Abstract
RepeatsDB (http://repeatsdb.bio.unipd.it/) is a database of annotated tandem repeat protein structures. Tandem repeats pose a difficult problem for the analysis of protein structures, as the underlying sequence can be highly degenerate. Several repeat types haven been studied over the years, but their annotation was done in a case-by-case basis, thus making large-scale analysis difficult. We developed RepeatsDB to fill this gap. Using state-of-the-art repeat detection methods and manual curation, we systematically annotated the Protein Data Bank, predicting 10 745 repeat structures. In all, 2797 structures were classified according to a recently proposed classification schema, which was expanded to accommodate new findings. In addition, detailed annotations were performed in a subset of 321 proteins. These annotations feature information on start and end positions for the repeat regions and units. RepeatsDB is an ongoing effort to systematically classify and annotate structural protein repeats in a consistent way. It provides users with the possibility to access and download high-quality datasets either interactively or programmatically through web services.
INTRODUCTION
A large portion of proteins contain repetitive motifs, which are generated by internal duplications and frequently correspond to structural and functional units of proteins. Many repetitions in protein sequences can be identified by using different approaches (1–4). A more difficult problem for identification is however posed by repeats in protein structure, which can be highly degenerate (5,6). In fact, it is possible for a protein to maintain a repetitive structure even in the presence of massive amounts of point mutations (7). Several repeat families have been studied so far due to their relevance in different biological processes such as health (8), neurodevelopment (9) and protein engineering (10–12), to name just a few.
Repeats have been previously divided into five broad classes, primarily as a function of repeat length (13,14). At the lower end of the repeat length spectrum, i.e. less than five residues, very short repeats can either form insoluble aggregates (crystallites, class I) or long and winding helices of fibrous structures like collagen and α-helical coiled-coils (class II). At the other end of the spectrum, repeats containing >∼50 residues appear to fold mostly as domains forming beads-on-a-string structures (class V). In between, for unit lengths of 5–40 residues, the known repeats can form either open elongated solenoids (class III) or closed toroids (class IV). Due to their fundamental functional importance, classes III and IV contain the most studied types of tandem repeat proteins. Solenoid folds appear to follow the distribution of repeat lengths rather closely, from all-beta (e.g. anti-freeze proteins) (15) to mixed alpha/beta (e.g. leucine-rich repeats) (16,17) to all-alpha structures (e.g. Armadillo and HEAT repeats) (18–20). They are characterized by some of the largest known autonomously folding domains, with 500 or more residues forming a single structure (21). Rapid addition or deletion of repeat units even between close homologs is of particular note for solenoid structures (22). Toroids on the other hand are restricted in overall size by their closed circular nature. Known toroid structures include the highly versatile TIM barrel and large outer membrane beta-barrels (23). Perhaps a more interesting fold is the beta propeller (e.g. WD repeats), which can accommodate variable numbers of repeat units while maintaining a closed circular structure (24,25).
An open question regarding repeat proteins is the existence of other common structures that may have gone undetected. After all, the most common way to detect repeat families so far was to manually annotate the sequence family first and only afterwards visually recognize their structural repetitiveness. Such an approach is obviously difficult when dealing with the entire Protein Data Bank (PDB) (26), especially considering the many uncharacterized protein structures deposited by the main structural genomics consortia (27). The systematic description of repeat structures becomes a question of using automated methods to detect them in protein structures. This field is relatively new, with only few available methods. One of the first attempts was made by the Thornton group (28), but is unfortunately no longer available. Some methods (4,29–33) were developed to detect internal symmetries in proteins, but these may be difficult to adapt to the systematic classification of repeats. Recently, our group has developed RAPHAEL (34) in an attempt to fill the gap for repeat detection from structure. Widely used structural classifications such as CATH (35) and SCOP (36) also do not explicitly annotate repeats in protein structures, although it may be possible to leverage individual annotations to find similar repeats. Some databases exist for the detection of repeats from sequence (37–39), but usually these are limited to short tandem repeats and do not take into account divergent repeats, such as solenoids or toroids. The main domain sequence databases such as Pfam (40) and SMART (41) do not excel at the annotation of these repeat types either, as coverage is rather low and many repeat units go undetected. For Pfam most of the largest clusters of human sequence regions not covered were recently found to be repeats (42). To the best of our knowledge, no database or classification is currently available for repeat structures. This is the motivation for our present work, and we introduce RepeatsDB as a way to fill this gap. The database was developed to provide a central resource for the systematic annotation and classification of repeats. Given the fact that the structure-based search and classification of repeat proteins is more complete than on the basis of sequences or key words, our database will allow more accurate assignment of proteins with repeats to the corresponding families. For example, it will be used to suggest a better subdivision of alpha-solenoid proteins where at present the boundaries between the structures with Armadillo, HEAT, TPR and other repeat types are frequently blurred.
DATABASE DESCRIPTION
Data curation
The initial dataset for RepeatsDB was extracted from the PDB (43). Repeat candidates were identified from the reduced PDB dataset with RAPHAEL (34), which uses a geometric approach imitating the work of a human curator (score cutoff ≥1). The resulting dataset consisted of >10 000 repeat candidates, stored in the database as ‘predicted’ entries, which underwent a classification and curation process.
The dataset of predicted repeats was manually curated using a two-level annotation system. The first manual annotation level (‘manually classified’) classifies an entry into structural repeat class and subclass. This classification is based on previous work (14), where five classes of repeat structures are proposed, which are then further divided into subclasses. Class assignment is based mainly on repeat unit length and subclass assignment on secondary and tertiary structure features. The second manual annotation level (‘detailed’) consists in providing information about the start and end positions of the repeat units, repeat regions and/or insertions. We define a repeat unit as the smallest structural building block that is repeated to form a repeat region. A repeat region is a group of at least three repeat units. Inclusion of proteins with two repeat units would significantly complicate classification because many typical globular domains have this type of architecture. Insertions are non-repeated segments of structure that occur either inside a repeat unit or between two of them. These are particularly interesting because they break the repeat symmetry, and represent a challenge both for automatic detection and for the analysis of repeat structures (34).
Several curators annotated each protein undergoing manual classification by consensus. For first-level annotations, at least 75% of the curators had to agree in order for a protein to be included, otherwise it would be excluded and placed on a reserve list for future annotation. The rationale for this choice is that ambiguous cases are generally difficult to classify but may occasionally represent a novel repeat class. For second-level annotations, the threshold for consensus was at least 65% agreement (typically two of three curators). In case of discrepancy, an expert would arbitrate the final annotation based on the alternative proposals. Proteins with detailed annotations were also used to search for similar sequences in proteins from the PDB. Any PDB chain with at least 40% sequence identity and a coverage of at least 80% of the classified protein, belonging to the initial list of predicted entries, is added to the ‘classified by similarity’ annotation level. The similarity thresholds were selected to exclude possible false-positives (data not shown).
Implementation
RepeatsDB was designed with a multi-tier architecture, using separate modules for data management, data processing and presentation functions. To simplify development and maintenance, all tiers handle the common JSON (JavaScript Object Notation) format, thereby eliminating the need for data conversion. The MongoDB database engine is used for data storage and Node.js as middleware between data and presentation. RepeatsDB exposes its resources through RESTful web services, by using the Restify library for Node.js. The Angular.js framework and Bootstrap library were selected to provide the overall look-and-feel. Angular.js to Bootstrap integration is available through the angular-ui project. A customized version of the BioJS (44) sequence component is used as sequence visualizer. Additional information is added to entries by querying the PDB web services at the structure and chain level. At the structure level, annotations like organism and experimental method used when resolving the structure are provided. At the chain level, secondary structure and links to other databases, among others. RepeatsDB offers users both graphical web interface access and RESTful web services from URL: http://repeatsdb.bio.unipd.it/
USING REPEATSDB
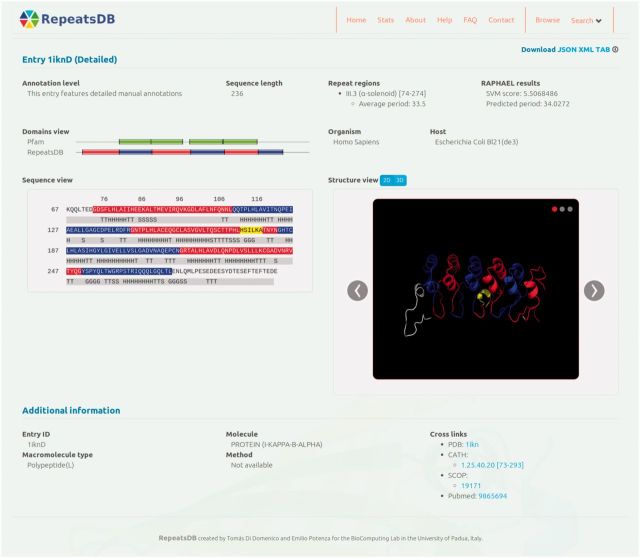
The user interface presents an intuitive tree-based browsing mechanism, where the root of the tree is the full database, second-level nodes repeat classes and third-level nodes subclasses. When clicking on a node, the user is presented with the list of RepeatsDB entries corresponding to the selected category. Each row of the list shows basic information about the entry, like its entry ID, title and organism. All annotated chains corresponding to an entry are displayed in a single page. The user interface presents a structure and sequence visualization widget (Figure 1). The user may choose to visualize the structure in four static images, or by using the 3D visualizer. If the entry features detailed annotations, the repeat regions, units and/or insertions are displayed using a combination of colours. The sequence visualization widget displays the sequence and secondary structure corresponding to the structure. It displays the same colour coding as the structure visualization widget, associating repeat annotations in the structure and sequence views. Additional information at the structure and chain levels is also provided.
Figure 1.
Screenshot of a sample RepeatsDB entry results page (PDB entry 1ikn). The sequence viewer and the structure viewer are shown in the middle of the page, towards the left and the right, respectively. Additional annotations at the structure and chain level are displayed, including links to other databases (above) and classifications (below).
The RepeatsDB search toolbar, available on top of every page, allows to search for entries either by database IDs or UniProt text query. The database ID search allows comma-separated PDB or UniProt IDs. The UniProt text search query uses the full UniProt search engine, see online documentation. RESTful web services are directly accessible through HTTP URLs. All data available on RepeatsDB are also available for programmatic access. Please refer to the ‘Help’ section of the website for details on using the RepeatsDB web services. Datasets can be downloaded in JSON, XML or text format using the browse function or RESTful web services.
Statistics
Analysis of the full PDB dataset yielded 10 745 repeats predicted by RAPHAEL, of which 2797 were finally classified into the RepeatsDB schema. Table 1 shows the distribution between classes and subclasses. The bulk of the annotations (∼90%) consist of entries belonging to classes III and IV. No effort was made to balance the distribution of entries between classes in this initial release. As coverage increases in the future, we expect the balance to approximate the real distribution more closely, although it may be necessary to fine-tune RAPHAEL. Of the classified entries, 321 representatives of the entire dataset were annotated in detail with information about the start and end of repeat regions, repeat units and/or insertions (Table 1). It is interesting to note the different distribution of insertions between classes. Apparently, some classes such as β-solenoid (class III.1) or TIM barrels (class IV.1) have stronger propensity to accommodate insertions.
Table 1.
Statistics for RepeatsDB
| Subclass | Name | Detailed | Classified (manually) | Classified (by similarity) | Predicted |
|---|---|---|---|---|---|
| I.1 | Poly-alanine β structure | 0 | 0 | 0 | 0 |
| II.1 | Collagen triple-helix | 0 | 5 | 0 | 0 |
| II.2 | α helical coiled coil | 23 | 38 | 69 | 0 |
| III.1 | β-solenoid | 43 | 113 | 21 | 0 |
| III.2 | α/β solenoid | 21 | 43 | 27 | 0 |
| III.3 | α-solenoid | 48 | 246 | 631 | 0 |
| III.4 | Trimer of β spirals | 7 | 0 | 13 | 0 |
| III.5 | Single layer anti-parallel β | 4 | 3 | 0 | 0 |
| IV.1 | TIM-barrel | 84 | 118 | 626 | 0 |
| IV.2 | β-barrel | 8 | 1 | 8 | 0 |
| IV.3 | β-trefoil | 20 | 0 | 29 | 0 |
| IV.4 | β-propeller | 40 | 182 | 227 | 0 |
| IV.5 | α/β prism | 0 | 17 | 0 | 0 |
| IV.6 | α-barrel | 6 | 0 | 0 | 0 |
| V.1 | α-beads | 2 | 1 | 0 | 0 |
| V.2 | β-beads | 29 | 12 | 71 | 0 |
| V.3 | α/β-beads | 3 | 3 | 1 | 0 |
| V.other | Unknown subclass | 3 | 0 | 4 | 0 |
| UA | Unassigned | 0 | 0 | 0 | 7948 |
| Total | 321 | 749 | 1727 | 7948 |
The subclass name is shown together with the number of entries on each of the four annotation levels. Note that ‘Unassigned’ entries are automatically predicted by RAPHAEL and therefore not assigned to a specific class.
CONCLUSIONS AND FUTURE WORK
RepeatsDB’s goal is to provide the community with a resource for high-quality tandem repeat protein structure annotations. The user can either interactively analyse his proteins of interest via the user interface, or create and download datasets for offline use. Far from being a static classification process, the annotation effort for the initial RepeatsDB dataset alone already motivated the extension of the original classification schema (14). Some of the curated structures, while clearly representing structural repeats, did not belong to any of the pre-defined subclasses. To allow them to be classified, subclasses IV.5 (α/β prism) and IV.6 (α-barrel) were added to the initial schema (14). Class V also underwent a re-classification according to the secondary structure content of the single domain repeats (‘beads’) to allow a broader classification range beyond individual repeat families, as the list of possible beads-on-a-string folds may be considerably larger than currently appreciated. The ‘other’ subclass was also added to allow collection of repeats that do not fit into the current classification scheme. RepeatsDB provides the community with a previously unavailable opportunity to easily create datasets of tandem repeat proteins. The detailed annotation subset further presents a unique opportunity to better understand the nature of tandem repeat proteins.
Beyond its initial release, RepeatsDB is a continuous effort to expand, revise and improve tandem protein repeat annotations. Predictions for new PDB structures are simple and fully automated, allowing regular database updates every 3 months. Manual curation of new entries for inclusion is also ongoing, aiming at regular and steady updates. Options to involve the community into the annotation process through crowd-sourcing tools are currently being analysed. A main goal for future versions is the extension of the annotation of repeats at the sequence level, starting from annotation for intrinsically disordered regions from MobiDB (45). We anticipate that RepeatsDB should prove valuable towards the understanding of the sequence–structure relationship in tandem repeat proteins and their evolutionary relationship.
FUNDING
FIRB Futuro in Ricerca [RBFR08ZSXY] and AIRC [MFAG 12740] (to S.T.); AIRC research fellow (to G.M.) and CONICET PhD student (to R.G.P.); Ministry of Education and Science of the Russian Federation, project 8831 (to A.V.K.) (in part). Funding for open access charge: FIRB Futuro in Ricerca [RBFR08ZSXY].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Diego Ferreiro for insightful discussions and to Manfred Sippl for his software tools.
REFERENCES
- 1.Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput. Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 2.Jorda J, Kajava AV. Protein homorepeats sequences, structures, evolution, and functions. Adv. Protein Chem. Struct. Biol. 2010;79:59–88. doi: 10.1016/S1876-1623(10)79002-7. [DOI] [PubMed] [Google Scholar]
- 3.Gribskov M, McLachlan AD, Eisenberg D. Profile analysis: detection of distantly related proteins. Proc. Natl Acad. Sci. USA. 1987;84:4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biegert A, Söding J. De novo identification of highly diverged protein repeats by probabilistic consistency. Bioinformatics. 2008;24:807–814. doi: 10.1093/bioinformatics/btn039. [DOI] [PubMed] [Google Scholar]
- 5.Schaper E, Kajava AV, Hauser A, Anisimova M. Repeat or not repeat?—statistical validation of tandem repeat prediction in genomic sequences. Nucleic Acids Res. 2012;40:10005–10017. doi: 10.1093/nar/gks726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buard J, Vergnaud G. Complex recombination events at the hypermutable minisatellite CEB1 (D2S90) EMBO J. 1994;13:3203–3210. doi: 10.1002/j.1460-2075.1994.tb06619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 2001;134:117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 8.Kajava AV, Steven AC. Beta-rolls, beta-helices, and other beta-solenoid proteins. Adv. Protein Chem. 2006;73:55–96. doi: 10.1016/S0065-3233(06)73003-0. [DOI] [PubMed] [Google Scholar]
- 9.De Wit J, Hong W, Luo L, Ghosh A. Role of leucine-rich repeat proteins in the development and function of neural circuits. Annu. Rev. Cell Dev. Biol. 2011;27:697–729. doi: 10.1146/annurev-cellbio-092910-154111. [DOI] [PubMed] [Google Scholar]
- 10.Main ER, Lowe AR, Mochrie SG, Jackson SE, Regan L. A recurring theme in protein engineering: the design, stability and folding of repeat proteins. Curr. Opin. Struct. Biol. 2005;15:464–471. doi: 10.1016/j.sbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Stefan N, Martin-Killias P, Wyss-Stoeckle S, Honegger A, Zangemeister-Wittke U, Plückthun A. DARPins recognizing the tumor-associated antigen EpCAM selected by phage and ribosome display and engineered for multivalency. J. Mol. Biol. 2011;413:826–843. doi: 10.1016/j.jmb.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 12.Javadi Y, Itzhaki LS. Tandem-repeat proteins: regularity plus modularity equals design-ability. Curr. Opin. Struct. Biol. 2013;23:622–631. doi: 10.1016/j.sbi.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Marcotte EM, Pellegrini M, Yeates TO, Eisenberg D. A census of protein repeats. J. Mol. Biol. 1999;293:151–160. doi: 10.1006/jmbi.1999.3136. [DOI] [PubMed] [Google Scholar]
- 14.Kajava AV. Tandem repeats in proteins: from sequence to structure. J. Struct. Biol. 2012;179:279–288. doi: 10.1016/j.jsb.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Bateman A, Murzin AG, Teichmann SA. Structure and distribution of pentapeptide repeats in bacteria. Protein Sci. 1998;7:1477–1480. doi: 10.1002/pro.5560070625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bella J, Hindle KL, McEwan PA, Lovell SC. The leucine-rich repeat structure. Cell. Mol. Life Sci. 2008;65:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 18.Tewari R, Bailes E, Bunting KA, Coates JC. Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 2010;20:470–481. doi: 10.1016/j.tcb.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Kajava AV, Gorbea C, Ortega J, Rechsteiner M, Steven AC. New HEAT-like repeat motifs in proteins regulating proteasome structure and function. J. Struct. Biol. 2004;146:425–430. doi: 10.1016/j.jsb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Andrade MA, Petosa C, O’Donoghue SI, Müller CW, Bork P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 21.Kobe B, Kajava AV. When protein folding is simplified to protein coiling: the continuum of solenoid protein structures. Trends Biochem. Sci. 2000;25:509–515. doi: 10.1016/s0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
- 22.Björklund AK, Ekman D, Elofsson A. Expansion of protein domain repeats. PLoS Comput. Biol. 2006;2:e114. doi: 10.1371/journal.pcbi.0020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Remmert M, Biegert A, Linke D, Lupas AN, Söding J. Evolution of outer membrane beta-barrels from an ancestral beta beta hairpin. Mol. Biol. Evol. 2010;27:1348–1358. doi: 10.1093/molbev/msq017. [DOI] [PubMed] [Google Scholar]
- 24.Jawad Z, Paoli M. Novel sequences propel familiar folds. Structure. 2002;10:447–454. doi: 10.1016/s0969-2126(02)00750-5. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhuri I, Söding J, Lupas AN. Evolution of the beta-propeller fold. Proteins. 2008;71:795–803. doi: 10.1002/prot.21764. [DOI] [PubMed] [Google Scholar]
- 26.Berman HM, Kleywegt GJ, Nakamura H, Markley JL. The future of the protein data bank. Biopolymers. 2013;99:218–222. doi: 10.1002/bip.22132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dessailly BH, Nair R, Jaroszewski L, Fajardo JE, Kouranov A, Lee D, Fiser A, Godzik A, Rost B, Orengo C. PSI-2: structural genomics to cover protein domain family space. Structure. 2009;17:869–881. doi: 10.1016/j.str.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray KB, Taylor WR, Thornton JM. Toward the detection and validation of repeats in protein structure. Proteins. 2004;57:365–380. doi: 10.1002/prot.20202. [DOI] [PubMed] [Google Scholar]
- 29.Parra RG, Espada R, Sánchez IE, Sippl MJ, Ferreiro DU. Detecting repetitions and periodicities in proteins by tiling the structural space. J. Phys. Chem. B. 2013;117:12887–12897. doi: 10.1021/jp402105j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsella L, Sirocco F, Trovato A, Seno F, Tosatto SC. REPETITA: detection and discrimination of the periodicity of protein solenoid repeats by discrete Fourier transform. Bioinformatics. 2009;25:i289–i295. doi: 10.1093/bioinformatics/btp232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szklarczyk R, Heringa J. Tracking repeats using significance and transitivity. Bioinformatics. 2004;20(Suppl. 1):i311–i317. doi: 10.1093/bioinformatics/bth911. [DOI] [PubMed] [Google Scholar]
- 32.Heger A, Holm L. Rapid automatic detection and alignment of repeats in protein sequences. Proteins. 2000;41:224–237. doi: 10.1002/1097-0134(20001101)41:2<224::aid-prot70>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 33.Abraham AL, Rocha EP, Pothier J. Swelfe: a detector of internal repeats in sequences and structures. Bioinformatics. 2008;24:1536–1537. doi: 10.1093/bioinformatics/btn234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh I, Sirocco FG, Minervini G, Di Domenico T, Ferrari C, Tosatto SC. RAPHAEL: recognition, periodicity and insertion assignment of solenoid protein structures. Bioinformatics. 2012;28:3257–3264. doi: 10.1093/bioinformatics/bts550. [DOI] [PubMed] [Google Scholar]
- 35.Sillitoe I, Cuff AL, Dessailly BH, Dawson NL, Furnham N, Lee D, Lees JG, Lewis TE, Studer RA, Rentzsch R, et al. New functional families (FunFams) in CATH to improve the mapping of conserved functional sites to 3D structures. Nucleic Acids Res. 2013;41:D490–D498. doi: 10.1093/nar/gks1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jorda J, Baudrand T, Kajava AV. PRDB: Protein Repeat DataBase. Proteomics. 2012;12:1333–1336. doi: 10.1002/pmic.201100534. [DOI] [PubMed] [Google Scholar]
- 38.Luo H, Lin K, David A, Nijveen H, Leunissen JA. ProRepeat: an integrated repository for studying amino acid tandem repeats in proteins. Nucleic Acids Res. 2011;40:D394–D399. doi: 10.1093/nar/gkr1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson AL, Bate MA, Androulakis SG, Bottomley SP, Buckle AM. PolyQ: a database describing the sequence and domain context of polyglutamine repeats in proteins. Nucleic Acids Res. 2011;39:D272–D276. doi: 10.1093/nar/gkq1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2011;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mistry J, Coggill P, Eberhardt RY, Deiana A, Giansanti A, Finn RD, Bateman A, Punta M. The challenge of increasing Pfam coverage of the human proteome. Database. 2013;2013:bat023. doi: 10.1093/database/bat023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose PW, Bi C, Bluhm WF, Christie CH, Dimitropoulos D, Dutta S, Green RK, Goodsell DS, Prlic A, Quesada M, et al. The RCSB Protein Data Bank: new resources for research and education. Nucleic Acids Res. 2013;41:D475–D482. doi: 10.1093/nar/gks1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gómez J, García LJ, Salazar GA, Villaveces J, Gore S, García A, Martín MJ, Launay G, Alcántara R, del-Toro N, et al. BioJS: an open source JavaScript framework for biological data visualization. Bioinformatics. 2013;29:1103–1104. doi: 10.1093/bioinformatics/btt100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Domenico T, Walsh I, Martin AJ, Tosatto SC. MobiDB: a comprehensive database of intrinsic protein disorder annotations. Bioinformatics. 2012;28:2080–2081. doi: 10.1093/bioinformatics/bts327. [DOI] [PubMed] [Google Scholar]