, respectively. Antibodies for detection of UTP10 (Prieto and McStay 2007), MPP10 (Turner et al. 2009), POLS (raised against the peptides CNRGHHQYNRTGWRRK and CDYRRWIKEKWGSKAH), and CSL4 (Sloan et al. 2013b) were used in Western blotting.
Pulse labeling
Cells were transfected with appropriate siRNAs and after 48 h were incubated in phosphate-free DMEM for 1 h, then phosphate-free DMEM supplemented with 15 μCi/mL 32P orthophosphate, as described previously (Sloan et al. 2013b). Next, cells were grown in standard DMEM for a further 3 h before harvesting and extracting RNAs (see below).
RNA extraction and Northern blotting
RNA was extracted using TRI reagent (Sigma-Aldrich) and separated by agarose-glyoxal gel or denaturing polyacrylamide gel electrophoresis, then transferred to nitrocellulose membrane. Northern blot analysis was performed (Sloan et al. 2013b). Pulse-labeled RNAs were analyzed using a phosphorimager. ETS1 and ETS2 probes have previously been described (Turner et al. 2009), and the ETS3 probe was prepared by random-prime labeling of a PCR product spanning between nucleotides 2068 and 2724 of the 5′ETS. 5′ end-labeled oligonucleotide probes were prepared using the following primers (5′ to 3′): 5′ITS1-(CCTCGCCCTCCGGGCTCCGTTAATGATC), ITS1 (AGGGGTCTTTAAACCTCCGCGCCGGAACGCGCTAGGTAC), 28S1302 (TGGTCCGTGTTTCAAGACGGGT), 28S1727 (CAAGACCTCTAATCATTCGCTT).
RNA analysis
S1 nuclease mapping was performed as described in Turner et al. (2009). For qPCR, cDNAs were prepared from RNA derived from siRNA-treated cells by reverse transcription using Superscript III (Invitrogen) from an oligo dT primer. qPCR analysis was performed using the LightCycle 480 SYBER Green I Master kit (Roche). The following primer pairs (5′ to 3′) were used for amplification: UTP12 (fwd-CAGTGAAAGCAGCTGAGAGGA; rev-GCCATTAGGATGGGGTTGCT), IMP3 (fwd-CGGCACGTGCTAGAGTACAA; rev-GCAAAGTGGGAGATCCGCTA), IMP4 (fwd-TCATTCCGGCACCATGTGTA; rev-GTGCCCAGACGGATCATGTA), UTP23 (fwd-GAATTCCCCCGGCAATTGTTT; rev-CTCCAGAAGCTGTCAGTCAGG), PAPD5 (fwd-GCCTTCATGCAATGGAAATGGT; rev-TCCAAGGGCTTCATTTTCTTCAG).
ACKNOWLEDGMENTS
We thank Brian McStay for providing the A′ S1 mapping probe and the anti-UTP10 antibody. We also thank Jens Kretschmer for his technical guidance in performing qPCR. This work was funded by the Wellcome Trust (N.J.W.), a Royal Society University Fellowship (UF100666; C.S.), and the Deutsche Forschungsgemeinschaft (BO 3442/1-1: M.T.B.).
Footnotes
Freely available online through the RNA Open Access option.
REFERENCES
- Armistead J, Khatkar S, Meyer B, Mark BL, Patel N, Coghlan G, Lamont RE, Liu S, Wiechert J, Cattini PA, et al. 2009. Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen-Conradi syndrome. Am J Hum Genet 84: 728–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnon P, Michaud J, Mitchell G, Mercier J, Marion JF, Drouin E, Rasquin-Weber A, Hudson TJ, Richter A 2002. A missense mutation (R565W) in Cirhin (FLJ14728) in North American Indian childhood cirrhosis. Am J Hum Genet 71: 1443–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright CA, Maxwell ES, Eliceiri GL, Sollner-Webb B 1996. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA 2: 1094–1099 [PMC free article] [PubMed] [Google Scholar]
- Freed EF, Prieto JL, McCann KL, McStay B, Baserga SJ 2012. NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet 8: e1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerczei T, Correll CC 2004. Imp3p and Imp4p mediate formation of essential U3–precursor rRNA (pre-rRNA) duplexes, possibly to recruit the small subunit processome to the pre-rRNA. Proc Natl Acad Sci 101: 15301–15306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P 1998. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J 17: 1476–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Serin G, Ghisolfi-Nieto L, Roger B, Libante V, Amalric F, Bouvet P 2000. Interaction of nucleolin with an evolutionarily conserved pre-ribosomal RNA sequence is required for the assembly of the primary processing complex. J Biol Chem 275: 18845–18850 [DOI] [PubMed] [Google Scholar]
- Granneman S, Gallagher JE, Vogelzangs J, Horstman W, van Venrooij WJ, Baserga SJ, Pruijn GJ 2003. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60–80S ribonucleoprotein complexes. Nucleic Acids Res 31: 1877–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Petfalski E, Tollervey D 2011. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. EMBO J 30: 4006–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T, Peng WT, Vanrobays E, Krogan N, Hiley S, Beyer AL, Osheim YN, Greenblatt J, Hughes TR, Lafontaine DL 2005. Esf2p, a U3-associated factor required for small-subunit processome assembly and compaction. Mol Cell Biol 25: 5523–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januszyk K, Liu Q, Lima CD 2011. Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA 17: 1566–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent T, Lapik YR, Pestov DG 2009. The 5′ external transcribed spacer in mouse ribosomal RNA contains two cleavage sites. RNA 15: 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox AA, McKeegan KS, Debieux CM, Traynor A, Richardson H, Watkins NJ 2011. A weak C′ box renders U3 snoRNA levels dependent on hU3-55K binding. Mol Cell Biol 31: 2404–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazdins IB, Delannoy M, Sollner-Webb B 1997. Analysis of nucleolar transcription and processing domains and pre-rRNA movements by in situ hybridization. Chromosoma 105: 481–495 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Tomecki R, Dziembowski A, Seraphin B 2008. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456: 993–996 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Baserga SJ 1999. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol Cell Biol 19: 5441–5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH 2011. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 43: 624–637 [DOI] [PubMed] [Google Scholar]
- McCann KL, Baserga SJ 2013. Genetics. Mysterious ribosomopathies. Science 341: 849–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougey EB, O'Reilly M, Osheim Y, Miller OL Jr, Beyer A, Sollner-Webb B 1993. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev 7: 1609–1619 [DOI] [PubMed] [Google Scholar]
- Mullineux ST, Lafontaine DL 2012. Mapping the cleavage sites on mammalian pre-rRNAs: Where do we stand? Biochimie 94: 1521–1532 [DOI] [PubMed] [Google Scholar]
- Narla A, Ebert BL 2010. Ribosomopathies: Human disorders of ribosome dysfunction. Blood 115: 3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M 2007. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol 27: 5414–5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Fernandez J, Martin-Marcos P, Dosil M 2011. Elucidation of the assembly events required for the recruitment of Utp20, Imp4 and Bms1 onto nascent pre-ribosomes. Nucleic Acids Res 39: 8105–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petfalski E, Dandekar T, Henry Y, Tollervey D 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol 18: 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps KR, Charette J, Baserga SJ 2011. The small subunit processome in ribosome biogenesis—progress and prospects. Wiley Interdiscip Rev RNA 2: 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti M, O'Donohue MF, Montel-Lehry N, Bortolin-Cavaille ML, Choesmel V, Gleizes PE 2013. Gradual processing of the ITS1 from the nucleolus to the cytoplasm during synthesis of the human 18S rRNA. Nucleic Acids Res 41: 4709–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JL, McStay B 2007. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev 21: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A 2009. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol 16: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Tollervey D 2013. Threading the barrel of the RNA exosome. Trends Biochem Sci 38: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Leung E, Brown J, Tollervey D 2009. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res 37: 1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG 2010. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep 11: 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Bohnsack MT, Watkins NJ 2013a. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep 5: 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan KE, Mattijssen S, Lebaron S, Tollervey D, Pruijn GJ, Watkins NJ 2013b. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J Cell Biol 200: 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava L, Lapik YR, Wang M, Pestov DG 2010. Mammalian DEAD box protein Ddx51 acts in 3′ end maturation of 28S rRNA by promoting the release of U8 snoRNA. Mol Cell Biol 30: 2947–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf CR, Ruggero D 2011. The cancerous translation apparatus. Curr Opin Genet Dev 21: 474–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL 2013. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Mol Cell 51: 539–551 [DOI] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. 2010. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J 29: 2342–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AJ, Knox AA, Prieto JL, McStay B, Watkins NJ 2009. A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol Cell Biol 29: 3007–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AJ, Knox AA, Watkins NJ 2012. Nucleolar disruption leads to the spatial separation of key 18S rRNA processing factors. RNA Biol 9: 175–186 [DOI] [PubMed] [Google Scholar]
- Venema J, Tollervey D 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet 33: 261–311 [DOI] [PubMed] [Google Scholar]
- Wang M, Pestov DG 2011. 5′-end surveillance by Xrn2 acts as a shared mechanism for mammalian pre-rRNA maturation and decay. Nucleic Acids Res 39: 1811–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT 2012. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 3: 397–414 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Luhrmann R 2004. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell 16: 789–798 [DOI] [PubMed] [Google Scholar]
- Zakrzewska-Placzek M, Souret FF, Sobczyk GJ, Green PJ, Kufel J 2010. Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res 38: 4487–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]