Abstract
The University of California Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu) offers online public access to a growing database of genomic sequence and annotations for a large collection of organisms, primarily vertebrates, with an emphasis on the human and mouse genomes. The Browser’s web-based tools provide an integrated environment for visualizing, comparing, analysing and sharing both publicly available and user-generated genomic data sets. As of September 2013, the database contained genomic sequence and a basic set of annotation ‘tracks’ for ∼90 organisms. Significant new annotations include a 60-species multiple alignment conservation track on the mouse, updated UCSC Genes tracks for human and mouse, and several new sets of variation and ENCODE data. New software tools include a Variant Annotation Integrator that returns predicted functional effects of a set of variants uploaded as a custom track, an extension to UCSC Genes that displays haplotype alleles for protein-coding genes and an expansion of data hubs that includes the capability to display remotely hosted user-provided assembly sequence in addition to annotation data. To improve European access, we have added a Genome Browser mirror (http://genome-euro.ucsc.edu) hosted at Bielefeld University in Germany.
INTRODUCTION
The University of California Santa Cruz (UCSC) Genome Browser (1,2) at http://genome.ucsc.edu is a web-based resource for the scientific, medical and academic research communities that provides timely, convenient access to a database of high-quality genome sequence and annotations. The Browser tools facilitate the visualization, comparison and analysis of both hosted and user-generated data sets ranging from a genome-wide perspective down to the base level.
The Genome Browser database contains genome sequence from GenBank (3) for a wide selection of organisms, many with multiple assembly versions. In September 2013 our database included 13 primates, 33 additional mammals, 17 non-mammalian vertebrates, 13 insects, 6 worms and 5 other invertebrates. Annotation data for each genome assembly are displayed graphically as ‘tracks’ aligned to the genomic sequence and grouped according to shared characteristics, such as gene predictions or comparative genomics. The level of annotation varies among organisms. At a minimum, most assemblies offer mapping and sequence annotation tracks describing assembly, gap and GC content, and alignments of mRNA, EST and RefSeq (3) genes (available on approximately one-half of the assemblies) from GenBank. Some assemblies provide additional gene annotation tracks, such as Ensembl Genes (4) and Human Proteins, as well as multiple sequence alignments (multiz) (5) and pairwise genomic alignments between assemblies to facilitate comparative and evolutionary investigations. The heavily annotated human genome offers extensive conservation and evolutionary comparisons, a large collection of gene models including the locally generated UCSC Genes track (6,7), regulation, expression, epigenetics and tissue differentiation, variation, phenotype and disease association data, and data that have been text-mined from publications. Much of our annotation data is obtained through external collaboration. When available, links are provided to the complementary annotations in the Ensembl and NCBI browsers, and to supplementary information on other websites.
The Genome Browser serves as the repository for human and mouse genome data that was contributed through September 2012 by the Encyclopedia of DNA Elements (ENCODE) Consortium (8,9). During the transition of the ENCODE Data Coordination Center role to a joint collaboration with Stanford University, the Genome Browser team has continued to add significant new content to the ENCODE data portal (http://encodeproject.org) and publish newly reprocessed ENCODE data sets (10).
In addition to the native data sets local to the UCSC servers, the Genome Browser offers several options to users for viewing their own sequence and annotations: track and assembly data hubs, custom tracks and sessions. Alternatively, the Genome Browser database and tools may be installed on a local server for customized use (see http://genome.ucsc.edu/license/ for more information). Instructions for downloading the data, software and source code may be found at http://hgdownload.soe.ucsc.edu/downloads.html.
The following sections highlight the genome assembly and annotation data sets added to the Genome Browser since the last update in this journal and describe the significant new features and capabilities of our data access tools.
GENOME BROWSER DATA SETS
New genome assemblies
During the past year the UCSC team added 35 vertebrate assemblies to the Genome Browser (Table 1), including the premier releases of 20 species. In line with our focus on primates and other vertebrates, the group of newly introduced species features 4 primates (baboon, mouse lemur, squirrel monkey and tarsier), 12 additional mammals (alpaca, dolphin, ferret, hedgehog, kangaroo rat, manatee, megabat, rock hyrax, shrew, sloth, southern white rhinoceros and tree shrew) and 4 additional vertebrates (American alligator, Atlantic cod, budgerigar and coelacanth). Several of the new assemblies were added to support the generation of the 60-species Conservation track released in 2012 on the GRCm38/mm10 mouse assembly, and many of these were originally sequenced and assembled for the Mammalian Genome Project (11). We plan to release a preliminary Browser with a minimal annotation set on the new GRCh38/hg38 human assembly in late 2013 or early 2014. Beginning with this new release, the numeric portion of the UCSC human assembly version name will match the Genome Reference Consortium version number to reduce confusion.
Table 1.
New and updated genome assemblies added to the Genome Browser since September 2012
| Common name | Scientific name | Sequencing center | UCSC ID | Seq. ctr ID |
|---|---|---|---|---|
| Primates | ||||
| Baboon | Papio hamadryas | Baylor College of Medicine HGSC | papHam1 | Pham_1.0 |
| Baboon | Papio anubis | Baylor College of Medicine HGSC | papAnu2 | Panu_2.0 |
| Bushbaby | Otolemur garnettii | Broad Institute | otoGar3 | OtoGar3 |
| Chimpanzee | Pan troglodytes | Chimpanzee Sequencing and Analysis Consortium | panTro4 | Build 2.1.4 |
| Gibbon | Nomascus leucogenys | Gibbon Genome Sequencing Consortium | nomLeu2 | Nleu1.1 |
| nomLeu3 | Nleu3.0 | |||
| Mouse lemur | Microcebus murinus | Broad Institute | micMur1 | MicMur1.0 |
| Rhesus macaque | Macaca mulatta | Beijing Genomics Institute | rheMac3 | CR_1.0 |
| Squirrel monkey | Saimiri boliviensis | Broad Institute | saiBol1 | SaiBol1.0 |
| Tarsier | Tarsius syrichta | Broad Institute | tarSyr1 | Tarsyr1.0 |
| Other mammals | ||||
| Alpaca | Vicugna pacos | Broad Institute | vicPac1 | VicPac1.0 |
| vicPac2 | VicPac2.0 | |||
| Armadillo | Dasypus novemcinctus | Baylor College of Medicine HGSC | dasNov3 | DasNov3 |
| Cat | Felis catus | International Cat Genome Sequencing Consortium | felCat5 | Felis_catus-6.2 |
| Dolphin | Tursiops truncatus | Baylor College of Medicine HGSC | turTru2 | Ttru_1.4 |
| Ferret | Mustela putorius furo | Ferret Genome Sequencing Consortium | musFur1 | MusPutFur1.0 |
| Hedgehog | Erinaceus europaeus | Broad Institute | eriEur1 | Draft_v1 |
| Kangaroo rat | Dipodomys ordii | Baylor College of Medicine HGSC, Broad Institute | dipOrd1 | DipOrd1.0 |
| Manatee | Trichechus manatus latirostris | Broad Institute | triMan1 | TriManLat1.0 |
| Megabat | Pteropus vampyrus | Broad Institute | pteVam1 | PteVap1.0 |
| Naked mole rat | Heterocephalus glaber | Broad Institute | hetGla2 | HetGla_female_1.0 |
| Pig | Sus scrofa | Swine Genome Sequencing Consortium | susScr3 | Sscrofa10.2 |
| Pika | Ochotona princeps | Broad Institute | ochPri2 | OchPri2 |
| Rock hyrax | Procavia capensis | Baylor College of Medicine HGSC | proCap1 | Procap1.0 |
| Shrew | Sorex araneus | Broad Institute | sorAra1 | SorAra1.0 |
| Sloth | Choloepus hoffmanni | Broad Institute | choHof1 | ChoHof1.0 |
| Southern white rhinoceros | Ceratotherium simum simum | Broad Institute | cerSim1 | cerSimSim1.0 |
| Squirrel | Spermophilus tridecemlineatus | Broad Institute | speTri2 | SpeTri2.0 |
| Tree shrew | Tupaia belangeri | Broad Institute | tupBel1 | Tupbel1.0 |
| Other vertebrates | ||||
| American alligator | Alligator mississippiensis | Int’l Crocodilian Genomes Working Group | allMis1 | allMis0.2 |
| Atlantic cod | Gadus morhua | Genofisk | gadMor1 | GadMor_May2010 |
| Budgerigar | Melopsittacus undulatus | Genome Institute at Wash. Univ. St. Louis | melUnd1 | v6.3 |
| Coelacanth | Latimeria chalumnae | Broad Institute | latCha1 | LatCha1 |
| Lamprey | Petromyzon marinus | Genome Institute at Wash. Univ. St. Louis | petMar2 | WUGSC 7.0 |
| Nile tilapia | Oreochromis niloticus | Broad Institute | oreNil2 | OreNil1.1 |
The ‘UCSC ID’ column shows the Genome Browser database designation for the genome assembly.
As the number of vertebrate assemblies deposited into GenBank increases, we continue to explore options for providing timely, maximum coverage of genome assemblies in the Genome Browser. Assembly data hubs (described below) offer a potential solution for streamlining our process for hosting genome assemblies, as well as providing our users with an easy way to visualize and share their own genome sequences in the Genome Browser.
New and updated annotations
We added many new annotation data sets to the Genome Browser in the past year, and several existing data sets underwent major revisions. Our human and mouse assemblies, which receive the bulk of attention from our user community, are the most richly annotated. This section highlights some of the new annotation tracks released this year. See Table 2 for a complete list of recent releases.
Table 2.
New and updated annotation data sets added to the Genome Browser between September 2012 and September 2013
| Annotation track | Assembly |
|---|---|
| Human genome | |
| 1000 Genomes Phase 1 Integrated Variant Calls | hg19 |
| 1000 Genomes Phase 1 Paired-end Accessible Regions | hg19 |
| Affymetrix CytoScan HD Array | hg19 |
| Coriell Cell Line Copy Number Variants | hg19 |
| Denisova: Modern Human Derived, Sequence Reads, Variant Calls, Variant Calls from 11 Modern Human Genome Sequences | hg19 |
| DGV: Structural Variation | hg18-19 |
| DNaseI Hypersensitivity Uniform Peaks— ENCODE/Analysis | hg19 |
| ENCODE Regulation: DNaseI HS Clusters, Transaction Factor ChiP-seq Clusters | hg19 |
| GENCODE Genes v14, v17 | hg19 |
| GeneReviews | hg18-19 |
| GRCh37 Patch 10 | hg19 |
| GWAS Catalog of Published Genome-Wide Association Studies | hg18-19 |
| Human Gene Mutation Database (HGMD) | hg19 |
| Leiden Open Variation Database (LOVD) | hg19 |
| Pfam domains in UCSC Genes | hg19 |
| Proteogenomics and GENCODE Mapping—ENCODE | hg19 |
| qPCR Primers | hg19 |
| Reactome v41 | hg17-19 |
| Retroposed Genes | hg19 |
| SNPs (Build 137): All SNPs, Common SNPs, Flagged SNPs, Mult. SNPs | hg19 |
| SNPs (Build 138): All SNPs, Common SNPs, Flagged SNPs, Mult. SNPs | hg19 |
| Transcription Factor ChIP-seq Uniform Peaks— ENCODE/Analysis | hg19 |
| UCSC Genes | hg19 |
| UniProt Mutations | hg19 |
| Mouse genome | |
| 60-species Conservation | mm10 |
| GRC Incident Database | mm10 |
| GRCm38 Patch Release 1 | mm10 |
| Mouse strain variants | mm10 |
| qPCR Primers | mm10 |
| Reactome v.41 | mm8-9 |
| SNPs (Build 137) | mm10 |
| UCSC Genes | mm10 |
| Cow genome | |
| NumtS Nuclear Mitochondrial Sequences | bosTau6 |
| Pig genome | |
| NumtS Nuclear Mitochondrial Sequences | susScr2 |
| Multiple genomes | |
| Ensembl Genes | Many |
| Human proteins | Many |
| Publications track | Many |
Gene annotations
The UCSC Genes track, which includes protein-coding genes and non-coding RNA genes from RefSeq, GenBank, CCDS (12), Rfam (13) and the tRNA Genes track (14), was updated on both the GRCh37/hg19 human and GRCm38/mm10 mouse assemblies. The human UCSC Genes set increased by 2038 transcripts to a total of 82 960 transcripts, 92% of which did not change between versions. The number of genes, defined as clusters of transcripts with overlapping exons on the same strand, increased by 621 genes to 31 848. In the mouse UCSC Genes set, the number of transcripts grew by 3702 transcripts to 59 121, with 88% remaining the same between versions. The number of genes increased by 2566 genes to 31 227. For more information on the latest methods used to generate the UCSC Genes data, refer to the description pages that accompany the tracks. We also updated the GENCODE Genes (15) track on the latest human assembly to version 17.
Variation data
We update our SNP annotations for the human and mouse (and occasionally for other species) whenever a new version is released by dbSNP. The latest human and mouse assemblies were updated to dbSNP Build 137 in 2012–13, and the human assembly SNP tracks were updated to dbSNP Build 138 in October 2013. The annotation includes an ‘All SNPs’ track that contains all mappings of reference SNPs to the human assembly, as well as three SNP subsets: Common SNPs (those with at least 1% minor allele frequency), Flagged SNPs (annotated by dbSNP as ‘clinical’) and Mult. SNPs (those that map to multiple genomic loci, and therefore should be viewed with suspicion). The updated tracks contain additional annotation data not included in previous dbSNP tracks, and offer coloring and filtering options for configuring the Genome Browser display.
This year we released three new tracks that describe human disease-associated genetic variation based on curated public data in the Leiden Open Variation Database (LOVD) (16), the Human Gene Mutation Database (HGMD) (17) and amino acid mutations in the UniProt database (18). We also added two annotation sets based on Phase 1 sequencing data from the 1000 Genomes Project (19). The integrated variant calls track, 1000G Ph1 Vars, shows single nucleotide variants (SNVs), indels and structural variants (SVs) that have been phased into independent haplotypes, which the Genome Browser clusters by local similarity for display. The paired-end accessible regions track, 1000G Ph1 Accsbl, shows which genome regions are more or less accessible to next-generation sequencing methods that use short, paired-end reads.
Comparative alignments
The 60-species multiple alignment and conservation track released in 2012 for the GRCm38/mm10 mouse assembly was the largest comparative alignment track generated by UCSC to date. In 2013, we undertook an ambitious project to produce a 100-species conservation track on the GRCh37/hg19 human assembly, released to the public in Nov. 2013. As part of this undertaking we have been evaluating software alternatives, such as Cactus (20), to extend the scalability of our multiple alignment pipeline, which has been challenged by the increasing number of species.
ENCODE data
In the past year UCSC has focused on improving the accessibility and usability of the ENCODE data hosted in the Genome Browser. The ENCODE Analysis Working Group (AWG) reprocessed the transcription factor ChIP-seq and DNaseI HS peak call data sets released through March 2012 using the uniform processing pipeline developed for the ENCODE Integrative Analysis effort. This reprocessing factored out many of the cross-lab differences, allowing the different data sets to be used more effectively in the same analyses. These reprocessed data sets were released on the Genome Browser as the Transcription Factor ChIP-seq Uniform Peaks track (within the ENC TF Binding super-track) and the DNaseI Hypersensitivity Uniform Peaks track (within the ENC DNase/FAIRE super-track). The new data sets that met a specific integrated quality metric defined by the AWG (http://genome.ucsc.edu/ENCODE/qualityMetrics.html) were then used to update the individual Transcription Factor ChIP-seq Clusters and Digital DNaseI Hypersensitivity Clusters tracks within the ENCODE Integrated Regulation super-track set, providing summary clustered views. ENCODE data hosted in the Browser has now been fully accessioned through the Gene Expressions Omnibus (GEO) repository (21) and cross-linked back to UCSC. We also added the ENCODE Integrative Analysis Data Hub to the Genome Browser public hubs page (http://genome.ucsc.edu/cgi-bin/hgHubConnect) to provide easy, integrated access to AWG data. Together with the Roadmap Epigenomic data track hub (22), the ENCODE data provide a comprehensive look at DNA landmarks across a large number of tissues.
The Genome Browser currently hosts a large amount of ChIP-seq data on transcription factors, many of which bind to specific DNA motifs. In late 2013, we plan to release an extension to this data type that displays the location of motifs within the peak and shows the sequence logo and matching score on the track details page for the peak.
Publications data
In 2012, we introduced a Publications track that shows mapped DNA and protein sequences, SNPs, cytogenetic bands and gene symbols that have been text-mined from biomedical articles in Elsevier, PubMed Central and other databases (23). In the past year we have doubled the number of research articles to more than 5 million, and now classify them into different categories (disease related, protein structure, cis-regulatory, etc.) depending on their keyword content. The categories are differentiated by color in the display, which can be filtered by categories or publishers.
Denisova data
In February 2013, we released a set of Denisova annotations tracks in conjunction with the publication of a paper by Meyer et al. (24). The sequence data were derived by applying a novel single-stranded DNA library preparation method to DNA previously extracted from 40 mg of a phalanx bone excavated from Denisova Cave in the Altai Mountains of southern Siberia. The Genome Browser tracks show mappings to the human reference sequence of high-coverage Denisova sequence reads, variant calls from sequence reads of 11 modern individuals and an archaic Denisovan individual, and mutations in the modern human lineage that rose to fixation or near fixation since the split from the last common ancestor with Denisovans, along with predicted functional effects from the Ensembl Variant Effect Predictor (25).
GENOME BROWSER SOFTWARE UPDATES
Track and assembly data hubs
In 2011 we introduced track data hubs (26), a means for users to import collections of their own locally hosted genome annotations into the Genome Browser where they may be organized, configured and viewed alongside native tracks. Track hubs now support four compressed binary indexed file formats: BigBed and BigWig (27), both developed at UCSC, BAM (28) and VCF/tabix (29). As genome sequencing becomes more accessible and cost-effective, we have faced a growing demand from researchers who wish to use the Genome Browser tools to browse and annotate genome sequences for which we do not host a database. In response to this need, we have extended the functionality of track data hubs to encompass entire assemblies that are not hosted natively on the Genome Browser. These ‘assembly data hubs’ enable researchers to import both the underlying reference sequence as well as data tracks annotating that sequence into the Genome Browser for display and analysis. The genome sequence is stored in the UCSC .2bit format and made available on the user’s remote web server, along with optional annotation data files stored in the same compressed binary formats supported by track data hubs. Track and assembly data hubs can be shared with others by providing the URL of the hub.txt file needed to load the hub. Hubs of general interest to the research community can be registered at UCSC for sharing on the Genome Browser website. We offer a growing collection of publicly shared track and assembly data hubs on the ‘Public Hubs’ tab on the Genome Browser Track Data Hubs web page (http://genome.ucsc.edu/cgi-bin/hgHubConnect), including data sets from the ENCODE AWG, the Roadmap Epigenomics Project (22) and the Blueprint Epigenome Project (30). For more information about creating and using assembly data hubs, refer to http://genomewiki.ucsc.edu/index.php/Assembly_Hubs and http://genome.ucsc.edu/goldenPath/help/hgTrackHubHelp.html.
Variant annotation integrator
To assist researchers in annotating and prioritizing thousands of variant calls from sequencing projects, we have developed a new software tool, the Variant Annotation Integrator (VAI). Given a set of variants uploaded as a custom track in either Personal Genome SNP (pgSnp) or VCF format, the VAI returns the predicted functional effect (e.g., synonymous, missense, frameshift, intronic) for each variant. The VAI can also provide several other types of relevant information, such as the dbSNP identifier if the variant is found in dbSNP, protein damage scores for missense variants from the Database of Non-synonymous Functional Predictions (dbNSFP) (31) and conservation scores computed from multiple-species alignments. Filters are available to focus results on the variants of greatest interest. The VAI can be accessed from the Genome Browser ‘Tools’ menu or through the VAI button on the ‘Manage Custom Tracks’ page that displays after a custom track is loaded into the Browser. For more information about the VAI, see http://genome.ucsc.edu/cgi-bin/hgVai.
Gene haplotype alleles
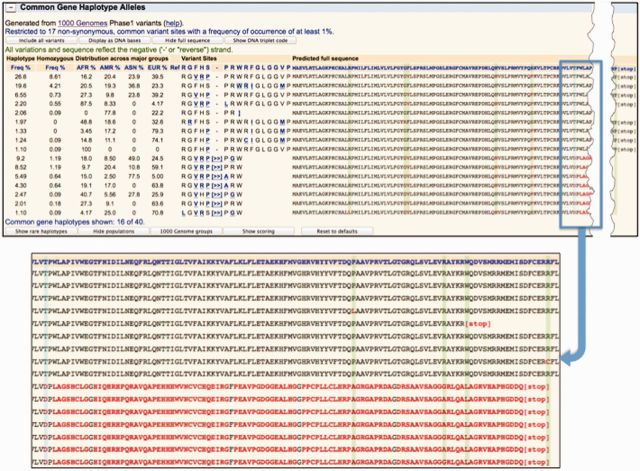
We have extended the protein-coding genes detail pages in the UCSC Genes track on the GRCh37/hg19 human assembly to include a section that displays and compares ‘gene haplotype alleles’ generated from phased chromosomal data from Phase 1 of the 1000 Genomes Project (19) (Figure 1). Each haplotype allele is a distinct set of variants found on at least one of the 1000 Genomes subject chromosomes. By default the common non-synonymous variants (those of at least 1% frequency) are displayed, although rare haplotypes are optionally available. The Browser shows the frequency of each haplotype in the 1000 Genomes populations and indicates the frequency with which it occurs homozygously. Unexpected frequencies of occurrence may be used to identify alleles that merit further study. Predicted protein sequence for common haplotypes can also be displayed, allowing differences among alleles to be used to identify differences at the amino acid level. To access the gene haplotype alleles information, go to the details page for any protein-coding gene in the UCSC Genes track (GRCh37/hg19 assembly) and click the ‘Gene Alleles’ link in the ‘Page Index’ matrix. For more information, see http://genome.ucsc.edu/goldenPath/help/haplotypes.html.
Figure 1.
The haplotype alleles display for the ABO gene, which encodes proteins related to the ABO blood group system. A large portion of the ‘Predicted full sequence’ section is truncated in the upper image for display purposes, and is shown in greater detail in the lower image. The leftmost columns of the top image indicate the frequency of each allele haplotype within the 1000 Genomes sample and the occurrence of homozygosity for each allele. In this instance the haplotype alleles display has been expanded to show the distribution of the haplotypes across the major 1000 Genomes population groups. The ‘Variant Sites’ columns summarize the non-synonymous variant sites that occur in at least 1% of the subject chromosomes, with the value from the reference genome (in this case GRCh37/hg19) indicated at the top of each variant column. In all but one case, the ‘O’ phenotype results from a common insertion (indicated by ‘-’ in the reference) causing a frameshift (indicated by ‘[≫]’) that results in a downstream premature stop codon, thus truncating the protein. Note that although certain haplotyes are more frequently found within one population, the insertion that gives rise to the majority of ‘O’ phenotypes is found across all populations, which may indicate that the insertion predates the most recent migration out of Africa. On the other hand, the haplotype in which the SNP variant introduces a stop codon at the variant site may have arisen in the Americas. The zoomed-in view of the ‘Predicted full sequence’ section in the bottom image shows the reference sequence (top row) and sequences incorporating the common non-synonymous variants. The residues corresponding to the variant sites are highlighted by green vertical bars, the site corresponding to the frameshift-causing insertion is highlighted by a blue bar and changes to the reference amino acid sequence are shown in red.
Updates to Browser display and navigation
During the past year, we have made several improvements to the Genome Browser web interface, many in response to requests from our users. We have updated the navigation menus for much of the website and simplified the background on the Genome Browser tracks page. The Browser now offers chromosome ideograms for genome assemblies that do not have a microscopically derived cytology. The drag-reorder feature in the Browser image now supports the vertical dragging of subtracks to any location in the image. We have also made display improvements to overlay wiggle tracks, and have improved the display speed of bigDataUrl custom tracks.
User training and mirror support
We continually update and expand our documentation and training materials, which offer extensive information on using the Genome Browser tools to explore UCSC-hosted data sets as well as custom sequence and annotation data hosted at user sites. We are broadening our onsite training program to include several additional geographical regions. To better support our Genome Browser mirror sites and source code users, we have rearchitected the software makefile system for our utilities and command-line tools to allow the compilation of specific tools independent of a full Browser installation. We have also adopted UDR (https://github.com/LabAdvComp/UDR), a new package that integrates rsync with the high-performance network protocol UDT, allowing quicker transfers of our large data sets to remote mirror sites.
FUTURE PLANS
During the upcoming year we will continue to add new and updated genome assemblies for vertebrate organisms as they become available in NCBI’s GenBank repository. We plan to release a preliminary Browser with a minimal annotation set on the new GRCh38/hg38 human assembly in late 2013 or early 2014. New annotation data display types and features will be added as required by new data sets. We plan to extend track hubs to support new file formats, such as the HAL hierarchical multiple alignment format (32), and to allow searching for tracks within a hub. The VAI will be expanded to include more input/upload options, output formats and annotation options.
CONTACTING US
To stay on top of the latest Genome Browser announcements, genome assembly releases, new software features, updates and training seminars, subscribe to the genome-announce@soe.ucsc.edu mailing list or follow @GenomeBrowser on Twitter. We have two public, moderated mailing lists for interactive user support: genome@soe.ucsc.edu for general questions about the Genome Browser and genome-mirror@soe.ucsc.edu for questions specific to the setup and maintenance of Genome Browser mirrors. Messages sent to these lists are archived on public, searchable Google Groups forums. You may also reach us privately at genome-www@soe.ucsc.edu, the preferred address for inquiring about mirror site licenses, reporting server errors or contacting us about confidential issues. You will find complete contact information, links to the browser’s Google Groups forums and access to our user suggestion box at http://genome.ucsc.edu/contacts.html.
FUNDING
This work was supported by the National Human Genome Research Institute [5U4 HG002371 to G.P.B., H.C., J.C., T.R.D., P.A.F., L.G., S.H., A.S.H., M.H., D.K., W.J.K., R.M.K., K.L., B.T.L., C.H.L, B.J.R., B.R., M.L.S. and A.S.Z.; 1U41HG006992 subcontract 60141508-106846-A to D.K., W.J.K., K.L., M.S.C., T.R.D., B.J.R., K.R.R., C.A.S. and A.S.Z]; National Institute of Dental and Craniofacial Research [5U01DE020057 subcontract 1000736806 to G.P.B. and R.M.K.]; National Cancer Institute [1U41HG007234 subcontract 2186-03 to M.D. and R.H.; 5U24CA143858 to M.C.]. European Molecular Biology Organization Long-Term Fellowship (in part) [ALTF 292-2011 to M.H.]; Howard Hughes Medical Institute fellow (to D.H.). Funding for open access charge: National Human Genome Research Institute.
Conflict of interest statement. G.P.B., H.C., M.D., T.R.D., P.A.F., L.G., D.H., R.A.H., S.H., A.S.H., D.K., W.J.K., R.M.K., K.L., C.H.L., B.J.R., B.R., K.R.R., C.A.S. and A.S.Z. receive royalties from the sale of UCSC Genome Browser source code licenses to commercial entities. W.J.K. works for Kent Informatics.
ACKNOWLEDGEMENTS
The authors would like to thank the many data contributors and collaborators whose work makes the Genome Browser possible, our Scientific Advisory Board for guiding our efforts, our users for their consistent support and valuable feedback, and our outstanding team of system administrators: Jorge Garcia, Erich Weiler and Gary Moro.
REFERENCES
- 1.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013;41:D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchette M, Diallo AB, Green ED, Miller W, Haussler D. Computational reconstruction of ancestral DNA sequences. In: Murphy WJ, editor. Methods in Molecular Biology: Phylogenomics. New York: Springer; 2007. pp. 171–184. [DOI] [PubMed] [Google Scholar]
- 6.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036––1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 7.Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.ENCODE Project Consortium. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mouse ENCODE Consortium. Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1506. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum. Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 17.Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database (HGMD®): 2008 Update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The UniProt Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paten B, Earl D, Nguyen N, Diekhans M, Zerbino D, Haussler D. Cactus: algorithms for genome multiple sequence alignment. Genome Res. 2011;21:1512–1528. doi: 10.1101/gr.123356.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haeussler M, Gerner M, Bergman CM. Annotating genes and genomes with DNA sequences extracted from biomedical articles. Bioinformatics. 2011;27:980–986. doi: 10.1093/bioinformatics/btr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. BMC Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raney BJ, Dreszer TR, Barber GP, Clawson H, Fujita PA, Wang T, Karolchik D, Kent WJ. Track Data Hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt637. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed data sets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams D, Altucci L, Antonarakis SE, Ballesteros J, Beck S, Bird A, Bock C, Boehm B, Campo E, Caricasole A, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat. Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hickey G, Paten B, Earl D, Zerbino D, Haussler D. HAL: a hierarchical format for storing and analyzing multiple genome alignments. Bioinformatics. 2013;29:1341–1342. doi: 10.1093/bioinformatics/btt128. [DOI] [PMC free article] [PubMed] [Google Scholar]