In this paper, the authors present a nonradioactive Northern blotting method for the detection of miRNAs. This protocol, which takes advantage of the previously developed splinted ligation approach for detecting small RNAs, also includes cross-linking with EDC, which improves capture of small RNAs and the use of a DIG-labeled probe to eliminate the need for radioactivity.
Keywords: miRNAs, DIG, splinted ligation, EDC cross-linking, Northern blots
Abstract
The discovery of microRNAs (miRNAs), which are ∼21–23 nucleotides that can regulate targeted mRNA by transcript cleavage or protein translation suppression, has changed the landscape of biomedical field greatly. At present, Northern blot analysis based on radioisotopes is still the most popular method on the detection of miRNAs for its high sensitivity. However, radioisotopes have been known for certain disadvantages, such as instability, expense, and safety; thus, developing a nonradioactive and highly sensitive method is needed. Here, we report a simple, nonradioactive, and sensitive method for miRNAs detection based on 5′-phos-3′-DIG–labeled probes prepared through splinted ligation and EDC cross-linking (DSLE). The method was more sensitive than traditional Northern blots with a DIG-labeled DNA probe and can detect as low as 2 fmol of miRNAs. The whole procedure can be completed within 6–8 h. DSLE method is very convenient, cost-effective, time-saving, and highly sensitive.
INTRODUCTION
MicroRNAs (miRNAs) are small noncoding RNAs with ∼21–23 nucleotides (nt) that can cause targeted mRNA degradation or protein translation suppression by base-pairing with a complementary sequence (Ambros 2003). Recent works suggest that miRNAs play important roles in cell differentiation, development, and various diseases including cancer (Brennecke et al. 2003; Lu et al. 2005).
The profiling of miRNA expression is important in understanding of their biological functions. Currently, the techniques on miRNAs detection are divided into two categories: one is based on probe hybridization, including Northern blots, microarray, nanogold labeling assay, and single molecule detection (Sempere et al. 2004; Grundhoff et al. 2006; Neely et al. 2006; Yang et al. 2008). Another is based on amplification, such as PCR amplification assay, RCA (rolling-circle amplification) assay (Jonstrup et al. 2006), primer invader assay, and clone sequencing assay (Allawi et al. 2004; Ambros and Lee 2004; Shi and Chiang 2005). Among all these methods for miRNAs detection, Northern blots analysis via radioisotopes is still the most widely used method, which is not only complicated and time-consuming but also expensive and unsafe to researchers and the environment. On the other hand, DIG-labeled system possesses the advantages of short exposure time, better stability, inexpensive cost, and better safety, yet with the disadvantage of relatively low sensitivity (Holtke and Kessler 1990; Ramkissoon et al. 2006). Nevertheless, a sensitive nonradioactive Northern blot method (LED) for miRNA detection with short exposure time (30 sec) has been developed by combining with LNA-modified oligonucleotide, DIG-labeled probe, and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) cross-linking (Kim et al. 2010). Unfortunately, the cost of LED is very high due to the use of LNA-modified oligonucleotides, which largely limits its application.
In view of the low sensitivity and overelaboration of traditional Northern blots, Maroney et al. (2007) developed a rapid and more sensitive method by applying splinted ligation incorporating a radioisotope-labeled probe (Maroney et al. 2007).
In traditional Northern blots, RNA was blotted to the membrane cross-linked using methods such as UV light. However, these methods are not effective for short RNAs like miRNAs (Pall et al. 2007; Pall and Hamilton 2008). It has been proven that using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) to cross-link small RNAs can improve the detection sensitivity up to 25- to 50-fold.
In order to improve the current methods on miRNAs detection, we took advantage of a DIG-labeled system, splinted ligation, and EDC cross-linking (DSLE) and combined them together. This method is very easy to follow and takes only 6–8 h to accomplish. Moreover, DSLE is much more sensitive than traditional DIG-labeled Northern blots, which is good enough to detect less abundant miRNAs.
MATERIALS AND METHODS
Cell lines
HeLa Cells were maintained in DMEM media supplemented with 10% (v/v) fetal calf serum. Giardia canis (Changchun strain) trophozoites infected with G. canis virus (GCV) were cultured as previously described (Lu et al. 1990).
RNA isolation
Total RNAs were extracted from HeLa Cells or Giardia trophozoites using TRIzol (Invitrogen) according to the manufacturer's protocol, with the last step modified to use 80% EtOH for a RNA wash instead of 75% EtOH to avoid the loss of small RNAs.
Oligonucleotides
DNA oligonucleotides were synthesized by Sangon Biotech. The probe was as follows: 5′-phos-CGCTTATGACATTC-DIG-3′. Each bridge oligonucleotide was designed according to target miRNA, as follows: hsa-miR-21 bridge, 5′-GAATGTCATAAGCGTCAACATCAGTCTGATAAGCTA-3′; hsa-miR-16 bridge, 5′-GAATGTCATAAGCGCGCCAATATTTACGTGCTGCTA-3′; hsa-miR-20a bridge, 5′-GAATGTCATAAGCGCTACCTGCACTATAAGCACTTTA-3′; and Giardia-miR2 bridge, 5′-GAATGTCATAAGCGGGACTATAGGGGCGGTGATTAGGCTG-3′.
The hsa-miR-21 RNA Oligonucleotide from TaKaRa was as follows: hsa-miR-21 synthetic RNA, 5′-phos-UAGCUUAUCAGACUGAUGUUGA-3′; hsa-miR-21 DIG probe, 5′-DIG-TCAACATCAGTCTGATAAGCTA-3′; and U6 DIG probe, 5′-DIG-ATATGGAACGCTTCACGAATT-3′.
The basic principle and procedure for miRNA detection by DSLE
Figure 1 is a schematic illustration of the basic principle and procedure of DSLE.
FIGURE 1.
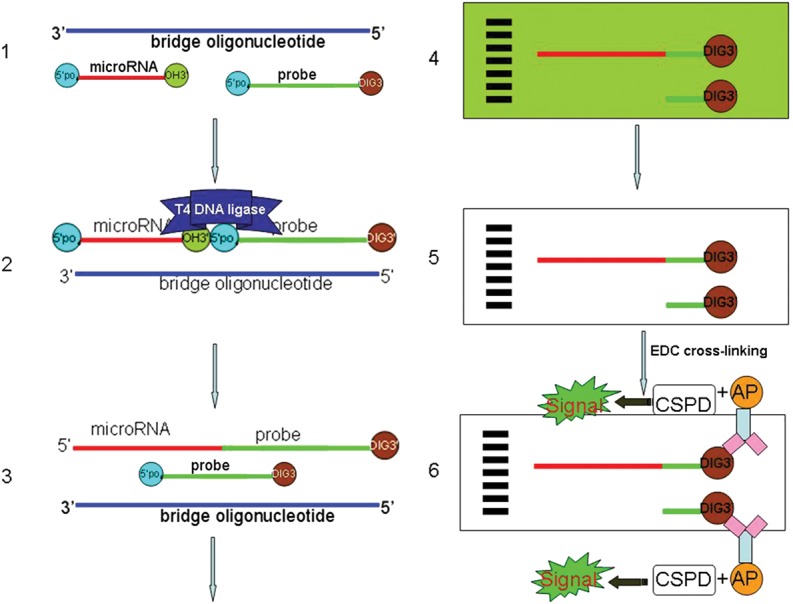
Schematic illustration of the protocol. (1) Annealing of the labeled probe, miRNA, and bridge oligonucleotide by slowly decreasing temperature after denaturation. (2) Filling in the nick with T4 DNA ligase. (3) Denaturing the double-stranded structure. (4) Fifteen percent urea–polyacrylamide gel electrophoresis. (5) Transferring the products to a membrane and cross-linking RNA by EDC. (6) Detecting the DIG-labeled miRNAs and free probes on the membrane using chemiluminescent substrates.
Briefly, the bridge oligonucleotide anneals with both the 5′-phosphate-3′-DIG–labeled DNA probe and target miRNA with a nick between them. Then, the nick between the 3′ hydroxyl group of the miRNA and the 5′ phosphate group from DNA probe was filled in by T4 DNA ligase. Therefore the target miRNA was labeled with DIG. Consequently, there would be two different-sized molecules labeled by DIG. One is the DNA probe of 14 nt, and the other is the probe plus miRNA. These two different molecules could be separated through 15% urea–polyacrylamide gels. The nucleotides were transferred from gels to blots and cross-linked by EDC. Finally, the two different-sized DIG-labeled molecules can be detected using chemiluminescent substrates.
miRNA analysis
miRNA was detected using DSLE. Splinted ligation was preformed according to a previously reported method (Maroney et al. 2007). Briefly, equal concentrations (0.1 μM) of oligonucleotide and probe were mixed with total RNA in a capture buffer (20 mM Tris-HCl at pH 8.0 and 75 mM KCl). The mixture was incubated for 1 min at 95°C, for 2 min at 65°C, and for 10 min at 37°C to allow the probe and miRNA to anneal with bridge oligonucleotide. T4 DNA ligase (40 units, NEB) was added to the mixture to fill in the nick between miRNA and probe molecules. The ligation reaction was incubated for 1 h at 30°C. The reaction was mixed with an equal amount of RNA loading buffer (TaKaRa), denatured for 5 min at 95°C, and loaded onto a denaturing 15% urea–polyacrylamide gel. Gels were transferred onto Immobilon-Ny+ Membrane by semi-dry transferring and cross-linked by EDC. Probe detection was performed using the DIG luminescent detection kit (Roche) according to manufacturer's protocol. Briefly, blots were blocked in blocking buffer for 0.5∼1 h at 25°C and incubated in antibody solution (anti-DIG, alkaline phosphatase conjugated antibody diluted in blocking buffer; 1:15,000) for 0.5 h at 25°C, followed by washing steps (twice, each 5 min at 25°C) with washing buffer (0.1 M maleic acid, 0.15 M NaCl at pH 7.5, 0.3% Tween 20 [v/v]). After equilibration in detection buffer (0.1 M Tris, 0.1 M NaCl), blots were incubated with chemiluminescent substrate chloro-5-substituted adamantyl-1,2-dioxetane phosphate (CSPD) at room temperature for 5 min and then exposed to films.
RESULTS
Detection of miRNAs with DSLE
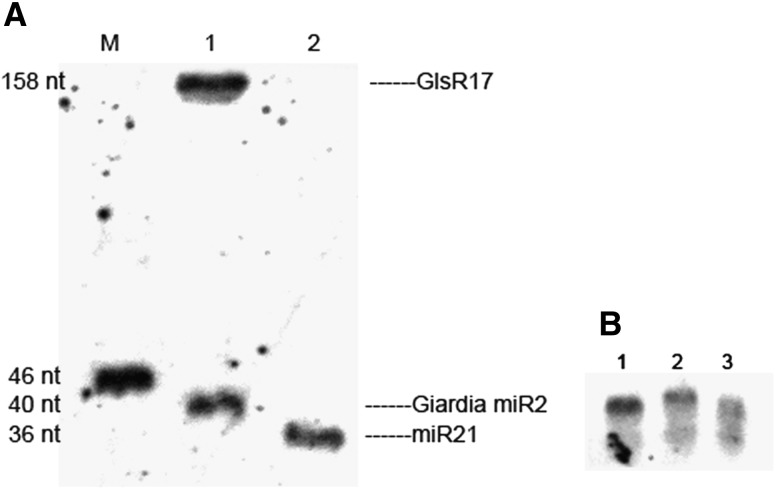
To examine the feasibility of this method on detecting miRNAs from total RNA, we performed analyses on hsa-miR-21 that is highly expressed in HeLa cells. A band with the prospective size (36 nt) was observed clearly (Fig. 2A). There were no other bands except for the target (36 nt) and the free probe (14 nt). The results indicated that this method can specifically detect abundant hsa-miR-21. We next chose the less abundant miRNA, Giardia miR2, which can be detected by using splinted ligation but not by traditional Northern blots, to investigate the specificity and sensitivity of this method. As expected, a strong band was observed, indicating that this method is very specific and sensitive (Fig. 2A). In addition, we have performed hsa-miR-16 (low quantities) and hsa-miR-20a detections with DSLE, and the results indicated that DSLE is sensitive enough to detect low-abundance miRNAs. The different mobility between hsa-miR-16 (22 nt) and hsa-miR-20a (23 nt) showed that DSLE is applicable for miRNA variant detection (Fig. 2B).
FIGURE 2.
Detection of miRNAs with DSLE. (A) Total RNAs were analyzed by DSLE for the detection of hsa-miR-21 and Giardia miR2: (lane M) 46 nt 5′-DIG-labeled oligonucleotide, (lane 1) 5 μg G. canis total RNA, and (lane 2) 2 μg HeLa RNA. (B) hsa-miR-21, hsa-miR-20a, and hsa-miR-16 were detected by DSLE using 4 μg HeLa RNA: (lane 1) hsa-miR-21, (lane 2) hsa-miR-20a, and (lane 3) hsa-miR-16.
In addition to the target and the free probe bands, a larger band was also observed, which might represent the precursor of miR2, as the miR2 bridge oligonucleotide is also complementary to the 3′ end of the precursor. The results showed that this method may be more sensitive than traditional Northern blots and can detect longer RNAs with 3′-hydroxyl-terminals as previously reported (Maroney et al. 2007).
Concentration optimization of DIG-labeled probe in DSLE
In splinted ligation, one molecule of 3′-DIG–labeled probe detects one molecule of miRNA. We inferred that more probes used in the reaction should yield stronger signals. Thus, we performed the assay with serial dilutions of 3′-DIG–labeled probe and bridge oligonucleotide (1 μM, 0.1 μM, 0.01 μM). As shown in Figure 3, the results were consistent with the conjecture. The strength of signals produced were correlated with the concentrations of probe in the assay. However, the results also indicated that higher probe concentrations may lead to apparently nonspecific bindings on the blot. Considering the balance between specificity and sensitivity, we recommend using 0.1 μM 3′-DIG–labeled probe and 0.1 μM bridge oligonucleotide for DSLE.
FIGURE 3.
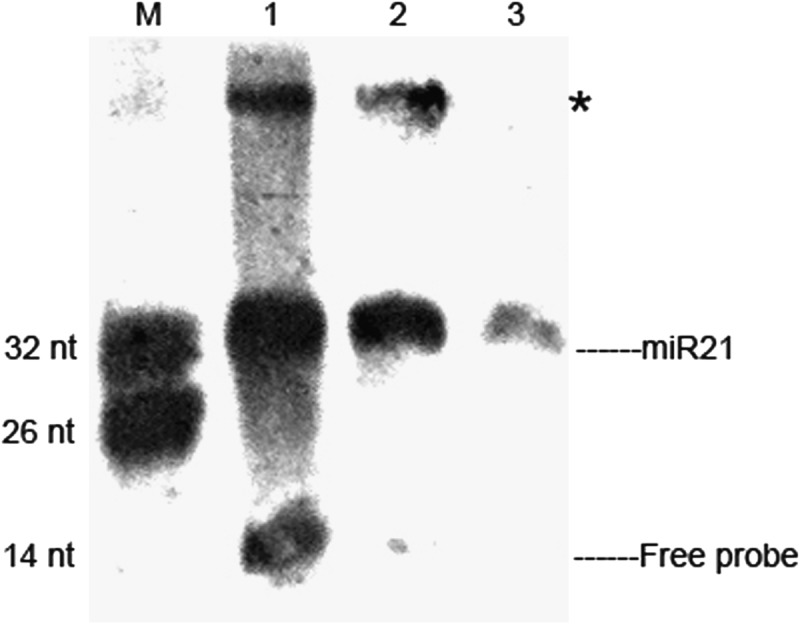
Concentration optimization of DIG-labeled probe in DSLE. Three different probe concentrations (1 μM, 0.1 μM, and 0.01 μM) were used to perform this assay. Ten micrograms of HeLa RNA was used for each probe concentration: (lane M) mixture of 26 nt and 32 nt 5′-DIG–labeled oligonucleotides, (lane 1) 1 μM probe concentration, (lane 2) 0.1 μM probe concentration, and (lane 3) 0.01 μM probe concentration. (*) Extra bands.
Comparison of DSLE to other methods
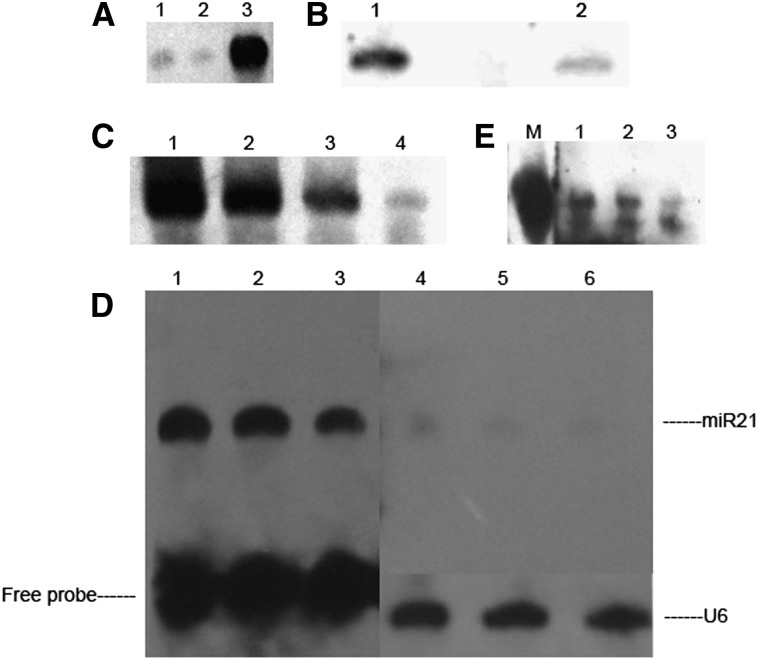
To assess DSLE, a synthetic 5′-phosphorylated RNA corresponding to mature hsa-miR-21 was used in the assay. We first compared the sensitivity of DIG-labeled splinted ligation to traditional DIG-labeled Northern blots. As expected, DIG-labeled splinted ligation was more sensitive than DIG-labeled Northern blots (Fig. 4A). To enhance the sensitivity, we performed the DIG-labeled splinted ligation based on EDC cross-linking instead of UV cross-linking. The results indicated that EDC cross-linking resulted in stronger signals (Fig. 4B). Next, we carried out the assay using synthetic hsa-miR-21 in serial dilutions from 200 fmol to 2 fmol to test the sensitivity of DSLE. As shown in Figure 4C, DSLE can detect at least 2 fmol miRNA, suggesting its high sensitivity. Finally, we contrasted the sensitivity of DSLE with traditional DIG-labeled Northern blots. As shown in Figure 4D, both methods can detect the mature hsa-miR-21 from HeLa cell RNA. However, our DSLE method displayed much stronger signals than traditional DIG-labeled Northern blots. Also, we examined the detection limit of DSLE using HeLa cell RNA. Mature hsa-miR-21 can still be detected by DSLE when using as little as 0.13 μg HeLa cell total RNA (Fig. 4E). In contrast to the data shown in Maroney et al. (2007), DSLE might be comparable with traditional 32p-labeled Northern blots.
FIGURE 4.
(A) Sensitivity comparison between DIG-labeled splinted ligation and traditional DIG-labeled Northern blots with both assays using UV cross-linking: (lane 1) 60 ng of synthetic hsa-miR-21 was assayed by traditional DIG-labeled Northern blots, (lane 2) 30 ng of synthetic hsa-miR-21 was assayed by traditional DIG-labeled Northern blots, and (lane 3) 30 ng synthetic hsa-miR-21 was assayed using DIG-labeled splinted ligation. (B) Sensitivity comparison between EDC cross-linking (lane 1) and UV cross-linking (lane 2) based on splinted ligation. We used 20 fmol synthetic hsa-miR-21 for splinted ligation with an equal amount of sample (15 μL) loaded on each lane. All steps were the same except for the cross-linking step. (C–E) Assessment of the effect of splinted ligation and EDC cross-linking on DSLE and the comparison between DSLE and other methods using hsa-miR-21. (C) Quantification of the absolute detective sensitivity of DSLE using synthetic hsa-miR-21: (lane 1) 200 fmol, (lane 2) 40 fmol, (lane 3) 20 fmol, and (lane 4) 2 fmol. (D) Comparison of the sensitivity of DSLE and traditional DIG-labeled Northern blots on miR21 detection: 7 μg (lanes 1,4), 4 μg (lanes 2,5), and 2 μg (lanes 3,6) HeLa RNA were used for each assay. Photo-exposure time was 10 min. (E) The detection limit assay of DSLE using total RNA. The concentration of RNA was as follows: (lane 1) 0.78 μg, (lane 2) 0.39 μg, and (lane 3) 0.13 μg. (Lane M) 30 ng of synthetic hsa-miR-21 was detected by DSLE.
DISCUSSION
In the present study, we combined DIG-labeled probe, splinted ligation with EDC cross-linking and developed a safe, convenient, cost-effective, and time-saving method to detect miRNAs. As seen from the DSLE protocol, the 3′-DIG–labeled probe was used instead of radioisotope-labeled probe, which makes this method safer and easier to handle. There is no need to change the 3′-DIG–labeled probe when detecting a different miRNA, only the nonlabeled bridge oligos were changed. The 3′-DIG–labeled probes are more stable and cheap compared with the radioisotope-labeled probe and LNA oligonucleotide probe (Valoczi et al. 2004). In addition, the whole procedure can be completed within 6–8 h.
More importantly, DSLE is much more sensitive than traditional DIG-labeled Northern blots, as suggested by the detection of low-abundance miRNAs, such as hsa-miR-16 and G. canis miR2. Although we only demonstrated its ability to detect miRNAs, DSLE could also be used to detect other small RNAs and even larger RNAs (i.e., 144 nt) (Fig. 2A) with a 3′-hydroxyl terminus. Also, it is possible that if more DIG molecules are conjugated to the 3′ end of the probe, the sensitivity of DSLE could be improved significantly.
Although the sensitivity of DSLE is lower than Nilsen's 32p-labeled splinted ligation method (0.01–0.02 fmol miRNAs) (Maroney et al. 2007), which is almost 200 times more sensitive than the 2 fmol level described in DLSE, DSLE is sensitive enough to detect low-abundance miRNAs hsa-miR-16 from as little as 4 μg of total RNA (see Fig. 2B), which indicated that DSLE could be an excellent alternative to traditional Northern blot when radioisotope-labeled or LNA probes are not available. The different sensitivities exhibited by DLSE and Nilsen's 32p-labeled splinted ligation method might be as follows: (1) Since no membrane transferring procedure involved in Nilsen's method, fewer RNAs were lost, which may lead to increased sensitivity; and (2) T4 DNA ligase is crucial in splinted ligation method, and the T4 DNA ligase used in our test (Takara/NEB) is different from Nilsen's method (USB). Our data indicated that 40 units of T4 DNA ligase from NEB in one reaction gives comparable results obtained with 350 units of T4 ligase from TaKaRa in one reaction, which suggested that T4 DNA ligase from NEB is more efficient. We believe that optimization for different parameters, such as annealing buffer and ligation efficiency, will further improve the sensitivity of this method.
In conclusion, a highly sensitive DSLE-on-miRNAs detection method was established. This method is environmentally safe, cost-effective, and time-saving on miRNA detection.
ACKNOWLEDGMENTS
The study is supported by National Science Foundation of China (no. 30970322). The experiments conducted in this study comply with the current laws of China. We thank all the persons who provided help to this work.
REFERENCES
- Allawi HT, Dahlberg JE, Olson S, Lund E, Olson M, Ma WP, Takova T, Neri BP, Lyamichev VI 2004. Quantitation of microRNAs using a modified invader assay. RNA 10: 1153–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V 2003. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 113: 673–676 [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC 2004. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol 265: 131–158 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36 [DOI] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS, Ganem D 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human γ-herpesviruses. RNA 12: 733–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtke HJ, Kessler C 1990. Non-radioactive labeling of RNA transcripts in vitro with the hapten digoxigenin (DIG); hybridization and ELISA-based detection. Nucleic Acids Res 18: 5843–5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonstrup SP, Koch J, Kjems J 2006. A microRNA detection system based on padlock probes and rolling circle amplification. RNA 12: 1747–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B 2010. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res 38: e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SQ, Wang ZY, Zhu H 1990. Establishment of an axenic culture of Giardia lamblia through preliminary passage in suckling gerbil. Chin Med J (Engl) 103: 583–587 [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. 2005. MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Maroney PA, Chamnongpol S, Souret F, Nilsen TW 2007. A rapid, quantitative assay for direct detection of microRNAs and other small RNAs using splinted ligation. RNA 13: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely LA, Patel S, Garver J, Gallo M, Hackett M, McLaughlin S, Nadel M, Harris J, Gullans S, Rooke J 2006. A single-molecule method for the quantitation of microRNA gene expression. Nat Methods 3: 41–46 [DOI] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ 2008. Improved Northern blot method for enhanced detection of small RNA. Nat Protoc 3: 1077–1084 [DOI] [PubMed] [Google Scholar]
- Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A 2007. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res 35: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramkissoon SH, Mainwaring LA, Sloand EM, Young NS, Kajigaya S 2006. Nonisotopic detection of microRNA using digoxigenin labeled RNA probes. Mol Cell Probes 20: 1–4 [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Chiang VL 2005. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39: 519–525 [DOI] [PubMed] [Google Scholar]
- Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z 2004. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res 32: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WJ, Li XB, Li YY, Zhao LF, He WL, Gao YQ, Wan YJ, Xia W, Chen T, Zheng H, et al. 2008. Quantification of microRNA by gold nanoparticle probes. Anal Biochem 376: 183–188 [DOI] [PubMed] [Google Scholar]