This study examines translational regulation of the GCN4 mRNA in Candida albicans. Surprisingly, unlike the homologous gene in budding yeast, a single upstream open reading frame is sufficient to regulate GCN4 translation in C. albicans. Under nonstress conditions, this microORF (uORF) represses translation. The effect of the uORF is eliminated upon stress.
Keywords: translational regulation, upstream open reading frame, Gcn4, Candida albicans
Abstract
Candida albicans is a major fungal pathogen that responds to various environmental cues as part of its infection mechanism. We show here that the expression of C. albicans GCN4, which encodes a transcription factor that regulates morphogenetic and metabolic responses, is translationally regulated in response to amino acid starvation induced by exposure to the histidine analog 3-aminotriazole (3AT). However, in contrast to the well-known translational control mechanisms that regulate yeast GCN4 and mammalian ATF4 expression via multiple upstream open reading frames (uORFs) in their 5′-leader sequences, a single inhibitory uORF is necessary and sufficient for C. albicans GCN4 translational control. The 5′-leader sequence of GCN4 contains three uORFs, but uORF3 alone is sufficient for translational regulation. Under nonstress conditions, uORF3 inhibits GCN4 translation. Amino acid starvation conditions promote Gcn2-mediated phosphorylation of eIF2α and leaky ribosomal scanning to bypass uORF3, inducing GCN4 translation. GCN4 expression is also transcriptionally regulated, although maximal induction is observed at higher concentrations of 3AT compared with translational regulation. C. albicans GCN4 expression is therefore highly regulated by both transcriptional and translational control mechanisms. We suggest that it is particularly important that Gcn4 levels are tightly controlled since Gcn4 regulates morphogenetic changes during amino acid starvation conditions, which are important determinants of virulence in this fungus.
INTRODUCTION
All cells must be able to maintain their intracellular homeostasis during exposure to diverse physiological and environmental stress conditions. This requires that they regulate their gene expression patterns to adapt to the altered conditions. A common regulatory mechanism is instigated by global inhibition of translation initiation (Proud 2005). Reducing the rate of protein synthesis not only prevents continued gene expression during potentially error-prone conditions but also allows for the turnover of existing mRNAs and proteins while gene expression is reprogrammed to deal with the stress. Inhibiting translation initiation via phosphorylation of the α subunit of translation initiation factor 2 (eIF2) is a common response to diverse stress conditions in most eukaryotic cells (Proud 2005; Shenton et al. 2006). eIF2 is an essential GTP-binding protein that interacts with the initiator methionyl-tRNA (Met-tRNAiMet) to form a ternary complex (TC) that is competent for translation initiation. Phosphorylation of eIF2α at a conserved serine residue (Ser51) blocks GDP-GTP exchange, resulting in reduced TC levels, which inhibit translation initiation (Pavitt et al. 1998; Harding et al. 2000).
Phosphorylation of eIF2α inhibits global translation initiation but also induces translation of specific mRNAs, such as that encoding the metazoan activating transcription factor 4 (ATF4) (Harding et al. 2000; Vattem and Wek 2004). Translational regulation of ATF4 requires two uORFs in the 5′-leader of the ATF4 mRNA. Translation of uORF1 promotes ribosomal reinitiation, which under normal growth conditions occurs at inhibitory uORF2. Reduced TC levels increase the time required for ribosomes to reinitiate following uORF1 translation, and more ribosomes bypass uORF2 to reinitiate translation at the ATF4 start codon. Activation of ATF4 is considered part of a stress response signaling pathway termed the integrated stress response (ISR) which is initiated by eIF2α phosphorylation (Harding et al. 2003). ATF4 ultimately regulates the expression of genes that protect against the inducing stress conditions. In the yeast Saccharomyces cerevisiae, Gcn2 phosphorylates eIF2α in response to amino acid starvation conditions. This not only reduces global protein synthesis but also enhances translation of the GCN4 mRNA (Hinnebusch 2005). Gcn4 is a transcription factor, which, analogous to the mammalian ISR, activates amino acid biosynthetic genes to overcome the imposed starvation that initially led to its translational control. Similar to ATF4, translation of the GCN4 mRNA is activated in response to low TC levels in a mechanism involving four short uORFs (Hinnebusch 2005). This translational control mechanism depends on the sequence context immediately surrounding the stop codons of the uORFs, which modulate the ability of ribosomes to reinitiate translation. During normal unstressed conditions, ribosomes translate uORF1 and can reinitiate at inhibitory uORFs2–4. Decreased TC levels mean that ribosomes take longer to bind the TC and are more likely to scan past uORF2–4 and reinitiate at GCN4 (Hinnebusch 2005).
Candida albicans is a major human fungal pathogen that causes opportunistic infections (Pfaller and Diekema 2007). Systemic C. albicans infections are life-threatening, particularly in immunocompromised patients undergoing organ transplantation and chemotherapy and in those with HIV/AIDS. This means that Candida infections are a leading cause of mortality arising from hospital-acquired bloodstream infections (Viudes et al. 2002; Wisplinghoff et al. 2004). Amino acid starvation conditions can promote morphogenetic changes in C. albicans, including hyphal differentiation and biofilm formation, which are important determinants of virulence (Tripathi et al. 2002; Rubin-Bejerano et al. 2003). C. albicans can mount a GCN (general amino acid control) response, where it induces the expression of most amino acid biosynthetic pathways in response to amino acid starvation, dependent on the Gcn4 transcription factor (Tripathi et al. 2002). Surprisingly, however, GCN4 expression was thought to mainly be regulated at the transcriptional level in C. albicans, and Gcn2 has been proposed to play only a minor role in the activation of general amino acid control (Tournu et al. 2005). We show here that C. albicans Gcn4 expression is subject to both transcriptional and translational controls, but the induction of GCN4 expression is predominantly regulated at the translational level in response to amino acid starvation conditions. Activation of GCN4 expression requires Gcn2-dependent phosphorylation of eIF2α, but in contrast to yeast GCN4 and mammalian ATF4, regulation is predominantly mediated by a single inhibitory uORF, which is necessary and sufficient to control GCN4 translation.
RESULTS
C. albicans Gcn2 is required for the response to amino acid starvation conditions
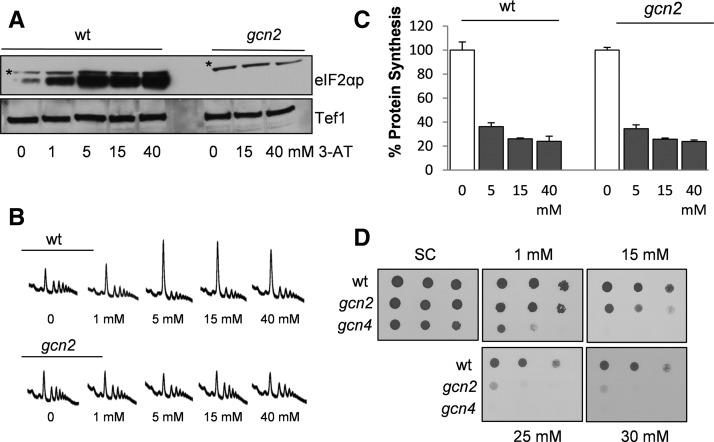
Amino acid starvation caused by exposure to the histidine analog 3-aminotriazole (3AT) has been extensively used to induce Gcn2-mediated phosphorylation of eIF2α in S. cerevisiae (Hinnebusch 2005). We first established amino acid starvation conditions using 3AT in C. albicans and examined eIF2α phosphorylation by immunoblot analysis. Exposure of C. albicans to 3AT at concentrations between 1 mM and 40 mM for 1 h caused a dose-dependent increase in eIF2α phosphorylation compared with untreated control cultures (Fig. 1A). No phosphorylation of eIF2α was detected in a gcn2 mutant, indicating that phosphorylation is entirely dependent on the Gcn2 protein kinase. Translational activity was analyzed by examining the distribution of polysomes under the same growth conditions. Polysomes are ribosomes that are actively translating mRNAs and can be separated and detected on sucrose density gradients. There was a shift of ribosomes from the polysomal region into the monosome peak following treatment with 3AT, which is indicative of decreased translation initiation (Fig. 1B). No inhibition of translation initiation was observed in a gcn2 mutant following exposure to 3AT. These data confirm that similar to other eukaryotes, C. albicans inhibits translation initiation in response to amino acid starvation conditions via Gcn2-mediated phosphorylation of eIF2α. The rate of protein synthesis was measured during the final 5 min of 3AT treatment by the incorporation of [35S]cysteine/methionine. Exposure of the wild-type strain to 5 mM 3AT inhibited translation by ∼70%, whereas 15 mM and 40 mM 3AT treatments caused ∼80% inhibition (Fig. 1C). The rate of protein synthesis was still inhibited to a similar extent in a gcn2 mutant that could not inhibit translation initiation. This is consistent with histidine starvation causing an additional Gcn2-independent inhibition of protein synthesis, presumably at the level of translation elongation.
FIGURE 1.
Regulation of C. albicans translation initiation by Gcn2. (A) The wild-type and gcn2 mutant strains were treated with the indicated concentrations of 3AT for 1 h. Immunoblots are shown probed with antibodies specific for phosphorylated eIF2α and translation elongation factor 1 (Tef1) as a loading control. (*) A nonspecific band recognized by anti-eIF2αP. (B) Polyribosome traces are shown for strains treated with the indicated concentrations of 3AT for 1 h. Peaks correspond to small ribosomal subunits (40S), large ribosomal subunits (60S), monosomes (80S), and increasing numbers of ribosomes bound to mRNAs (polysomes). (C) Protein synthesis was measured by pulse labeling with [35S]-cysteine/methionine for 5 min. Data are shown for untreated cultures (100%) and following treatments with different concentrations of 3AT. (D) Strains were grown to stationary phase and diluted cultures (A600 = 1.0, 0.1, and 0.01) spotted onto SCD agar plates containing the indicated concentrations of 3AT.
Loss of GCN2 was reported to cause only a minor decrease in the resistance of C. albicans to 3AT, suggesting that the C. albicans GCN response is not Gcn2 dependent (Tournu et al. 2005). To further examine this finding, we compared the sensitivity of gcn2 and gcn4 mutants to a range of 3AT concentrations. In agreement with previous observations (Tripathi et al. 2002; Tournu et al. 2005), gcn4 mutants are hypersensitive to amino acid starvation conditions and were unable to grow on plates containing 1 mM 3AT (Fig. 1D). In contrast, gcn2 mutants were less sensitive and could grow on plates containing 15 mM 3AT. However, gcn2 mutants are sensitive to 3AT since they could not grow at 3AT concentrations >25 mM, a concentration of 3AT that did not affect the growth of the wild-type strain. These data indicate that C. albicans Gcn2 is required for tolerance to amino acid starvation conditions.
Transcriptional and translational regulation of C. albicans GCN4 expression
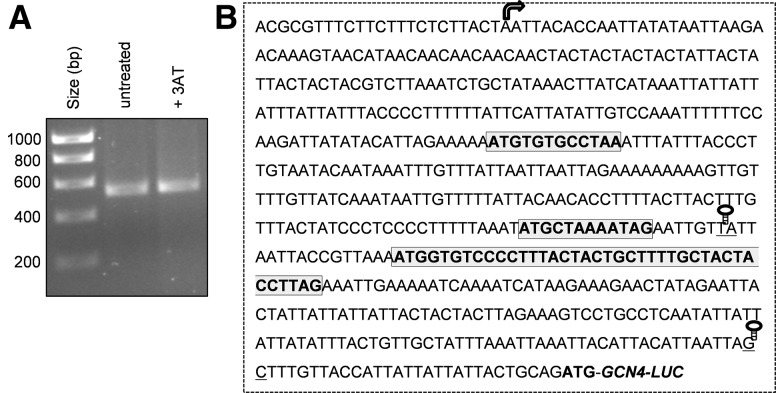
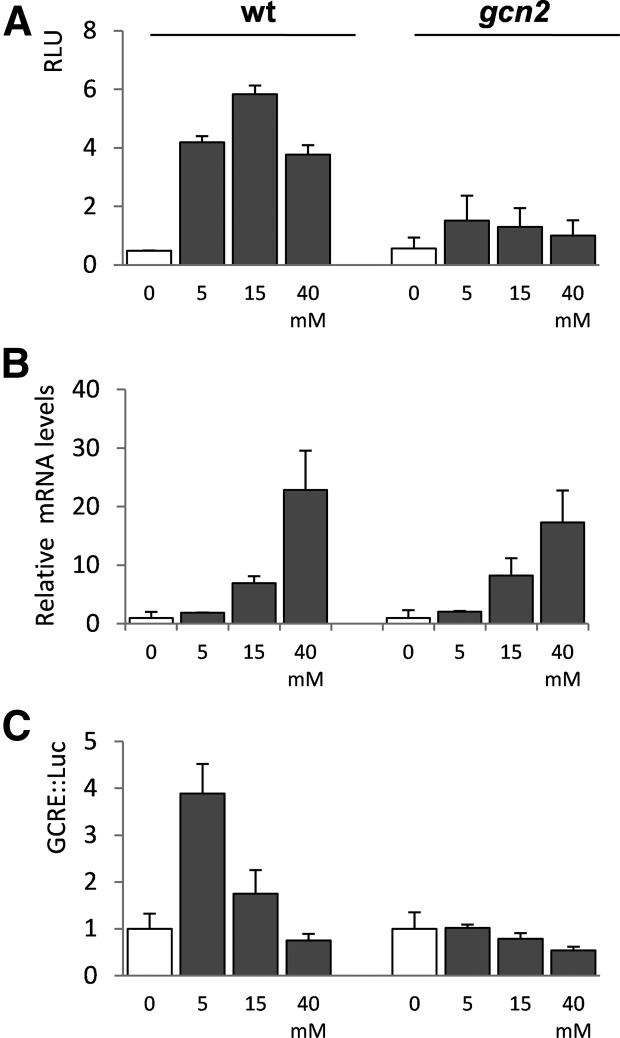
The transcription start site of GCN4 was mapped by 5′-RACE and DNA sequencing. This analysis identified a similar 577-nt 5′-leader sequence in the GCN4 mRNA under both nonstressed and 3AT stress conditions (Fig. 2A,B). To examine the translational activity of the GCN4 mRNA, its 5′-leader sequence was cloned upstream of a luciferase reporter gene. The expression of GCN4::Luc was induced following exposure to 3AT with a maximal increase of about 10-fold following treatment with 15 mM 3AT for 3 h. (Fig. 3A). In contrast, the induction of GCN4::Luc expression was largely abrogated in a gcn2 mutant, supporting a translational control mechanism for increasing GCN4 expression. A much reduced two- to threefold increase in GCN4::Luc expression was still observed in the gcn2 mutant, which presumably arises as a consequence of increased transcriptional activity. We used quantitative RT-PCR analysis to confirm that GCN4 mRNA levels are elevated in response to 3AT exposure (Fig. 3B). GCN4-Luc mRNA levels were increased in response to 3AT, with maximal induction observed at 40 mM 3AT exposure in both the wild-type and gcn2 mutant strains, consistent with transcriptional regulation of GCN4 expression.
FIGURE 2.
Mapping the transcription start site of GCN4 by 5′-RACE. (A) The transcription start site of GCN4 was mapped by 5′-RACE under both nonstressed and following 15 mM 3AT stress for 3 h. (B) The transcription start site is indicated with an arrow on the leader sequence of GCN4. uORF1, uORF2, and uORF3 are indicated with boxes. The positions of the stem–loop structures introduced upstream of and downstream from uORF3 are indicated.
FIGURE 3.
Transcriptional and translational regulation of C. albicans GCN4 expression. (A) The wild-type and gcn2 mutant strains were treated with the indicated concentrations of 3AT for 3 h, and GCN4-Luc activity was measured (RLU). (B) The wild-type and gcn2 mutant were treated with 3AT as above, and GCN4-Luc mRNA levels are expressed relative to GAPDH. (C) GCRE-Luc activity is shown for the wild-type and gcn2 mutant treated with 3AT as above.
To further examine the importance of Gcn2-mediated translational regulation of GCN4 expression, we used a previously described Gcn4-response element (GCRE) reporter construct that can be used to measure Gcn4-mediated transcriptional activation (Tripathi et al. 2002; Tournu et al. 2005). This reporter construct contains five copies of the GCRE cloned in a basal promoter upstream of a luciferase reporter gene. The expression of GCRE-Luc was increased in response to 3AT treatment with maximal increases observed at lower concentrations of 3AT (Fig. 3C). This increased activation of the GCRE reporter was largely abrogated in a gcn2 mutant, confirming that GCN4 activity is predominantly regulated at the translational level in response to amino acid starvation conditions.
A single uORF is required for translational regulation of GCN4 expression
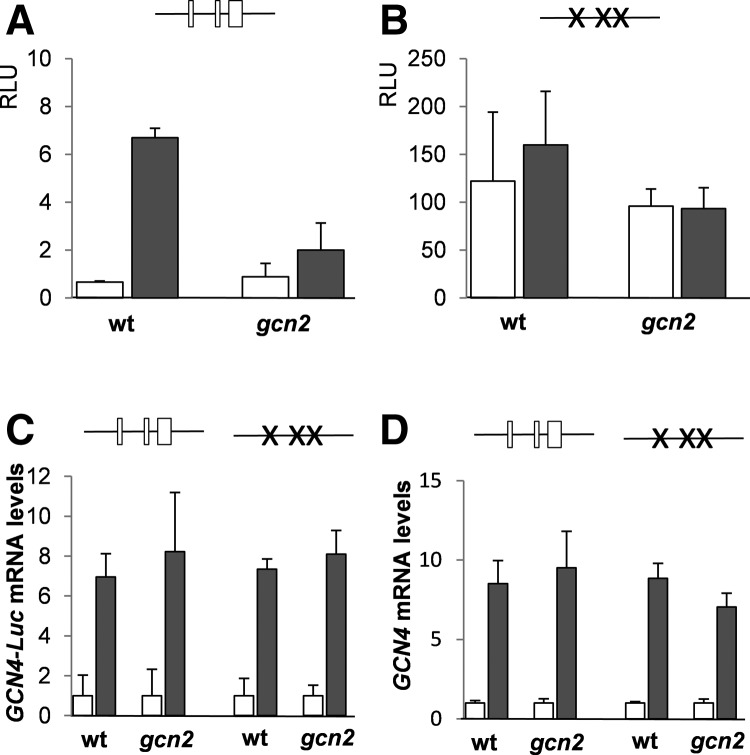
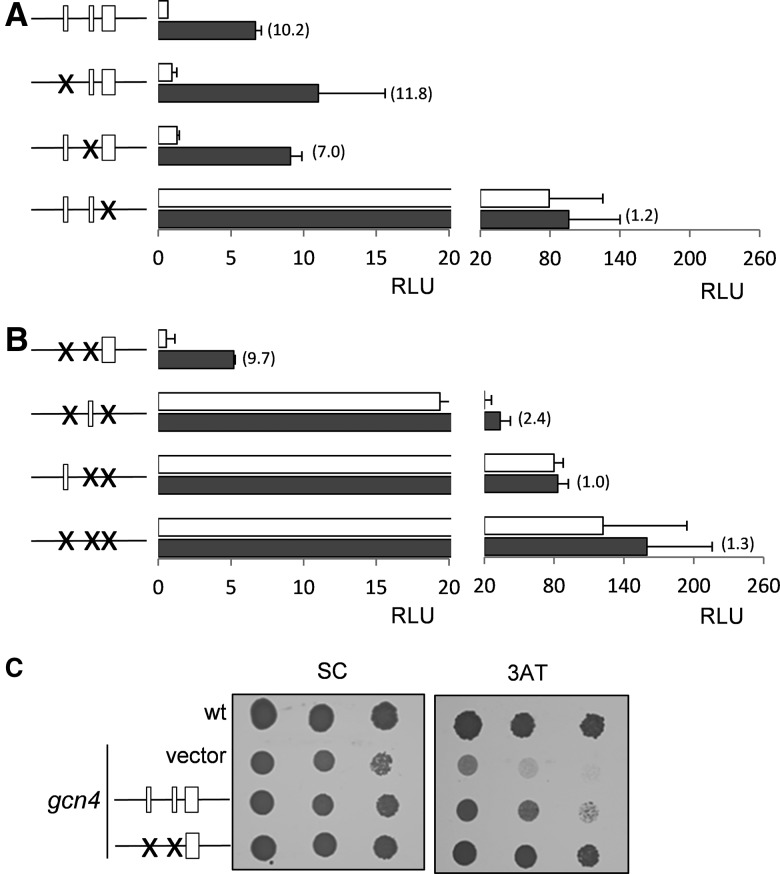
The 5′-leader sequence of GCN4 is predicted to contain three uORFs preceding the GCN4 coding region (Fig. 2B). uORF1 and uORF2 encode small three amino acid polypeptides, whereas uORF3 encodes a larger 12-amino-acid polypeptide. By analogy with yeast GCN4 and mammalian ATF4, these uORFs would be expected to play a role in regulating the translational activation of GCN4 expression. We therefore mutated the initiation codon of each uORF to prevent its translation and to determine the role of each uORF in regulating GCN4 expression. GCN4::Luc expression was measured following treatment of cells with 15 mM 3AT, which increased the expression of the wild-type GCN4 construct by about 10-fold in a largely Gcn2-dependent manner (Fig. 4A). Simultaneous mutation of uORF1, uORF2, and uORF3 resulted in a greater than 100-fold increase in GCN4-Luc expression, indicating that these uORFs normally act to inhibit GCN4 translation as expected (Fig. 4B). Quantitative RT-PCR analysis indicated that GCN4-Luc mRNA levels are induced by approximately sevenfold in response to 3AT stress conditions for both constructs in wild-type and gcn2 mutant cells, confirming that translational alterations account for the differences in GCN4-Luc expression (Fig. 4C). Similar increases in GCN4 mRNA levels were observed in response to 3AT compared with GCN4-Luc mRNA levels, confirming that the GCN4::Luc constructs are transcriptionally regulated similar to endogenous GCN4.
FIGURE 4.
Regulation of GCN4 translation by uORFs. (A) The wild-type and gcn2 mutant strains containing a wild-type GCN4-Luc construct were treated with 15 mM 3AT for 3 h. The uORFs in GCN4 are indicated as boxes in the schematic. (B) The wild-type and gcn2 mutant strains containing a GCN4-Luc construct where uORF1–3 were mutated were treated with 15 mM 3AT for 1 h. Mutations in the uORFs of GCN4 are indicated as crosses in the schematic. (C) Quantitative RT-PCR analysis of GCN4-Luc mRNA levels relative to GAPDH mRNA levels. (D) Quantitative RT-PCR analysis of GCN4 mRNA levels relative to GAPDH mRNA levels. White-colored bars denote values obtained from nonstressed cells; black-colored bars, values obtained from cells treated with 15 mM 3AT for 3 h.
We next examined the requirement for each uORF in regulating GCN4 expression. Surprisingly, mutation of uORF1 did not significantly affect GCN4 translational regulation. Basal expression was moderately increased by 1.5-fold, but GCN4 expression was still induced by about 12-fold in the uORF1 mutant in response to 3AT stress (Fig. 5A). This is unexpected since the 5′-proximal uORFs of yeast GCN4 and mammalian ATF4 play positive roles in promoting ribosomal scanning and reinitiation to promote GCN4/ATF4 expression under amino acid starvation conditions (Vattem and Wek 2004; Hinnebusch 2005). Mutation of uORF2 increased the basal expression levels of GCN4 by approximately twofold, but GCN4-Luc expression was still induced by sevenfold in response to 3AT stress. Mutation of uORF3 had the most dramatic effect on GCN4 expression; basal GCN4-Luc expression was increased by 120-fold, and little or no further induction was observed in response to 3AT stress (Fig. 5A). Taken together, these data indicate that uORF3 appears to play the predominant role as an inhibitory uORF reducing GCN4 translation.
FIGURE 5.
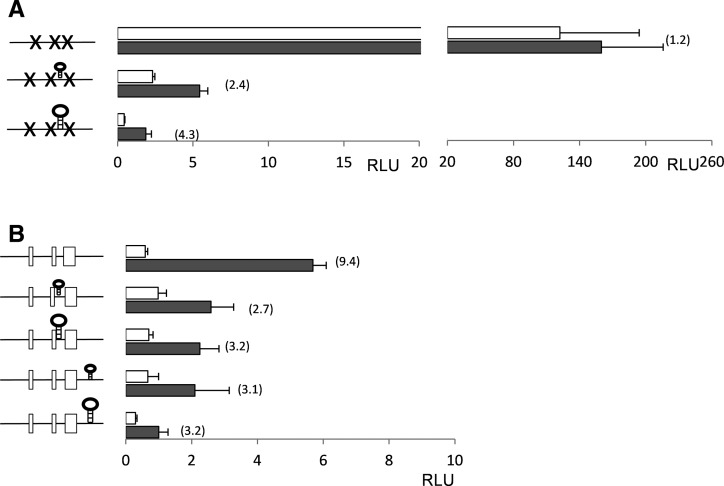
uORF3 is required for translational regulation of GCN4 expression. (A) The start codons of each individual uORF were mutated in GCN4-Luc and are indicated as crosses in the schematic. Fold-induction is indicated in brackets. (B) Pairs of uORFs were mutated as indicated. White-colored bars denote values obtained from nonstressed cells; black-colored bars, values obtained from cells treated with 15 mM 3AT for 3 h. (C) The 3AT sensitivity of a gcn4 mutant is complemented by reintroduction of wild-type GCN4 and a GCN4 construct containing uORF3 alone. The wild-type strain and gcn4 mutant are shown containing the empty vector. Strains were grown to stationary phase and diluted cultures (A600 = 1.0, 0.1, and 0.01) spotted onto SCD agar plates containing 1 mM 3AT.
To further define the role of uORFs in regulating GCN4 expression, we mutated each uORF in pairs. Mutants lacking combinations of uORF1 and uORF3 or of uORF2 and uORF3 displayed significant increases in basal GCN4 expression and were largely unable to induce GCN4 expression further in response to 3AT stress (Fig. 5B). In contrast, regulation was still largely maintained in a construct lacking both uORF1 and uORF2, indicating that uORF3 is necessary and largely sufficient to control GCN4 translation in response to amino acid starvation conditions (Fig. 5B). To confirm that the GCN4-Luc reporter is a faithful model of the authentic GCN4 gene, a wild-type Gcn4 construct and a Gcn4 construct containing uORF3 alone were reintroduced into a gcn4 deletion strain. Both constructs were able to complement the 3AT sensitivity of the gcn4 mutant, confirming that uORFs1–2 are dispensable for regulation and uORF3 alone is sufficient to maintain GCN4 function (Fig. 5C).
Regulation of GCN4 expression depends on a leaky scanning mechanism at uORF3
DNA segments encoding stem–loop structures were inserted into the GCN4 mRNA leader to confirm that regulation of GCN4 translation requires a scanning mechanism rather than internal initiation. In one set of constructs, the sequence 5′-GAATTCCCATCTTGGGAATTC-3′ (ΔG = −8.7 kcal) was introduced 16 nt upstream of uORF3 or 57 nt downstream from uORF3 (Fig. 2B). The secondary structure provided by this sequence has previously been shown to inhibit yeast GCN4 expression (Abastado et al. 1991). In a second set of constructs, the sequence 5′-CTGCAGCCACCACGGCCCCCAAGCTTGGGCCGTGGTGGCTGCAG-3′ (ΔG =−41 kcal), which has previously been shown to inhibit mammalian ATF4 expression (Vattem and Wek 2004), was introduced at the same locations. Both stem–loop structures were initially introduced into a construct lacking uORF1–3, where they were found to significantly decrease GCN4-Luc expression by about 50-fold and 200-fold, respectively (Fig. 6A). Interestingly, GCN4-Luc expression was induced by two- to fourfold in these constructs, suggesting that scanning ribosomes may be better able to scan past secondary structures during conditions of limiting TC availability. Alternatively, increased transcriptional induction may account for this increase in GCN4-Luc expression under 3AT starvation conditions. Both stem–loop structures also decreased GCN4-Luc expression in a wild-type construct, supporting a mechanism where ribosomes scan the leader sequence past uORF3 under starvation conditions to initiate GCN4 translation (Fig. 6B). These stem–loop structures were also less inhibitory under derepressing conditions in a construct containing uORF1–3 (Fig. 6B). The reasons for this difference are unknown at present but may, again, suggest that C. albicans ribosomes are better able to scan past secondary structures during conditions of limiting TC availability.
FIGURE 6.
Regulation of GCN4 expression depends on a leaky scanning mechanism at uORF3. Two different stem–loop structures (ΔG = −8.7 kcal and −41 kcal) were introduced into GCN4-Luc lacking uORFs (A) or containing uORFs (B) as indicated. Fold-induction is indicated in brackets. White-colored bars denote values obtained from nonstressed cells; black-colored bars, values obtained from cells treated with 15 mM 3AT for 3 h.
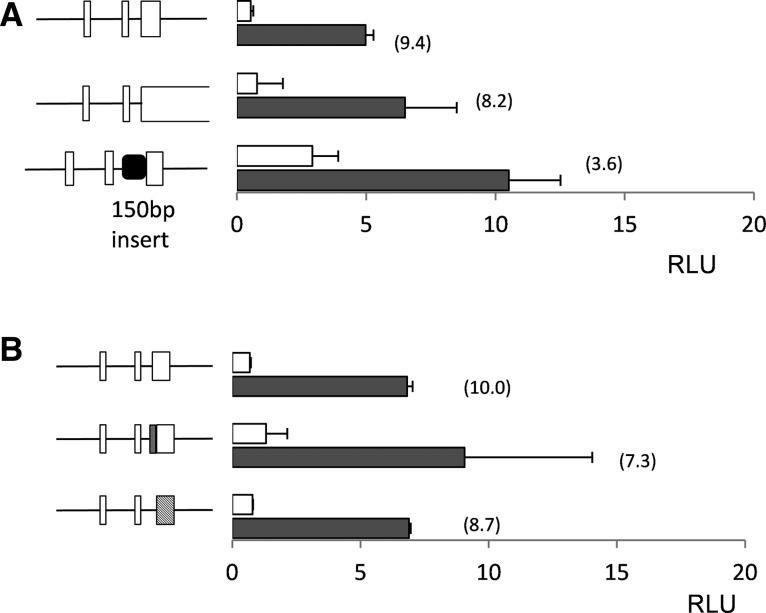
Extensive overlap between uORF3 and GCN4 coding sequences and increasing the spacing between uORF2 and uORF3 has little effect on GCN4 expression
To rigorously test the model that ribosomes leaky scan past uORF3 under starvation conditions, we extended uORF3 such that it terminates downstream from the GCN4 ATG codon. This was done by mutating the uORF3 stop codon and removing a subsequent in-frame stop codon. This creates an extended 67-codon version of uORF3 that terminates 30 nt downstream from the start codon of GCN4. This alteration had little or no effect on the expression of the GCN4-LUC reporter consistent with a model where ribosomes leaky scan past uORF3 under starvation conditions to reach GCN4 (Fig. 7A). As a further test to rule out any requirement for a positively acting uORF that promotes scanning and reinitiation at GCN4, we inserted a 150-nt sequence upstream of uORF3. When a similar insertion was made in S. cerevisiae GCN4 increasing the separation between uORF1 and uORF4, it did not affect expression under nonstarvation conditions but significantly reduced expression under derepressing conditions (Abastado et al. 1991). This is consistent with the idea that S. cerevisiae uORF1 is required to promote reinitiation at GCN4 under starvation conditions. In contrast, increasing the separation between uORF1–2 and uORF3 in C. albicans GCN4 did not reduce GCN4 expression under starvation conditions (Fig. 7A). Basal expression under nonstarvation conditions was somewhat increased, suggesting that increasing the separation between uORF2 and uORF3, or the sequence introduced, somehow decreased the ability of ribosomes to recognize and initiate translation at the uORF3 start codon. Nevertheless, these data are inconsistent with the S. cerevisiae GCN4 model where ribosomes initiate translation at an uORF and control depends on their ability to scan past an inhibitory uORF in order to reinitiate at GCN4.
FIGURE 7.
Regulation of GCN4 expression. (A) Extensive overlap between uORF3 and GCN4 coding sequences has little effect on GCN4 expression. The uORF3 stop codon and a subsequent in-frame stop codon were removed. This creates an extended 67-codon version of uORF3 that terminates 30 nt downstream from the start codon of GCN4. Increasing the spacing between uORF2 and uORF3 does not reduce GCN4 expression under starvation conditions. A 150-nt sequence, normally found between uORF3 and GCN4, was inserted between uORF2 and uORF3 as indicated. (B) The start codon context of uORF3 is not important for GCN4 translational control. The start codon context of uORF3 (GTTAAAATGG) was changed to more closely match the Kozak consensus sequence (GCCACCATGG), indicated as a shaded box in the schematic. The coding sequence of uORF3 is not important for GCN4 translational control. The peptide sequence encoded by uORF3 (MVSPLLLLLLLP) was changed (MAVPFTTAFATT), indicated as a hatched box in the schematic. White-colored bars denote values obtained from nonstressed cells; black-colored bars, values obtained from cells treated with 15 mM 3AT for 3 h.
The sequence of uORF3 is not important for regulating GCN4 expression
Codon context plays an important role in the recognition of AUG start codons, and one possibility is that ribosomes scan past uORF3 to translate GCN4 under amino acid starvation conditions. However, the start codon context of uORF3 (GTTAAAATGG) is similar to the Kozak consensus sequence 5′-gcc(A/G)ccATGG-3′, where the A/G at −3 and the G at +4 are thought to be the most critical residues (Kozak 1986). Nevertheless, we changed the sequence around the AUG codon of uORF3 to match the Kozak consensus sequence. However, the induction of GCN4 expression in response to amino acid starvation conditions was largely maintained, although there was a moderate increase in expression under both normal and stressed conditions (Fig. 7B). The peptide sequence encoded by uORF3 (MVSPLLLLLLLP) is unusual since it contains a run of seven Leu residues. We therefore tested the requirement for the coding sequence of uORF3 by inserting a C residue at +5 and deleting a C residue at +35 to generate an unrelated uORF3 peptide sequence (MAVPFTTAFATT). Replacing uORF3 with an uORF encoding an unrelated peptide sequence did not significantly alter GCN4 translational regulation, indicating that the peptide sequence of uORF3 is not important for control (Fig. 7B).
DISCUSSION
Our data indicate that C. albicans GCN4, which encodes a central regulator of amino acid biosynthetic genes, is regulated at both the transcriptional and post-transcriptional levels in response to amino acid starvation. Yeast GCN4 is minimally regulated at the transcriptional level and has long served as a model of translational gene regulation (Hinnebusch 2005), whereas mammalian ATF4 gene expression is subject to both transcriptional and translational regulation (Dey et al. 2010). Regulation of C. albicans GCN4 is therefore more similar to mammalian ATF4 than to yeast GCN4. Combining transcriptional and translational regulation is thought to provide flexibility in controlling ATF4 expression, which is important in the response to differing stress conditions as part of the ISR (Dey et al. 2010). Amino acid starvation conditions can promote morphogenetic changes in C. albicans, including hyphal differentiation and biofilm formation, which are important determinants of virulence (Tripathi et al. 2002; Rubin-Bejerano et al. 2003; Garcia-Sanchez et al. 2004). GCN4 is implicated in these morphogenetic changes, and it is therefore important that C. albicans GCN4 expression is carefully controlled.
Maximal transcriptional induction of GCN4 was observed at higher concentrations of 3AT compared with translational induction. Translational regulation of GCN4 expression therefore appears to be the first response to low levels of amino acid starvation. Increasing 3AT concentrations induced GCN4 transcription, which may reflect the need to increase GCN4 mRNA levels under conditions where global translation is strongly inhibited. Indeed, protein synthesis measured by radiolabelling was strongly inhibited in wild-type and gcn2 mutant cells at higher concentrations of 3AT. This inhibition of protein synthesis at the post-initiation phase of translation may explain why no increase in GCN4-Luc expression is observed in a gcn2 mutant despite the strong transcriptional induction that is observed following exposure to 40 mM 3AT. It is interesting that induction of the GCRE-Luc reporter, which provides a measure of gene activation by Gcn4 (Tripathi et al. 2002), was mainly induced at low concentrations of 3AT compared with both the transcriptional and translational induction observed in response to 3AT. Additional regulatory inputs are known to regulate yeast Gcn4-target gene expression, including transcription factors and coactivators, which may account for this difference (Takemaru et al. 1998; Patil et al. 2004).
In contrast to both ATF4 and yeast GCN4, which are translationally regulated via mechanisms that require multiple uORFs in their 5′-leader sequences, a single inhibitory uORF is sufficient for C. albicans GCN4 translational control. uORF3 appears to play the predominant role as an inhibitory uORF reducing GCN4 translation. This is similar to yeast GCN4, where the 3′-proximal uORF4 plays the predominant role as an inhibitory uORF (Abastado et al. 1991). GCN4 translational regulation is maintained in a construct containing a single uORF3, which raises the question as to the requirement for uORF1 and uORF2. The presence of any positively acting short uORFs that might be initiated by a near-cognate start codon is unlikely since inserting a large 150-nt sequence upstream of uORF3 did not alter GCN4 translational control. Inserting a similar-sized sequence between uORF1 and uORF4 in S. cerevisiae GCN4 significantly reduced GCN4 expression under derepressing conditions, consistent with the idea that the increased scanning time enabled ribosomes to reacquire TC and to reinitiate at inhibitory uORF4 (Abastado et al. 1991).
Comparison of the construct lacking any uORFs with constructs containing uORF1 and uORF2 alone indicates that they inhibit GCN4 translation by ∼35% and 85%, respectively. One possibility is that uORF1 and uORF2 act to moderate GCN4 expression by regulating ribosomal scanning on the GCN4 mRNA. This may be important in order to respond to different levels of amino acid starvation and eIF2α phosphorylation. It is not clear why uORF3 is more inhibitory than uORF1 and uORF2. Codon context plays an important role in the recognition of AUG start codons and, for example, is important in the regulation of translation at mammalian uORFs (Kolitz and Lorsch 2010; Palam et al. 2011). The start codon contexts of uORF1 (GAAAAAATGT) and uORF2 (TTAAATATGC) may be less optimal than uORF3 since they do not contain a G at the +4 position. Additionally, uORF3 may be more inhibitory since it is longer, encoding a 12-amino-acid polypeptide, compared with uORF1 and uORF2, which encode short three-amino-acid polypeptides. Interestingly, uORF1 does appear to somewhat mitigate the inhibitory effect of uORF2 under nonstarvation conditions based on the comparison of constructs containing both uORF1 and uORF2 with constructs containing uORF1 or uORF2 alone (Fig. 5). These data suggest that uORF1 and uORF2 may be able to play regulatory roles under certain growth conditions.
Changing the start codon at uORF3 to match the Kozak consensus sequence did not abrogate GCN4 translational control. It should be stressed, however, that little is known regarding the importance of start codon context in C. albicans, and it is possible that the sequence around the uORF3 start codon is important for regulating C. albicans GCN4 translation. Many fungal systems contain frequent uORFs in the 5′-leader regions of diverse genes (Hood et al. 2009). Although it is thought that a number of these may play a regulatory role by controlling ribosomal scanning to their downstream coding regions, few examples have been examined experimentally. Regulation of C. albicans GCN4 translation involves a scanning mechanism to reach the GCN4 coding region since the introduction of stem–loop structures around uORF3 decreased the induction of GCN4 expression normally seen in response to amino acid starvation conditions. A scanning mechanism is also consistent with the finding that the uORF3 reading frame could be extended such that it overlapped the start of GCN4 without affecting translational regulation in response to amino acid starvation conditions.
Our model for translational regulation of C. albicans GCN4 is that under normal growth conditions, most ribosomes initiate translation at uORF3 and, to a lesser extent, at uORF1 and uORF2, preventing scanning to the GCN4 ORF. Hence, inserting stem–loop structures into a wild-type construct did not have the same large impact on scanning to GCN4 compared with a construct lacking uORFs since most scanning to GCN4 is already blocked by uORF3. It is possible that the low-basal level of translation seen in the presence of stem–loop structures (or absence of Gcn2) does not require scanning from the predominant 5′ end of the mRNA. This may be explained by proposing the existence of a small percentage of transcripts with 5′ ends located between the stem–loop insertion points and the GCN4 start codon, which would be below the detection of the 5′-RACE analysis. Under amino acid starvation conditions, phosphorylation of eIF2α increases leaky scanning past uORF3 and promotes GCN4 translation. This is similar to the bypass of uORFs with poor initiation codon context, which has been observed in response to phosphorylation of eIF2α (Palam et al. 2011). One possibility is that under conditions of limiting TC levels, ribosomes are able to leaky scan past small uORFs such as uORF3, but large structured coding regions like the GCN4 ORF are able to “catch” these ribosomes, initiating GCN4 translation. It should be emphasized that the majority of scanning ribosomes (>80%) still initiate at uORF3, and only a modest decrease in translation at uORF3 accounts for the 10-fold increase in GCN4 translation. Hence, uORF3 appears to act as a moderator of C. albicans GCN4 expression, regulating the number of scanning ribosomes that initiate GCN4 translation in response to amino acid starvation conditions.
MATERIALS AND METHODS
Strains and growth conditions
The C. albicans gcn2 and gcn4 mutants were isogenic derivatives of CAI-4 (ura3::λ imm434/ura3::λ imm434) as described previously (Tournu et al. 2005). Strains were grown in complex YEPD (2% w/v glucose, 2% w/v bactopeptone, 1% w/v yeast extract) or synthetic complete (SC) medium (2% w/v glucose, 0.67% w/v yeast nitrogen base, 0.185% w/v complete amino acid supplement [Formedium] mixtures). Uridine was supplemented to a final concentration of 25 µg/mL. Strains were grown at 30°C and 180 rpm. Media were solidified by the addition of 2% (w/v) agar. Stress sensitivity was determined by growing cells to stationary phase and spotting diluted cultures (A600 = 1.0, 0.1 and 0.01) onto agar plates containing various concentrations of 3AT. Liquid cultures were treated with 3-AT by growing cells to exponential phase and treating with different concentrations of 3-AT as indicated.
Plasmids and strain construction
The 600 bp immediately upstream of the C. albicans GCN4 coding sequence were synthesized by Life Technologies. Sequences were also synthesized containing single point mutations (ATG to TTG) at the start codons of uORF1, uORF2, and uORF3. An extended uORF3 construct was synthesized by changing the normal uORF3 stop codon (TAG) to TAC and a subsequent in-frame TAG codon (33 nt downstream from uORF3) to TAC. The separation between uORF2 and uORF3 was increased by inserting a 150-nt sequence normally found between uORF3 and GCN4, 16 nt upstream of uORF3. Constructs containing multiple uORF start codon mutations were made using the QuikChange method (Agilent). Similarly, the 6 nt immediately upstream of uORF3 was changed to match the mammalian kozak consensus sequence (GCCACCATGG), and the uORF3 coding sequence was changed by inserting a C residue at +5 and deleting a C residue at +35 using the QuikChange method. Two sequences with the potential to form stem–loop structures (5′-GAATTCCCATCTTGGGAATTC-3′) and (5′-CTGCAGCCACCACGGCCCCCAAGCTTGGGCCGTGGTGGCTGCAG-3′) were introduced 16 nt upstream of uORF3 and 57 nt downstream from uORF3 using the Q5 site-directed mutagenesis kit (New England Biolabs). Constructs were ligated immediately upstream of RLUC in the pCRW3N basal vector using MluI and PstI restriction sites as described previously (Tripathi et al. 2002). Plasmids were integrated at the ADE2 locus of wild-type and gcn2 mutant strains, and three independent correct colonies were used for each experiment. The reporter construct containing five copies of the GCRE cloned in a basal promoter upstream of a luciferase reporter gene has been described previously (Tripathi et al. 2002; Tournu et al. 2005). To check for complementation of the gcn4 mutant, wild-type GCN4 and GCN4 with mutated uORF1 and uORF2 was PCR amplified and cloned in the pCRW3N vector using PstI and Xho1 restriction sites. Plasmids were integrated at the ADE2 locus.
Transcription start site mapping of GCN4
Total RNA was extracted using the RNEasy mini kit (Qiagen). Five micrograms of total RNA was used to identify the transcriptional start site of GCN4 using the FirstChoice RLM-RACE Kit (Ambion) according to the manufacturer’s instructions. Two rounds of nested PCR were performed using 5′-RACE outer primer (forward 5′-GCTGATGGCGATGAATGAACACTG-3′), GCN4 outer primer (reverse 5′-TGCTGAAACCCGACTCCAAT-3′), 5-RACE inner primer (forward 5′-CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG-3′), and GCN4 inner primer (reverse 5′-TTGAGGCAGGACTTTCTAAGTAGT-3′). PCR products were run in 1.5% agarose gels and purified using a PCR purification kit (Macherey-Nagel). Purified PCR products were cloned using a TA cloning kit (Invitrogen), and transcription start sites were identified by DNA sequencing.
Quantitative RT-PCR analysis
RNA was extracted using the RNeasy mini kit (Qiagen) and treated with recombinant DNase I (Ambion). Two hundred nanograms of total RNA was reverse transcribed into cDNA using Oligo(dT)20 primer and iScript reverse transcriptase (Biorad). RT-PCR was performed using iTaq Universal SYBR green supermix (Biorad) in a CFX connect real-time PCR detection system (Biorad). Primers used for detection of RLUC mRNA forward and reverse primers were 5′-GGAATTATAATGCTTATCTACGTGC-3′ and 5′-CTTGCGAAAAATGAAGACCTTTTAC-3′; GAPDH mRNA forward and reverse primers were 5′-CGAAGGTGCTCAAAAACACA-3′ and 5′-TGTACCACCAACTGTTTGGC-3′; and GCN4 mRNA forward and reverse primers were 5′-CCAGAAATGCAAAAGGCTTC-3′ and 5′-GACTTTGGCTCCGTCCATAA-3′.
Analysis of protein synthesis
For the analysis of ribosome distribution on sucrose density gradients, cultures were grown to exponential phase and treated with 3AT as indicated. Extracts were prepared in the presence of 1 mg/mL cycloheximide/mL and layered onto 15%–50% sucrose gradients. The gradients were sedimented via centrifugation at 40,000 rpm in a Beckman ultracentrifuge for 2.5 h, and the A254 was measured continuously to give the traces shown, as described previously (Shenton et al. 2006). Monosome and polysome peaks were quantified using the National Institutes of Health Image J software (http://rsb.info.nih.gov/ij/). The rate of protein synthesis was measured in cells treated with various concentrations of 3AT. Cells were treated with 3AT for 1 h and pulse-labeled for the last 5 min of the treatment with 85 µM L-[35S] cysteine/methionine as described previously (Shenton and Grant 2003).
Western blot analysis
Protein extracts were electrophoresed under reducing conditions on SDS-PAGE minigels and electroblotted onto PVDF membrane (Amersham Pharmacia Biotech). Blots were probed using phosphospecific eIF2α antibodies as described previously (Holmes et al. 2004). Antibodies raised against yeast elongation factor 1 (Tef1) were used as a loading control.
Reporter assays
Luciferase analysis was performed essentially as explained previously by Srikantha et al. (1996). Briefly, cells were harvested and extracted using RLUC buffer (0.5 M NaCl, 0.1 M K2HPO4 at pH 6.7, 1 mM Na2 EDTA, 0.6 mM sodium azide, 1 mM phenylmethylsulfonyl fluoride, 0.02% bovine serum albumin). Luciferase assays were started by adding 1.25 µM coelenterazine h (Promega) to cell extracts, and activity was measured using a GloMax 20/20 luminometer (Promega). Luciferase activity (RLU) is expressed as relative luminescence per 10 sec/mg protein.
ACKNOWLEDGMENTS
We thank Professor Al Brown (Aberdeen University) for strains and plasmids and helpful discussions. A.S. is funded by the Ministry of Higher Education, Malaysia.
REFERENCES
- Abastado JP, Miller PF, Jackson BM, Hinnebusch AG 1991. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol 11: 486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Baird TD, Zhou D, Palam LR, Spandau DF, Wek RC 2010. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J Biol Chem 285: 33165–33174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanchez S, Aubert S, Iraqui I, Janbon G, Ghigo JM, d’Enfert C 2004. Candida albicans biofilms: A developmental state associated with specific and stable gene expression patterns. Eukaryot Cell 3: 536–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450 [DOI] [PubMed] [Google Scholar]
- Holmes LE, Campbell SG, De Long SK, Sachs AB, Ashe MP 2004. Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol Cell Biol 24: 2998–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood HM, Neafsey DE, Galagan J, Sachs MS 2009. Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu Rev Microbiol 63: 385–409 [DOI] [PubMed] [Google Scholar]
- Kolitz SE, Lorsch JR 2010. Eukaryotic initiator tRNA: Finely tuned and ready for action. FEBS Lett 584: 396–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292 [DOI] [PubMed] [Google Scholar]
- Palam LR, Baird TD, Wek RC 2011. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem 286: 10939–10949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil CK, Li H, Walter P 2004. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol 2: E246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD, Ramaiah KV, Kimball SR, Hinnebusch AG 1998. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine–nucleotide exchange. Genes Dev 12: 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ 2007. Epidemiology of invasive candidiasis: A persistent public health problem. Clin Microbiol Rev 20: 133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG 2005. eIF2 and the control of cell physiology. Semin Cell Dev Biol 16: 3–12 [DOI] [PubMed] [Google Scholar]
- Rubin-Bejerano I, Fraser I, Grisafi P, Fink GR 2003. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc Natl Acad Sci 100: 11007–11012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton D, Grant CM 2003. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J 374: 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM 2006. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem 281: 29011–29021 [DOI] [PubMed] [Google Scholar]
- Srikantha T, Klapach A, Lorenz WW, Tsai LK, Laughlin LA, Gorman JA, Soll DR 1996. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J Bacteriol 178: 121–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru K, Harashima S, Ueda H, Hirose S 1998. Yeast coactivator MBF1 mediates GCN4-dependent transcriptional activation. Mol Cell Biol 18: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournu H, Tripathi G, Bertram G, Macaskill S, Mavor A, Walker L, Odds FC, Gow NA, Brown AJ 2005. Global role of the protein kinase Gcn2 in the human pathogen Candida albicans. Eukaryot Cell 4: 1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi G, Wiltshire C, Macaskill S, Tournu H, Budge S, Brown AJ 2002. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J 21: 5448– 5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, Wek RC 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci 101: 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viudes A, Peman J, Canton E, Ubeda P, Lopez-Ribot JL, Gobernado M 2002. Candidemia at a tertiary-care hospital: Epidemiology, treatment, clinical outcome and risk factors for death. Eur J Clin Microbiol Infect Dis 21: 767–774 [DOI] [PubMed] [Google Scholar]
- Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB 2004. Nosocomial bloodstream infections in US hospitals: Analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39: 309–317 [DOI] [PubMed] [Google Scholar]