Abstract
The eukaryotic linear motif (ELM http://elm.eu.org) resource is a hub for collecting, classifying and curating information about short linear motifs (SLiMs). For >10 years, this resource has provided the scientific community with a freely accessible guide to the biology and function of linear motifs. The current version of ELM contains ∼200 different motif classes with over 2400 experimentally validated instances manually curated from >2000 scientific publications. Furthermore, detailed information about motif-mediated interactions has been annotated and made available in standard exchange formats. Where appropriate, links are provided to resources such as switches.elm.eu.org and KEGG pathways.
INTRODUCTION
In recent years, our understanding of the nature of protein–protein interactions has changed dramatically. Intrinsically disordered protein regions (IDRs) have been established as key facilitators of protein functionality (1–4), and consequently, globular domains no longer prevail as the sole purveyors of protein function. Short linear motifs (SLiMs), a class of compact, degenerate and convergently evolvable interaction modules, are the predominant functional modules found in intrinsically disordered regions (5–7). Interactions mediated by SLiMs, also referred to as linear motifs or MiniMotifs, have been shown to direct many diverse processes, such as controlling cell cycle progression, tagging proteins for proteasomal degradation, modulating the efficiency of translation, targeting proteins to specific sub-cellular localizations and stabilizing scaffolding complexes. Undoubtedly, more functions will be revealed in the future as additional SLiM instances are characterized.
SLiMs are represented by a limited number of constrained affinity- and specificity-determining residues within peptides that are typically between 3 and 11 amino acids in length (5,7,8). The compactness of a SLiM results in low-affinity binding (typically in the low micromolar 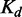 range) (7,9–12), and consequently, SLiMs often mediate transient, dynamic and reversible interactions. As a result of the limited number of binding determinants in a short linear motif, novel SLiMs can readily evolve de novo, adding functionality to a protein. The ease of evolution of motifs has resulted in the proliferation of SLiMs that encode functions of broad utility and many motif classes are ubiquitous, occurring in tens or hundreds of proteins. Many pathogens have also taken advantage of the intrinsic evolutionary plasticity of SLiMs by mimicking host motifs to deregulate and repurpose host pathways (13,14).
range) (7,9–12), and consequently, SLiMs often mediate transient, dynamic and reversible interactions. As a result of the limited number of binding determinants in a short linear motif, novel SLiMs can readily evolve de novo, adding functionality to a protein. The ease of evolution of motifs has resulted in the proliferation of SLiMs that encode functions of broad utility and many motif classes are ubiquitous, occurring in tens or hundreds of proteins. Many pathogens have also taken advantage of the intrinsic evolutionary plasticity of SLiMs by mimicking host motifs to deregulate and repurpose host pathways (13,14).
On a higher regulatory level, short linear motifs often exhibit complex switching behavior by co-operating with each other and with post-translational modifications to facilitate switching between different functional states of a protein, and thus, SLiMs function as key regulatory modules that allow for context-dependent, integrative regulatory decision-making (15–17).
THE EUKARYOTIC LINEAR MOTIF (ELM) RESOURCE
The ELM resource was established in 2003 with the mission to collect, annotate, classify and detect short linear motifs (18). It consists of two main modules: A relational database that stores all annotations and a prediction tool that uses the stored data to detect instances of short linear motifs in query sequences submitted by the user. The annotated data are manually curated from literature and made freely available to the scientific community.
At its core, the ELM database consists of ELM classes (grouped hierarchically into ELM types, see below) and ELM instances: An ELM class describes the specificity of a peptide-binding domain or domain family, usually summarized in the syntax of regular expressions. For example, the ELM class DEG_SCF_TRCP1_1 describes the pattern 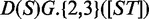 , whereby the first three amino acid positions (D,S,G) are fixed positions, followed by a flexible gap of either two or three amino acids of any type, followed by either an S or T residue. In addition, the round brackets around the second and last position indicate that these positions have to be phosphorylated to have a fully functional motif. The website for this ELM class (http://elm.eu.org/elms/elmPages/DEG_SCF_TRCP1_1.html) summarizes current literature on the motif, providing information about biological context, taxonomic distribution, a set of representative class instances, interacting protein domain(s), as well as links to primary literature and additional resources.
, whereby the first three amino acid positions (D,S,G) are fixed positions, followed by a flexible gap of either two or three amino acids of any type, followed by either an S or T residue. In addition, the round brackets around the second and last position indicate that these positions have to be phosphorylated to have a fully functional motif. The website for this ELM class (http://elm.eu.org/elms/elmPages/DEG_SCF_TRCP1_1.html) summarizes current literature on the motif, providing information about biological context, taxonomic distribution, a set of representative class instances, interacting protein domain(s), as well as links to primary literature and additional resources.
Most ELM classes have at least one ELM instance annotated, whereby an instance describes the experimentally determined match of a regular expression of an ELM class in a protein sequence. During annotation of instances, focus is put on the curation of experimental methods. Well-validated instances have been shown by more than one method in more than one publication and ideally include structural data and interaction information in addition to cell assays. Transient over-expression experiments on their own are not very trustworthy and have led to many false positive motifs being reported (19).
The ELM resource has been updated significantly since the last release (20), with 26 new ELM classes and 689 new ELM instances, raising the total count to 197 ELM classes and 2404 ELM instances (Tables 1 and 2).
Table 1.
Summary of data stored in the ELM databasea
| Functional sites | ELM classes | ELM instances | PDB structures | GO terms | PubMed Links | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | 127 | 197 | 2404 | 290 | 419 | 2132 | ||||
| By category | LIG | 103 | Human | 1391 | ||||||
| MOD | 30 | Mouse | 211 | Biological process | 217 | From ELM class | 976 | |||
| TRG | 23 | Rat | 115 | |||||||
| DEG | 15 | Yeast | 86 | Cell compartment | 95 | From instance | 1558 | |||
| DOC | 15 | Fly | 77 | |||||||
| CLV | 11 | Other | 524 | Molecular function | 107 | |||||
aAs of September 2013.
Table 2.
Novel ELM classes and number of associated instances (middle column) that have been added since the last ELM releasea
| Identifier | Numbers | Description |
|---|---|---|
| CLV_C14_Caspase3-7 | 39 | Caspase3 and Caspase7 cleavage site. |
| CLV_Separin_Fungi | 4 | Separase cleavage site, best known in sister chromatid separation. |
| CLV_Separin_Metazoa | 5 | |
| DEG_APCC_TPR_1 | 22 | This short C-terminal motif is present in co-activators, the Doc1/APC10 subunit and some substrates of the APC/C and mediates direct binding to TPR-containing APC/C core subunits. |
| DEG_CRL4_Cdt2_1 | 6 | This degron overlaps a PCNA interaction protein (PIP) box and is recognised by the CRL4_Cdt2 ubiquitin ligase in a PCNA- and chromatin-dependent manner. |
| DEG_CRL4_Cdt2_2 | 1 | |
| DEG_SCF_COI1_1 | 9 | Degron motif binding to the COI1 F-Box protein of the SCF E3 ubiquitin ligase in a jasmonate- dependent manner. |
| DEG_SCF_Skp2-Cks1_1 | 3 | This phosphodegron uniquely requires a pre-assembled target recognition site composed of Skp2 and Cks1. |
| DEG_SCF_TIR1_1 | 24 | Degron motif present in Aux/IAA transcriptional repressor proteins binding to TIR1/AFB F-box proteins of the SCF E3 ubiquitin ligase in an auxin-dependent manner. |
| LIG_APCC_Cbox_1 | 3 | Motif in APC/C co-activators that mediates binding to the APC/C core. |
| LIG_APCC_Cbox_2 | 2 | |
| LIG_CAP-Gly_2 | 1 | Short, partly aromatic carboxy terminal sequence found in the SLAIN group of microtubule-associated-proteins. |
| LIG_EABR_CEP55_1 | 6 | This proline-rich motif binds to the EABR domain of Cep55 and is involved in both cytokinesis of somatic cells and intercellular bridge formation in differentiating germ cells. |
| LIG_MYND_2 | 3 | Motif that mediates the interaction between MYND domain of AML1/ETO and co-repressors SMRT and N-CoR. |
| LIG_MYND_3 | 2 | A variant MYND binding motif found in the HSP90 co-chaperones p23 and FKBP38 interacting with PHD2 MYND domain. |
| LIG_NBox_RRM_1 | 2 | Amino terminal region on Far Upstream Element (FUSE) binding protein, which mediates the interaction with FIR in order to recruit FIR to FUSE DNA. |
| LIG_OCRL_FandH_1 | 3 | The F and H motif describes a 10-13-mer peptide sequence determined by a highly conserved phenylalanine and histidine residue surrounded by hydrophobic amino acids. A complex of ASH and RhoGAP-like domain binds the F and H motif within a hydrophobic pocket. |
| LIG_PAM2_2 | 4 | Peptide ligand motif that directly binds to the MLLE/PABC domain found in poly(A)-binding proteins and HYD E3 ubiquitin ligases, mainly via a common central core region and a complementary C-terminal region. |
| LIG_SPRY_1 | 2 | Peptide motif binding to the members of the SSB (or SPSB) family (SPRY domain- and SOCS box-containing protein). |
| LIG_SUMO_SBM_1 | 39 | Motif that mediates binding to SUMO proteins non-covalently. |
| LIG_SUMO_SBM_2 | 8 | Inverted version of LIG_SUMO_SBM_1 that mediates binding to SUMO proteins non-covalently. |
| MOD_LATS_1 | 23 | The LATS phosphorylation motif is recognised by the LATS kinases for Ser/Thr phosphorylation. Substrates are often found toward the end of the Hippo signaling pathway. |
| MOD_NEK2_1 | 3 | NEK2 phosphorylation motif; NEK protein kinases play a critical role in cell cycle control, interacting with and phosphorylating several centrosomal proteins. |
| MOD_NEK2_2 | 0 | |
| TRG_PEX_2 | 2 | Motifs present in peroxisomal import receptors important for binding to peroxisomal membrane proteins (PMPs) or other peroxisomal import receptors. |
| TRG_PEX_3 | 1 |
aAs of September 2013.
More updates and changes are described in the following sections.
NEW TYPES FOR ELM CLASSES
ELM classes were originally categorized into four different types based on the function of the motif: Proteolytic cleavage sites (CLV), general ligand binding sites (LIG), sites for post-translational modification (MOD) and sub-cellular targeting sites (TRG) [see Table 1 in Gould et al. (21)].
The annotation of many additional ELM classes made it both possible and necessary to introduce novel ELM types to categorize motif classes in more detail. Ligand binding classes describing docking sites or destruction motifs have been grouped together as two new types, DOC and DEG, respectively, raising the number of individual ELM types to six. Docking motifs (DOC) can be described as motifs that recruit a modifying enzyme using a site that is distinct from the active site (22), whereas a degron motif (DEG) is a specific region of a protein sequence that directs protein polyubiquitylation and targets the protein to the proteasome for degradation (23). Technically, all docking sites and destruction motifs belong to the ‘ligand binding sites (LIG)’ type; however, grouping together motif classes of similar function adds an additional level of discrimination.
NEW FEATURES
Interactions
For all ELM classes, the corresponding interacting domain that recognizes the particular short linear motif (SLiM) has been annotated (24). In addition, links have been provided to Pfam (25) or SMART (26), where more detailed information about the respective domain can be found. Where possible, the community annotation feature of Pfam has been used to curate each interaction domain present in ELM as an ‘external link’ in Pfam/Wikipedia to allow the user to easily jump between these resources.
Furthermore, for >700 ELM instances, the interacting protein has been annotated and, if possible, the position of the interacting domain as well as the affinity of the interaction has been curated. This information is presented in the ELM instance detail page (see Figure 1) and can be downloaded in either PSI-MI TAB or PSI-MI XML 2.5 format (16,27) (see links section on the ELM website).
Figure 1.
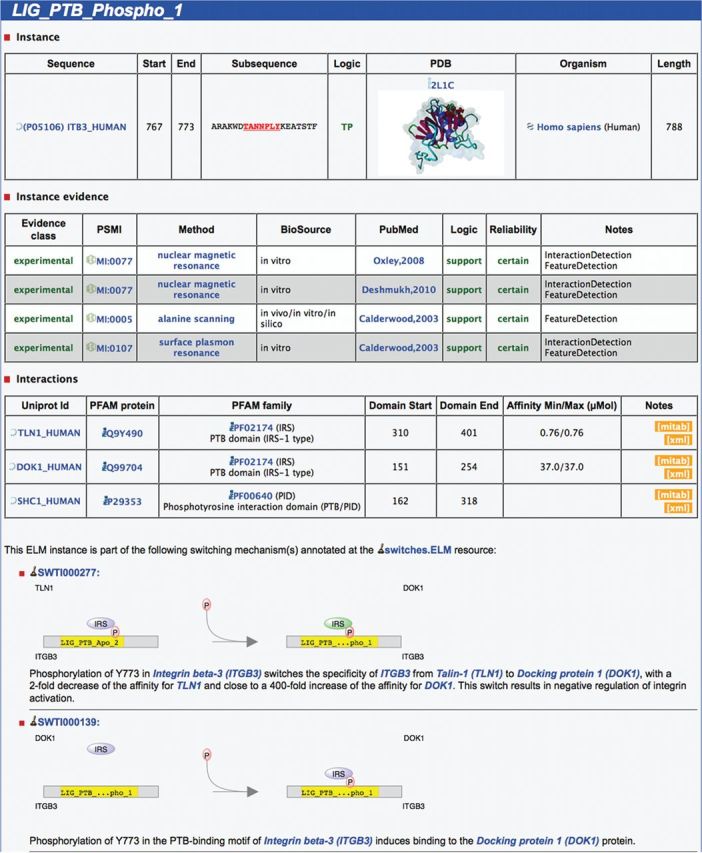
Screenshot of the ELM website showing details for an instance of the ELM class LIG_PTB_Phospho_1 in the human protein Integrin beta-3 at position 767–773. Details about the instance are depicted on top, including a representation of the 3D structure PDB:2LIC showing the instance bound by ‘SHC-transforming protein 1’. Below the instance evidence, which holds details about the methods used in the article, information regarding the interaction between the linear motif and the domain can be found. Here, three interaction partners containing phosphotyrosine-binding domains (PTB) are annotated: ‘talin-1’, ‘docking protein 1’ and ‘SHC-transforming protein 1’. Finally, the two schematics at the bottom illustrate the involvement of this motif instance in molecular switching mechanisms.
ELMs involved in molecular switches
As key regulatory interaction modules, linear motifs are tightly controlled and many motifs are conditionally turned ‘on’ and ‘off’ depending on cell state. Pre-translational addition or removal of a SLiM-containing exon, post-translational modification of the SLiM-containing peptide, allosteric SLiM inhibition or activation and SLiM binding site competition are amongst the most common mechanisms to regulate linear motifs. The switches. ELM database (15) is a resource dedicated to the curation and representation of experimentally validated motif-based molecular switches. It provides a categorized repository of >700 manually curated, experimentally validated instances of SLiM-based switch mechanisms collected from literature.
Each ELM instance that is part of a switching mechanism annotated by the switches.ELM resource has additional information shown on the ELM instance detail page as indicated in Figure 1: A short description of the switching mechanism is displayed with links to all participants as well as an illustrative scheme of the switching type. More details can be found at the switches.ELM page (http://switches.elm.eu.org) by clicking on the illustration.
MOTIFS–COMPLEXES–NETWORKS
Motif-mediated interactions play an important role in complex formation as exemplified by the multi-protein complex ‘anaphase promoting complex or cyclosome’ (APC/C) E3 ubiquitin ligase. This complex is represented by the novel ELM classes LIG_APCC_Cbox_1 and DEG_APCC_TPR_1 complementing the existing entries DEG_APCC_DBOX_1 and DEG_APCC_KENBOX_2. APC/C controls the progression through the cell cycle by ubiquitylation of cell cycle regulators and consists of at least 13 subunits assembled into the major subunit and two subcomplexes: One subcomplex consists mainly of tetratricopeptide repeat (TPR) domain-containing proteins (28), while the other subcomplex includes the catalytic core with the cullin domain-containing subunit Apc2 and the RING domain-containing subunit Apc11 (29). APC/C can ubiquitylate substrates only in the presence of the WD40 repeat-containing co-activator proteins Cdc20 or Cdh1, which are active at distinct phases of the cell cycle. Binding of Cdh1 to the APC/C is mediated by at least two motifs (Figure 2), a C-Box possibly binding to the catalytic Apc2 subunit (31) and a C-terminal IR-tail motif (32) (DEG_APCC_TPR_1) binding to the tetratricopeptide repeat (TPR) region of one subunit of the Cdc27 dimer and possibly additional uncharacterized interfaces.
Figure 2.
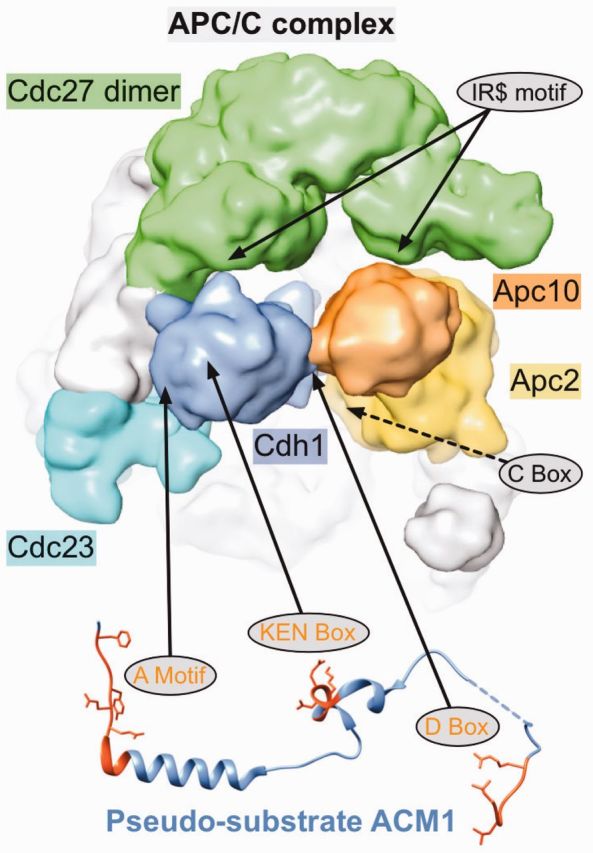
Motif-mediated regulation of APC/C function. Structure of the yeast APC/C complex [EMD-1815, determined by Cryo-EM. Figure generated with chimera (30)] with confirmed (full arrows) or putative (dashed arrows) motif-binding sites indicated. Binding of the co-activator Cdh1 (blue) to the APC/C is mediated by two motifs: The C-terminal IR motif binds to the tetratricopeptide repeat (TPR) region of one subunit of the Cdc27 dimer (green) and the C-Box binds to the catalytic Apc2 subunit (yellow). The Apc10 (orange) subunit also contains a C-terminal IR motif, which binds to the TPR domain of the second Cdc27 subunit (green). Recruitment of substrates or additional regulators such as the pseudo-substrate ACM1 (PDB:4BH6) also depends on motifs. The A Motif, KEN-Box and D-Box bind to different sites on the WD40 domain of Cdh1. In addition, the D-Box also contacts Apc10, which functions as a co-receptor for this degron (31).
Destruction of substrate proteins is facilitated by interactions with the bound co-activators, also mediated via short linear motifs (called degradation motifs or degrons), such as the well-characterized D-Box (33) and KEN-Box motifs (34,35). Strikingly, Cdc20 itself contains a KEN-Box, which is therefore recognized by Cdh1, ensuring the temporal degradation of Cdc20 (36).
Several pseudosubstrates for APC/C have been identified, which—while being able to bind to the APC/C complex—are not ubiquitylated and thus function as inhibitors of the APC/C complex (37). One example is illustrated in Figure 2: The yeast ACM1 protein was identified as a stable binding partner of Cdh1 and an inhibitor of APC/C-Cdh1 activity, containing three motifs mediating binding to Cdh1 (38).
Taken together, these motif-mediated interactions of the APC/C complex ensure the timely turnover of numerous cell-cycle regulatory proteins, thus governing eukaryotic cell cycle progression (39). We consider APC/C to be an excellent paradigm for the domain-motif interplay that pervades cell regulation.
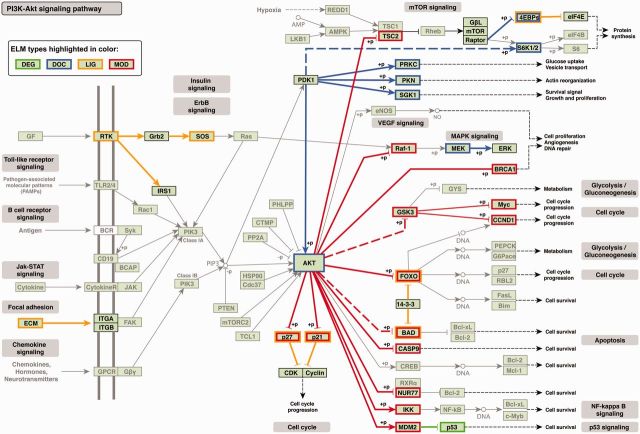
With the wealth of information available in the ELM compendium, it is also possible to map these data onto higher-level information systems such as the KEGG resource (40). Figure 3 illustrates the human phosphatidylinositol-3′-kinase-(PI3K)–Akt signaling pathway taken from KEGG (pathway-id: hsa04151). All interactions that are motif-mediated and annotated in the ELM resource have been mapped onto this pathway, with colours indicating the type of ELM class. Although it is likely that not all motif-mediated interactions are shown (simply because of the fact that they have not yet been annotated in ELM), the sheer amount of motif involvement is compelling. Mappings of annotated motifs onto other resources as shown in Figure 3 for a KEGG pathway can broaden our understanding of the importance of short linear motifs by allowing us to investigate motifs on a broader scale and to inspect their role in different regulatory pathways.
Figure 3.
Motif-mediated interactions annotated in the ELM resource mapped onto the KEGG (40) human Phosphatidylinositol-3′-kinase-(PI3K)–Akt signaling pathway (hsa04151). The direction of arrows denotes pathway direction. A colored border around a protein name indicates a motif within this protein is responsible for mediating the interaction to another protein in this pathway, also highlighted by a colored edge—docking motifs (blue), degrons (green), ligand binding motifs (orange) and modification sites (red). Colored dotted lines represent motif-mediated interactions mapped by homology. Phosphorylation/dephosphorylation events are indicated as ‘+p’/‘−p’ next to a node, respectively.
Webservices update
To allow users programmatic access to query the ELM server, SOAP/XML web services had previously been implemented (21). This system has been updated to a REST service model (41), whereby the communication uses the HTTP protocol/features to deliver structured data. Users now no longer need a special client to talk to the server but instead can use any browser or text-mode client (such as ‘curl’ or ‘wget’). Libraries to request and process HTTP queries are an integral part of most programming languages, consequently programmatically retrieving and utilizing the information provided by the ELM server should be straightforward for bioinformaticians of any skill level. A detailed list of URLs that can be used to query ELM via REST can be found at the ‘downloads’ page (http://elm.eu.org/downloads.html). We encourage users to send us feedback on the new methods as well as suggestions for possible future features.
PERSPECTIVE
Over the 10 years of ELM availability, the ELM resource has proven to be a valuable source of information for bench biologists (42–46) as well as for bioinformaticians performing computational analyses (47–51). It is now very clear that linear motifs are central to the understanding of cell regulation. Their co-operative interactions are to be found in all regulatory protein complexes of the cell. It does remain computationally challenging to discover new motifs by in silico methods, although progress is being made. Experimentalists continue to report new motif discoveries. Because there seems to be no drop in the rate of motif discovery, it can be extrapolated that the ∼200 motif classes listed in ELM is surely well short of the true number in eukaryotic proteomes. We shall need to keep on counting for the next 10 years and beyond.
FUNDING
European Commission Seventh Framework Programme [FP7-HEALTH-2009] [242129 SyBoSS to K.V.R.]; Federal Government Department of Education and Science [FKZ01GS0862 DigToP to M.S.]; Medical Research Council [U105185859 to R.J.W.] and ERASysBio+ [GRAPPLE to R.J.W.]; Career Investigator from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) (to L.B.C. and I.E.S.); Type I Graduate fellowship from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) (to J.G.); Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) [PICT 2010-1052 to I.E.S]; EMBL International PhD Program (to B.U.). Funding for open access charge: EMBL.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank the users of the ELM resource for their longstanding support as well as colleagues and annotators for their contributions and feedback to the ELM database.
REFERENCES
- 1.Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem. Sci. 2012;37:509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 3.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 4.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 5.Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 6.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 7.Neduva V, Russell RB. Linear motifs: evolutionary interaction switches. FEBS Lett. 2005;579:3342–3345. doi: 10.1016/j.febslet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson T. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- 9.Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ. Attributes of short linear motifs. Mol. Biosyst. 2012;8:268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- 10.Ladbury JE, Lemmon MA, Zhou M, Green J, Botfield MC, Schlessinger J. Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proc. Natl Acad. Sci. USA. 1995;92:3199–3203. doi: 10.1073/pnas.92.8.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mészáros B, Tompa P, Simon I, Dosztányi Z. Molecular principles of the interactions of disordered proteins. J. Mol. Biol. 2007;372:549–561. doi: 10.1016/j.jmb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Haslam NJ, Shields DC. Peptide-binding domains: are limp handshakes safest? Sci. Signal. 2012;5:pe40. doi: 10.1126/scisignal.2003372. [DOI] [PubMed] [Google Scholar]
- 13.Davey NE, Travé G, Gibson TJ. How viruses hijack cell regulation. Trends Biochem. Sci. 2011;36:159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Kadaveru K, Vyas J, Schiller MR. Viral infection and human disease–insights from minimotifs. Front. Biosci. 2008;13:6455–6471. doi: 10.2741/3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Roey K, Dinkel H, Weatheritt RJ, Gibson TJ, Davey NE. The switches.ELM resource: a compendium of conditional regulatory interaction interfaces. Sci. Signal. 2013;6:rs7. doi: 10.1126/scisignal.2003345. [DOI] [PubMed] [Google Scholar]
- 16.Van Roey K, Orchard S, Kerrien S, Dumousseau M, Ricard-Blum S, Hermjakob H, Gibson TJ. Capturing cooperative interactions with the PSI-MI format. Database (Oxford) 2013;2013:bat066. doi: 10.1093/database/bat066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Roey K, Gibson TJ, Davey NE. Motif switches: decision-making in cell regulation. Curr. Opin. Struct. Biol. 2012;22:378–385. doi: 10.1016/j.sbi.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Puntervoll P, Linding R, Gemúnd C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DMA, Ausiello G, Brannetti B, Costantini A, et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson TJ, Seiler M, Veitia RA. The transience of transient overexpression. Nat. Methods. 2013;10:715–721. doi: 10.1038/nmeth.2534. [DOI] [PubMed] [Google Scholar]
- 20.Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A, et al. ELM–the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40:D242–D251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gould CM, Diella F, Via A, Puntervoll P, Gemúnd C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2010;38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reményi A, Good MC, Lim WA. Docking interactions in protein kinase and phosphatase networks. Curr. Opin. Struct. Biol. 2006;16:676–685. doi: 10.1016/j.sbi.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat. Chem. Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weatheritt RJ, Jehl P, Dinkel H, Gibson TJ. iELM–a web server to explore short linear motif-mediated interactions. Nucleic Acids Res. 2012;40:W364–W369. doi: 10.1093/nar/gks444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon–level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labit H, Fujimitsu K, Bayin NS, Takaki T, Gannon J, Yamano H. Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. EMBO J. 2012;31:3351–3362. doi: 10.1038/emboj.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters J-M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat.Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 30.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.da Fonseca PCA, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Passmore LA, McCormack EA, Au SW, Paul A, Willison KR, Harper JW, Barford D. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 2003;22:786–796. doi: 10.1093/emboj/cdg084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 34.Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 35.Michael S, Travé G, Ramu C, Chica C, Gibson TJ. Discovery of candidate KEN-box motifs using cell cycle keyword enrichment combined with native disorder prediction and motif conservation. Bioinformatics. 2008;24:453–457. doi: 10.1093/bioinformatics/btm624. [DOI] [PubMed] [Google Scholar]
- 36.Listovsky T, Oren YS, Yudkovsky Y, Mahbubani HM, Weiss AM, Lebendiker M, Brandeis M. Mammalian Cdh1/Fzr mediates its own degradation. EMBO J. 2004;23:1619–1626. doi: 10.1038/sj.emboj.7600149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi E, Dial JM, Jeong D-E, Hall MC. Unique D box and KEN box sequences limit ubiquitination of Acm1 and promote pseudosubstrate inhibition of the anaphase-promoting complex. J. Biol. Chem. 2008;283:23701–23710. doi: 10.1074/jbc.M803695200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez JS, Jeong D-E, Choi E, Billings BM, Hall MC. Acm1 is a negative regulator of the CDH1-dependent anaphase-promoting complex/cyclosome in budding yeast. Mol. Cell Biol. 2006;26:9162–9176. doi: 10.1128/MCB.00603-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 40.Arakawa K, Kono N, Yamada Y, Mori H, Tomita M. KEGG-based pathway visualization tool for complex omics data. In Silico Biol. 2005;5:419–423. [PubMed] [Google Scholar]
- 41.Fielding RT, Taylor RN. Principled design of the modern Web architecture. ACM Trans. Internet Technol. 2002;2:115–150. [Google Scholar]
- 42.de Oliveira EAG, Romeiro NC, Ribeiro EDS, Santa-Catarina C, Oliveira AEA, Silveira V, de Souza Filho GA, Venancio TM, Cruz MAL. Structural and functional characterization of the protein kinase Mps1 in Arabidopsis thaliana. PLoS One. 2012;7:e45707. doi: 10.1371/journal.pone.0045707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 2011;7:e1001350. doi: 10.1371/journal.pgen.1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang R, Li K-M, Zhou C-H, Xue J-L, Ji C-N, Chen J-Z. Cdc20 mediates D-box-dependent degradation of Sp100. Biochem. Biophys. Res. Commun. 2011;415:702–706. doi: 10.1016/j.bbrc.2011.10.146. [DOI] [PubMed] [Google Scholar]
- 45.Lauck M, Sibley SD, Lara J, Purdy MA, Khudyakov Y, Hyeroba D, Tumukunde A, Weny G, Switzer WM, Chapman CA, et al. A novel hepacivirus with an unusually long and intrinsically disordered NS5A protein in a wild old world primate. J. Virol. 2013;87:8971–8981. doi: 10.1128/JVI.00888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perfetto L, Gherardini PF, Davey NE, Diella F, Helmer-Citterich M, Cesareni G. Exploring the diversity of SPRY/B30.2-mediated interactions. Trends Biochem. Sci. 2013;38:38–46. doi: 10.1016/j.tibs.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Rigden DJ, Woodhead DD, Wong PWH, Galperin MY. New structural and functional contexts of the Dx[DN]xDG linear motif: insights into evolution of calcium-binding proteins. PLoS One. 2011;6:e21507. doi: 10.1371/journal.pone.0021507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luck K, Charbonnier S, Travé G. The emerging contribution of sequence context to the specificity of protein interactions mediated by PDZ domains. FEBS Lett. 2012;586:2648–2661. doi: 10.1016/j.febslet.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 49.Stavropoulos I, Khaldi N, Davey NE, O’Brien K, Martin F, Shields DC. Protein disorder and short conserved motifs in disordered regions are enriched near the cytoplasmic side of single-pass transmembrane proteins. PLoS One. 2012;7:e44389. doi: 10.1371/journal.pone.0044389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kobe B, Boden M. Computational modelling of linear motif-mediated protein interactions. Curr. Top. Med. Chem. 2012;12:1553–1561. doi: 10.2174/156802612802652439. [DOI] [PubMed] [Google Scholar]
- 51.Trabuco LG, Lise S, Petsalaki E, Russell RB. PepSite: prediction of peptide-binding sites from protein surfaces. Nucleic Acids Res. 2012;40:W423–W427. doi: 10.1093/nar/gks398. [DOI] [PMC free article] [PubMed] [Google Scholar]