Abstract
Antimicrobial peptides (AMPs) are gaining importance as anti-infective agents. Here we describe the updated Collection of Antimicrobial Peptide (CAMP) database, available online at http://www.camp.bicnirrh.res.in/. The 3D structures of peptides are known to influence antimicrobial activity. Although there exists databases of AMPs, information on structures of AMPs is limited in these databases. CAMP is manually curated and currently holds 6756 sequences and 682 3D structures of AMPs. Sequence and structure analysis tools have been incorporated to enhance the usefulness of the database.
INTRODUCTION
Antimicrobial peptides (AMPs) are widely studied as potential alternatives for antibiotics. Surge in research on AMPs has led to the development of several databases and prediction tools. Some of these are general databases such as APD2 (1), DAMPD (2) and LAMP (3), whereas others are specialized databases like—AMSdb (http://www.bbcm.units.it/∼tossi/pag1.htm) that contains AMPs from only plant and animal sources; RAPD (4) provides information on recombinant methods to generate AMPs; PhytAMP (5) and BACTIBASE (6) are databases dedicated to AMPs from plant and bacterial sources, respectively; Defensins knowledgebase (7) and PenBase (8) are devoted to AMPs from defensin and penaeidin families, respectively; Peptaibol Database (9) is a database of peptaibols (unusual class of peptides); BAGEL (10) is a database of bacteriocins; and HIPdb (11) is a database of experimentally validated HIV-inhibiting peptides. The enormous amount of data on AMPs had motivated us to develop a general database, Collection of Antimicrobial Peptides (CAMP) (12), which included a sequence-based prediction tool for AMPs.
While all these databases provide comprehensive information on sequences of AMPs, information on structures of AMPs is limited. The topological features of peptides play a crucial role in dictating antimicrobial activity (13). Although many sequence-based prediction algorithms are available, the knowledge of 3D structural features of known AMPs has not been exploited to develop prediction algorithms. The lack of structural databases of AMPs is probably one of the main impediments in this direction. Presently, there are several AMPs whose structural information is available in the Protein Data Bank (PDB) (14). However, retrieving information on structures of AMPs from the structural databases such as PDB is not a trivial task; for example, the structures may have additional chains that are non-AMPs, and these have to be filtered out by manual curation. The structures may also not be easily retrieved from structure databases based on simple keyword searches such as ‘antibacterial’, ‘antifungal’, etc. To address these shortcomings, the current release of CAMP has been developed.
MATERIALS AND METHODS
Data collection and organization
Sequence and structural information of AMPs was retrieved from protein databases of NCBI, UniProtKB (15) and PDB using combination of keywords like ‘antimicrobial’, ‘antibacterial’, ‘antifungal’, ‘antiviral’ and ‘antiparasitic’. Manually curated information related to sequence, structure, protein definition, accession numbers, reference literature, activity, taxonomy of the source organism, target organisms with minimum inhibitory concentration (MIC) values, hemolytic activity of the peptide, functional and structural classifications, protein family descriptions and links to external databases like UniProtKB, PDB, PubMed and other AMP databases is made available to the users.
Database architecture
The updated CAMP database is built on Apache HTTP server 2.0.59. MySQL Server 5.0 is used at the back-end, whereas the front-end is built using PHP, HTML, JavaScript, Perl and Open Flash Chart 2.
Below is a brief description of the user interface of CAMP:
Home: The CAMP database along with its various features is described in this section.
Databases: Data are sectioned into sequence, structure and patent databases.
- Tools: The following analysis tools are available to the users.
- AMP prediction: Users can predict AMPs and/or scan for antimicrobial regions within the peptides using Support Vector Machine (SVM), Random Forests (RF) and Artificial Neural Network (ANN).
- Feature calculator: Amino acid composition, secondary structural propensities and physicochemical properties such as net charge, hydrophobicity, etc of the peptides can be calculated.
- BLAST: Users can use BLAST (16) tool against the sequence or structure database of CAMP to find homologous sequences or structures, respectively.
- ClustalW: Multiple sequence alignment of the peptides can be obtained using ClustalW (17) tool from EMBL-EBI.
- Vector Alignment Search Tool: Similar protein structures can be identified using this NCBI tool (18).
- Helical wheel: Alpha-helical AMPs can be studied using the helical wheel Java applet created by Edward K. O'Neil and Charles M. Grisham (University of Virginia in Charlottesville, Virginia).
Search: Users can search for sequences and/or structures of AMPs using basic and advanced search options.
Links to other available AMP databases have been provided.
Statistics: Coverage of the database based on the nature of data, taxonomy of source organism and activity has been depicted using pie charts and Venn diagram.
Help: A detailed explanation about the features and tools available in the database has been provided in this section.
Prediction algorithm
Dataset creation
The positive dataset constituted of 3010 AMP sequences. These were obtained from the patent and experimentally validated datasets of CAMP, after removing sequences that (i) are redundant (100% similarity cut-off), (ii) have non-standard amino acids and (iii) have length >100. CD-HIT server was used for removing redundant sequences (23).
The negative dataset consists of 4011 sequences, generated in our previous work (12). It includes experimentally proven non-antimicrobial sequences, arbitrary sequences generated using random numbers and protein sequences retrieved from the UniProt database without annotation as ‘antimicrobial’. The sequences had length approximately in the same range as the positive dataset. The CD-HIT program (23) was used to eliminate sequences with >90% identity. These datasets were randomly divided into training (70%) and test (30%) datasets.
Model generation
Sixty-four best peptide descriptors based on the RF Gini score were used for developing SVM-, RF- and ANN-based prediction models. All the models were evaluated using Matthews correlation coefficient (MCC), prediction accuracy and 10-fold cross-validation accuracy on training and test datasets. For developing the prediction models, implementation of SVM, RF and ANN in R (version 2.15.3) was used (24).
SVM
Kernlab package in R was used to train the SVM classifier (25). In this study, we have used polynomial kernel function. The values of the hyper parameters were set as follows: degree = 4, scale = 0.01 and offset = 1.
RF
‘randomForest’ package was used to train the RF classifier with a maximum of 1500 trees (26).
ANN
‘nnet’ package in R was used for building the ANN-based prediction model (27).
RESULTS AND DISCUSSION
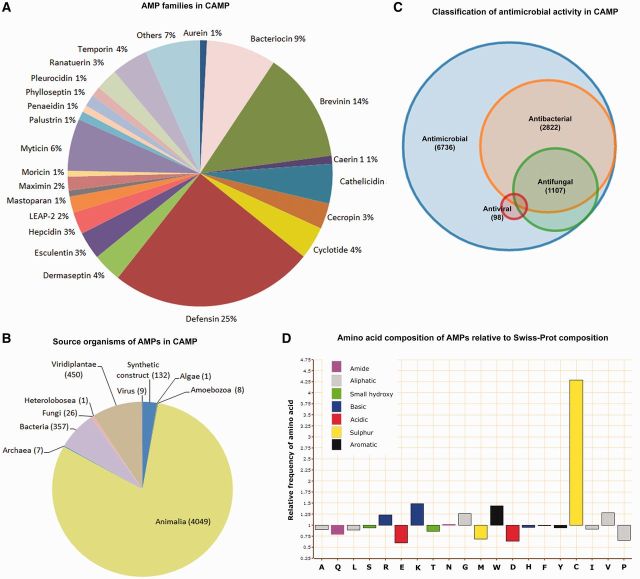
The updated CAMP is a comprehensive database on sequences and structures of AMPs. It currently holds 6756 sequences of AMPs (experimentally validated (2602), predicted (2438) and patents (1716)), which include 2736 recently identified AMP sequences. The information on the sequence, AMP family, source, target organism and activity is captured in the database. As can be seen in Figure 1A–C, CAMP has a wide coverage on the above fields.
Figure 1.
(A) Pie chart of AMP families in CAMP, (B) Pie chart of source organisms of AMPs in CAMP, (C) Venn diagram of classification of AMP activity in CAMP and (D) Relative amino acid composition of experimentally validated and predicted sequences of AMPs in CAMP as compared with Swiss-Prot composition.
CAMP presently contains 682 AMP structures. Multiple structures of AMPs, if available in PDB, are also integrated in the database. Although structural information on AMPs is available in databases such as APD2, LAMP, etc, the structures can be directly viewed using Jmol viewer in CAMP. Direct viewing of structures is also available in Defensins knowledgebase, PhytAMP, HIPdb and BACTIBASE. However, these databases cater to specific class of AMPs.
Another interesting feature of the current release of CAMP is that users can selectively retrieve information on specific families of AMPs of their interest; e.g. cathelicidins, defensins and cecropins. The AMP family information for the peptides has been annotated manually using information from Pfam (28), InterPro (29) and associated literature. The distribution of the AMP families in the database can be seen in Figure 1A.
The prediction algorithm for AMPs has been modified using the updated sequence information. Supplementary Table S1 shows the prediction accuracy, MCC and cross-validation accuracy of the prediction models. Users can predict the antimicrobial activity of proteins and/or scan regions (with user-defined lengths) within proteins for antimicrobial activity.
Tools that aid in sequence and structure analysis such as feature calculator, PRATT, ClustalW, Vector Alignment Search Tool, BLAST and PDB2PQR have also been incorporated in CAMP. Effect of mutations on the structure of AMPs and/or their analogs can be visualized using the Jmol visualizer integrated in the database. Helicity is known to influence antimicrobial activity (30) and therefore, tool for helical wheel projection is also available. AMPs are known to be rich in hydrophobic and cationic amino acids. The ratio of the percentage frequency of amino acids in CAMP to the percentage frequency of amino acids in UniProtKB/Swiss-Prot protein knowledgebase (Release 2013_08 of 24 July 2013) is plotted in Figure 1D. As expected, AMPs were observed to be enriched in positively charged and hydrophobic residues such as Arg, Lys, Gly, Cys, Trp and Val residues.
CONCLUSIONS
CAMP holds a massive update on AMP sequences and incorporates several tools relevant to design of AMPs. The 3D conformations of peptides are known to be critical determinants of antimicrobial activity. The prominent feature of the current release of CAMP is the addition of experimentally derived structures of AMPs, which can be directly viewed using the Jmol viewer. The update also facilitates family-based study on AMPs. A detailed comparison of CAMP with the existing databases on AMPs is presented in Table 1. The information, present in an easily searchable and downloadable form, is envisaged to accelerate sequence–structure–activity studies on AMPs.
Table 1.
Comparison of CAMP with existing AMP databases
| Features | Database |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RAPD | PhytAMP | BACTIBASE second release | Defensins knowledg- ebase | PenBase | Peptaibol database | AMSDb | HIPdb | APD2 | DAMPD | LAMP | CAMP | |
| Type | Specific (Recombinantly produced AMPs only) | Specific (Plant AMPs only) | Specific (Bacteriocins only) | Specific (Defensin family AMPs only) | Specific (Penaeidin family AMPs only) | Specific (Peptaibols only) | Specific (Eukaryotic AMPs only) | Specific (HIV inhibiting peptides only) | General | General | General | General |
| Total number of entries | 179 | 273 | 220 | 566 | 28 | 317 | 895 | 1068 | 2307 | 1232 | 5547 | 7438 |
| Prediction algorithm | Absent | Present | Present | Absent | Absent | Absent | Absent | Absent | Present | Present | Absent | Present |
| Structural information | Absent | Present | Present | Present | Absent | Presenta | Presenta | Present | Presenta | Presenta | Presenta | Present |
| Search based on AMP family | Present | Present | Absent | Present | Absent | Absent | Present | Present | Absent | Present | Absent | Present |
| MIC values | Absent | Present | Present | Present | Absent | Absent | Present | Present | Present | Present | Present | Present |
| Separate searches for experimental and predicted datasets | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Present | Present |
| Tools | DNA translator, peptide calculator, DNA sequence convertor | BLAST, FASTA, Smith-Waterman search, ClustalW, muscle, physiochemical profile | BLAST, FASTA, Smith-Waterman search, ClustalW, Muscle, T-coffee, physiochemical profile, MODELLER | BLAST and ClustalW | BLAST and ClustalW | Absent | HydroMCalc and HydroPlot | HIPdb map, HIPdb BLAST | AMP designer | BLAST, ClustalW, NJPLOT, HMMER, hydrocalulator, signalp, graphical views. | BLAST | ClustalW, PRATT, helical wheel, vector alignment search tool , BLAST, PDB2PQR, Feature calculator |
aThe PDB IDs are available. Structures cannot be directly viewed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work [RA/18-09/2013] was supported by grants from Department of Science and Technology, Government of India [SB/S3/CE/028/2013]; and Indian Council of Medical Research. Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Dr Smita D. Mahale (PI of Biomedical Informatics Centre) for all the help and support. They also acknowledge the assistance provided by Ms Shaini Joseph and Ms Pratima Gurung in data collection.
REFERENCES
- 1.Wang G, Li X, Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seshadri Sundararajan V, Gabere MN, Pretorius A, Adam S, Christoffels A, Lehväslaiho M, Archer JA, Bajic VB. DAMPD: a manually curated antimicrobial peptide database. Nucleic Acids Res. 2012;40:D1108–D1112. doi: 10.1093/nar/gkr1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao X, Wu H, Lu H, Li G, Huang Q. LAMP: a database linking antimicrobial peptides. PLoS One. 2013;8:e66557. doi: 10.1371/journal.pone.0066557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Chen Z. RAPD: a database of recombinantly produced antimicrobial peptides. FEMS Microbiol. Lett. 2008;289:126–129. doi: 10.1111/j.1574-6968.2008.01357.x. [DOI] [PubMed] [Google Scholar]
- 5.Hammami R, Ben Hamida J, Vergoten G, Fliss I. PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 2009;37:D963–D968. doi: 10.1093/nar/gkn655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammami R, Zouhir A, Le Lay C, Ben Hamida J, Fliss I. BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol. 2010;10:22. doi: 10.1186/1471-2180-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seebah S, Anita S, Zhuo SW, Yong HC, Chua H, Chuon D, Beuerman R, Verma CS. Defensins knowledgebase: a manually curated database and information source focused on the defensins family of antimicrobial peptides. Nucleic Acids Res. 2006;35:D265–D268. doi: 10.1093/nar/gkl866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gueguen Y, Garnier J, Robert L, Lefranc MP, Mougenot I, De Lorgeril J, Janech M, Gross PS, Warr GW, Cuthbertson B, et al. PenBase, the shrimp antimicrobial peptide penaeidin database: sequence-based classification and recommended nomenclature. Dev. Comp. Immunol. 2005;30:283–288. doi: 10.1016/j.dci.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Whitmore L, Wallace BA. The Peptaibol database: a database for sequences and structures of naturally occurring peptaibols. Nucleic Acids Res. 2004;32:D593–D594. doi: 10.1093/nar/gkh077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong A, van Heel AJ, Kok J, Kuipers OP. BAGEL2: mining for bacteriocins in genomic data. Nucleic Acids Res. 2010;38:W647–W651. doi: 10.1093/nar/gkq365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qureshi A, Thakur N, Kumar M. HIPdb: a database of experimentally validated HIV inhibiting peptides. PLoS One. 2013;8:e54908. doi: 10.1371/journal.pone.0054908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas S, Karnik S, Barai RS, Jayaraman VK, Idicula-Thomas S. CAMP: a useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010;38:D774–D780. doi: 10.1093/nar/gkp1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sitaram N, Nagaraj R. Host-defense antimicrobial peptides: importance of structure for activity. Curr. Pharm. Des. 2002;8:727–742. doi: 10.2174/1381612023395358. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein data bank: a computer-based archival file for macromolecular structures. Arch. Biochem. Biophys. 1978;185:584–589. doi: 10.1016/0003-9861(78)90204-7. [DOI] [PubMed] [Google Scholar]
- 15.The UniProt Consortium. Update on activities at the universal protein resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 18.Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 19.Jonassen I, Collins JF, Higgins D. Finding flexible patterns in unaligned protein sequences. Protein Sci. 1995;4:1587–1595. doi: 10.1002/pro.5560040817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonassen I. Efficient discovery of conserved patterns using a pattern graph. Comput. Appl. Biosci. 1997;13:509–522. doi: 10.1093/bioinformatics/13.5.509. [DOI] [PubMed] [Google Scholar]
- 21.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup, execution, and analysis of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R Development Core Team. R Foundation for statistical computing; 2009. R: A Language and Environment for Statistical Computing. Vienna, Austria. [Google Scholar]
- 25.Karatzoglou A, Smola A, Hornik K, Zeileis A. Kernlab - an S4 package for Kernel methods. R. J. Stat. Softw. 2004;11:1–20. [Google Scholar]
- 26.Liaw A, Wiener M. Classification and regression by random forest. R News. 2002;2:18–22. [Google Scholar]
- 27.Venables WN, Ripley BD. Modern Applied Statistics with S. 4th edn. New York: Springer; 2002. ISBN 0-387-95457-0. [Google Scholar]
- 28.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen HC, Brown JH, Morell JL, Huang CM. Synthetic magainin analogues with improved antimicrobial activity. FEBS Lett. 1988;236:462–426. doi: 10.1016/0014-5793(88)80077-2. [DOI] [PubMed] [Google Scholar]