Abstract
Hemolytik (http://crdd.osdd.net/raghava/hemolytik/) is a manually curated database of experimentally determined hemolytic and non-hemolytic peptides. Data were compiled from a large number of published research articles and various databases like Antimicrobial Peptide Database, Collection of Anti-microbial Peptides, Dragon Antimicrobial Peptide Database and Swiss-Prot. The current release of Hemolytik database contains ∼3000 entries that include ∼2000 unique peptides whose hemolytic activities were evaluated on erythrocytes isolated from as many as 17 different sources. Each entry in Hemolytik provides comprehensive information about a peptide, like its name, sequence, origin, reported function, property such as chirality, types (linear and cyclic), end modifications as well as details pertaining to its hemolytic activity. In addition, tertiary structure of each peptide has been predicted, and secondary structure states have been assigned. To facilitate the scientific community, a user-friendly interface has been developed with various tools for data searching and analysis. We hope, Hemolytik will be useful for researchers working in the field of designing therapeutic peptides.
INTRODUCTION
During the past decade, there has been a renewed interest in peptides and peptide-based therapeutics and diagnostics. Peptide-based therapeutics have several advantages, including high specificity, high tissue penetration ability, easy to modify, etc, over small molecules and antibody-based therapeutics (1,2). Despite tremendous therapeutic potential, so far only limited number of peptides has been commercialized as drugs. Development of peptide-based drugs is very challenging and time-consuming process. High toxicity and poor stability are the two key concerns while developing peptide-based drugs (1,3). Most of the peptides, despite their high therapeutic potential, do not reach the clinical trials because of their toxicity (hemolytic activity), and because of the difficulties in their manufacturing.
Toxicity of therapeutic peptides against normal eukaryotic cells is usually first checked by testing their hemolytic activity against red blood cells (RBCs) because RBCs provide a model system, which is physiologically relevant and significantly easier to work with as compared with other systems such as liposomes. In general, peptides having high hemolytic activity are not suitable for therapeutic use. Thus, it is essential to reduce the hemolytic potency of peptide without compromising its therapeutic activity. Presently, there is a paucity of information that can help in designing therapeutic peptides without exhibiting hemolytic activity. A systematic analysis of hemolytic and non-hemolytic peptide sequences is necessary to delineate the features of peptides responsible for their hemolytic activity, which can be taken care while designing a therapeutic peptide.
Over the past decades, a plethora of articles describing therapeutic peptides (e.g. antimicrobial, anticancer, antiviral, cell penetrating peptides) and their hemolytic potential has been published. This information is scattered in the literature and is thus difficult to access. To the best of authors’ knowledge, to date, no heed has been paid to collect and compile the information pertinent to the hemolytic peptides and their potencies. In the present study, for the first time, a systematic attempt has been made to collect this scattered information of experimentally determined hemolytic and non-hemolytic peptides. This information is compiled in the form of a database called Hemolytik. We hope that this database will be useful for researchers working in the field of peptide therapeutics.
SYSTEM AND METHODS
Data acquisition
Data were manually collected from published literature and various databases, including the Antimicrobial Peptide Database (4), Dragon Antimicrobial Peptide Database (5), Collection of Antimicrobial Peptides (6) and Swiss-Prot (7). Hemolytic peptides were included in the database if those were found to be evaluated experimentally using hemolysis assay. Specific searches were carried out to collect all the research articles describing experimentally determined hemolytic peptides. In PubMed, advanced search using keyword such as ‘hemolytic peptides’ (in abstract/title) resulted into ∼900 research articles. Similar searches were also carried out in Swiss-Prot and other related databases using keywords ‘hemolytic/hemolysis’. All research articles were downloaded and compiled systematically. Comprehensive information, including peptide sequences, chirality, end modifications, hemolytic potency, source of RBCs used in the assay, etc, were extracted and compiled for experimentally determined hemolytic and non-hemolytic peptides.
We have made multiple entries of hemolytic peptides if same peptide has been tested against RBCs of different sources (e.g. human, sheep, rabbit). Therefore, the number of total entries in ‘Hemolytik’ is 2970, but unique hemolytic and non-hemolytic sequences are 1750 and 295, respectively.
Database architecture and web interface
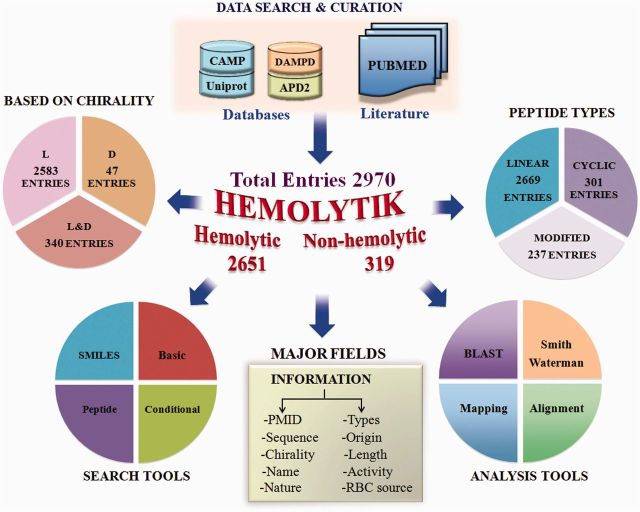
After the collection and compilation of all the information, database was built on an Apache HTTP Server with MySQL server. MySQL is an object-relational database management system (RDBMS), and it works at the backend. It provides commands to retrieve and store the data into the database. HTML, PHP and JAVA scripts were used to develop the front-end web interface. All common gateway interface and database interfacing scripts were written in the PHP and PERL programming language. Because Apache, MySQL and PHP technology are platform-independent and open-source software, these were preferred to develop the database. The architecture of Hemolytik database is shown in Figure 1.
Figure 1.
Architecture of Hemolytik database.
Organization of data
In Hemolytik database, each peptide is assigned a unique entry number, and detailed information about each peptide has been provided. Each entry contains following major fields: (i) name of peptide, (ii) amino acid sequence of peptide, (iii) chirality/conformation of peptide (L/D and linear/cyclic), (iv) details of modified amino acids (e.g. ornithine, β-alanine), (v) function/activity (e.g. antimicrobial) of peptide, (vi) source of peptide (e.g. snake venom), (vii) hemolytic activity, (viii) modifications at N- and C-termini of peptide (e.g. acetylation/amidation) and (ix) source of RBCs used in the assay (e.g. human, mouse).
One of the unique features of Hemolytik database is that it provides structural information of peptides. Structure-function analysis of various therapeutic peptides suggests that a defined secondary structure, with amphipathic distribution of hydrophobic and hydrophilic residues, is the requisite features for their membranolytic activity (8,9). Therefore, understanding of tertiary structure of these peptides is of a considerable interest. However, most of the hemolytic peptides and their analogs’ structures have not been determined and thus are not available in Protein Data Bank (PDB). To provide structural information of peptides, we have predicted tertiary structures of all the peptides having natural amino acids using software PEPstr (10), which is a state-of-the-art method for predicting structure of bioactive peptides. For a given peptide sequence, PEPstr first predicts beta-turn types followed by PSIPRED (11)-predicted secondary structure states and integrates this information along with energy minimization and molecular dynamics using AMBER 11 (12) to predict tertiary structure of peptides. Using DSSP software (13), eight secondary structure states have also been assigned for each peptide.
In Hemolytik database, most of the derivatives of existing therapeutic peptides contain non-natural/modified amino acids (e.g. d-amino acids, ornithine, β-alanine) and non-proteinogenic moieties (e.g. 6-amino hexanoic acid, p-Hydroxy cinnamic acid). To the best of authors’ knowledge, there is no web server that can predict the tertiary structures of peptides having non-natural and modified amino acids. Therefore, we have predicted structures of peptides having natural amino acids, d-amino acids, end modifications like acetylation/amidation as well as peptides having ornithine as modified amino acid by extending the use of AMBER 11 (12) in PEPstr algorithm. For changing the stereochemistry of a residue in d-form, flip command of AMBER 11 was used. For non-natural residues like ornithine (14), force field library for that residue was used in AMBER 11. In our database, we maintain tertiary structure of peptides in PDB format.
Data statistics
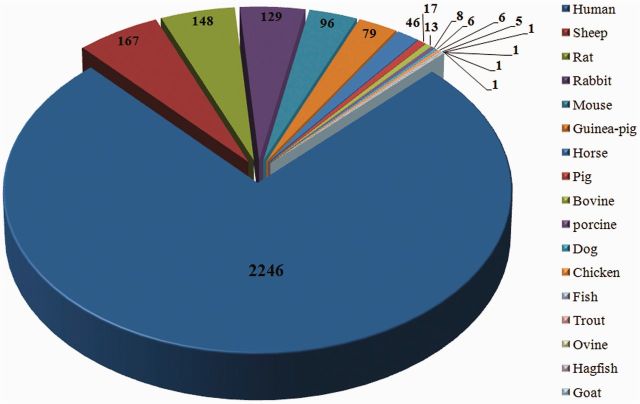
The current release of Hemolytik database contains 2970 entries (Figure 1) based on hemolysis assays carried out on RBCs isolated from different organisms. During data curation and compilation, we have observed that many peptides have shown differential hemolytic activity on different RBCs isolated from different sources (e.g. human, sheep, rat). Therefore, we have incorporated information of as many as 17 RBC sources (Figure 2). In most of the entries, hemolytic potencies of peptides were evaluated on human RBCs (2246) followed by sheep (167) and rat RBCs (148, Figure 2). We have made multiple entries of single peptide if the same peptide has been found to be tested either on different RBCs (e.g. human and rabbit RBCs) or at different concentrations (e.g. at 25 µM and 50 µM). Because the information related to experimentally determined non-hemolytic peptides is equally important, we have extracted the information related to non-hemolytic peptides, and 319 entries of non-hemolytic peptides (unique 295) were compiled. In addition, there are few entries where same peptides have been reported to be hemolytic as well as non-hemolytic. This is probably due to the different experimental conditions (e.g. type of RBC source, incubation temperature, concentration of peptides). As we do not want to lose any information, we have incorporated this information as well.
Figure 2.
Classification of peptides based on sources of RBCs used to evaluate the hemolytic potencies.
The peptides in Hemolytik database belong to diverse classes of therapeutic peptides and have different functions that include antimicrobial, antiparasitic, anticancer, cell penetrating, antiviral peptides, etc. However, most of the entries in Hemolytik belong to antimicrobial peptides followed by anticancer peptides. We have also categorized peptides based on their conformation (linear/cyclic) and chirality, i.e. L/D/Mix (both L and D). Hemolytik contains 2669 entries of linear peptides, while 301 entries have information about cyclic peptides. We have compiled 2583 entries of peptides containing only l-amino acids, 47 entries of peptides consist of only d-amino acids and 340 entries of peptides have both l- and d-amino acids. In addition, we have also collected and compiled information related to chemical modifications in peptides like modified amino acids (α-aminoisobutyric acid, norleucine, ornithine, etc) or non-proteinogenic moieties (tetrahydroisoquinoline carboxylic acid, octahydro-1H-indole-2-carboxylic acid, γ-aminobutyric acid) and modified peptide chemistry (reduced amide bond ψ[CH2NH]). A total of 237 peptide entries have been made, which provide information about hemolytic peptides having chemical modifications.
Integration of web tools
Many user-friendly tools have been integrated in Hemolytik for extraction and analysis of peptides. Following are the main tools provided with the Hemolytik database.
Search facility
We have implemented four search tools that include basic search, conditional search, peptide search and SMILES search. Basic search option allows users to perform a search on any field of the database like PubMed ID, peptide name, peptide sequence, chirality of peptide, origin, nature of peptide, RBC source, etc. User can display any or all fields for selected searched records. Conditional search facility allows users to perform multiple queries at a time. Under conditional search option, users can perform complex search by adding any number of queries. In addition, user can select conditions (e.g. AND and OR) between queries. In peptide search, user can search a peptide sequence in the database. There are two options for peptide search: (i) containing peptide: it is for searching user-defined peptide sequence in Hemolytik. Users can search whether their peptide of interest is present (partially or complete) in any hemolytic peptide or not; and (ii) exact search: it allows users to search hemolytic peptides, which are identical to user’s peptide. SMILES search option provides facility to search SMILES notation of a given peptide against Hemolytik peptide database in SMILES format.
Categorization
Categorization is basically a powerful browsing facility that allows users to browse data on the four major fields that include (i) source of RBCs used in the assay, (ii) chirality of peptides, (iii) function of peptides and (iv) length of peptides. In Hemolytik, we have covered 17 different sources of RBCs on which peptides have been tested. These peptides have shown different hemolytic potencies on different RBCs. Using this field, users can browse peptides tested on RBCs isolated from a particular source. In the chirality field, three types of peptides have been compiled: (i) peptides having all l-amino acids, (ii) peptides having all d-amino acids and (iii) peptides having both l- and d-amino acids (mixed). User can browse this information using chirality field. In addition, user can browse peptides on the basis of their function. We have covered peptides with >15 types of functions, including anticancer, antimicrobial, cell-penetrating, antiviral, anti-parasitic, etc. Peptides based on their length can also be browsed. In Hemolytik, peptides’ length varies from 2 to 104 amino acids. However, most of the peptides have length between 11 and 15 amino acids.
BLAST search
This search tool facilitates users to perform similarity-based search against hemolytic and non-hemolytic peptides. Users can submit peptide sequences in FASTA format and select different parameters like weight matrix and expectation value for performing BLAST search (15).
Smith–Waterman algorithm
Because Smith–Waterman algorithm (16) performs similarity search more effectively in case of small peptides, we have integrated this tool. This option allows users to search hemolytic or non-hemolytic peptides in the database that are similar to their peptides. Users can search multiple peptide sequences at a time by submitting sequences in FASTA format.
Alignment
This tool allows users to align their sequences with the peptides in Hemolytik database. A user can input multiple FASTA sequences in the sequence box and peptide IDs of Hemolytik database in the ID box, and get aligned sequences. User also has the option to upload a file in PDB format and align its structure with the structure of the peptide whose ID is provided in the box.
Mapping
It allows the users to map hemolytic peptides on their peptide sequence. It allows the user to run a sub-search and super-search. In sub-search, a given peptide is mapped against all peptides of Hemolytik database, whereas super-search returns similar peptides of our database against a protein sequence given as query. It also allows user to submit protein or polypeptide sequence to identify segments that are identical to hemolytic peptides.
DISCUSSION
The field of therapeutic peptides is growing very rapidly due to substantial technological progress (1,17). Literature on therapeutic peptides is rapidly adding (18), and therefore in the past few years only, many comprehensive databases of various therapeutic peptides including antimicrobial peptides (5,6), cell-penetrating peptides (19), tumor-homing peptides (20) and quorum-sensing peptides (21) have been developed. In addition, an interesting and useful resource of blood–brain barrier peptides—Brainpeps—(22) has also been developed recently. Brainpeps not only provides information of blood–brain barrier peptides but also gives comprehensive information related to many experimental techniques used to study the peptide penetration with various parameters. These databases have demonstrated the increasing popularity of peptides as therapeutic candidates. However, despite several advantages, peptides often suffer from poor in vivo stability and high toxicity toward eukaryotic cells (18), which is often judged by their hemolytic activities. Therefore, most of the research on therapeutic peptides is currently being focused on designing peptide derivatives having low/no hemolytic activities while retaining their therapeutic activity (23,24). Therefore, tremendous data related to hemolytic peptides and their derivatives with their hemolytic potencies have been reported in the past. This information may be very useful for researchers/scientists to design therapeutic peptides without hemolytic potencies. However, no attempt has been made to catalog this information, and thus, it is currently difficult to access this useful information. Hemolytik is a first comprehensive database of its kind that provides experimental information related to hemolytic peptides and their potencies.
Besides collection of hemolytic peptides and their potencies, various web-based tools have been integrated in Hemolytik database, which facilitate various types of analysis. Users can make the best use of Hemolytik in the following ways: (i) while designing therapeutic peptides, users can check whether their peptides of interest are already reported to be hemolytic or not; (ii) because Hemolytik also contains experimentally determined non-hemolytic peptides, users can select the least hemolytic or non-hemolytic peptides for further therapeutic applications; and (iii) users can exploit the predicted structural information of hemolytic peptides for docking or molecular dynamics of the peptide-membrane complex.
The accurate prediction of therapeutic activities of peptides may expedite peptide-based drug discovery. In this context, Hemolytik database will be useful for developing novel in silico prediction, as well as designing methods for hemolytic or non-hemolytic peptides. In the recent past, few prediction and quantitative structure activity relationship models on various therapeutic peptides, including antimicrobial peptides (25–27), cell-penetrating peptides (28–31), antioxidant peptides (32,33), have been developed, which will be helpful to reduce the costs and efforts involved in laboratory screening. The optimized structures and SMILES of hemolytic peptides available in the Hemolytik database can also be used to develop quantitative structure activity relationship models for rapid screening of therapeutic peptides with less hemolytic activity. In conclusion, Hemolytik would be very useful to the scientific community working in the field of therapeutic peptides.
UPDATE OF HEMOLYTIK
The web interface provides an option for users to submit a new entry of experimentally determined hemolytic and non-hemolytic peptides by filling HTML form. To maintain the high standard of quality, we will confirm the validity of new entry before including in Hemolytik. In addition, our team will also update this database regularly.
LIMITATIONS AND FUTURE DEVELOPMENTS
Besides a collection of hemolytic and non-hemolytic peptide sequences, Hemolytik database also provides structural information of peptides. In Hemolytik, there are many peptides that have modified amino acids (e.g. 4-nitrophenylalanine)/non-proteinogenic moieties. We have made an attempt to predict the tertiary structure of peptides with non-natural (d-form) and modified amino acids (ornithine). However, a limitation of this database is that structures of few modified peptides (with complex modifications) could not be predicted. In the future, as more and more published force field libraries of individual modified residues will be available, the prediction of peptides having such residues will be feasible.
AVAILABILITY AND REQUIREMENTS
Hemolytik is available at http://crdd.osdd.net/raghava/hemolytik/
FUNDING
Council of Scientific and Industrial Research Projects: Open Source Drug Discovery and GENESIS BSC0121. Department of Biotechnology (Project: BTISNET), Government of India. Funding for open access charge: Projects: Open Source Drug Discovery and GENESIS BSC0121.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sato AK, Viswanathan M, Kent RB, Wood CR. Therapeutic peptides: technological advances driving peptides into development. Curr. Opin. Biotechnol. 2006;17:638–642. doi: 10.1016/j.copbio.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Vlieghe P, Lisowski V, Martinez J, Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug Discov. Today. 2010;15:40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chem. Biol. Drug Des. 2013;81:136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 4.Wang G, Li X, Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seshadri Sundararajan V, Gabere MN, Pretorius A, Adam S, Christoffels A, Lehvaslaiho M, Archer JA, Bajic VB. DAMPD: a manually curated antimicrobial peptide database. Nucleic Acids Res. 2012;40:D1108–D1112. doi: 10.1093/nar/gkr1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas S, Karnik S, Barai RS, Jayaraman VK, Idicula-Thomas S. CAMP: a useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010;38:D774–D780. doi: 10.1093/nar/gkp1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The UniProt Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peck-Miller KA, Blake J, Cosand WL, Darveau RP, Fell HP. Structure-activity analysis of the antitumor and hemolytic properties of the amphiphilic alpha-helical peptide, C18G. Int. J. Pept. Protein Res. 1994;44:143–151. doi: 10.1111/j.1399-3011.1994.tb00569.x. [DOI] [PubMed] [Google Scholar]
- 9.McInnes C, Kondejewski LH, Hodges RS, Sykes BD. Development of the structural basis for antimicrobial and hemolytic activities of peptides based on gramicidin S and design of novel analogs using NMR spectroscopy. J. Biol. Chem. 2000;275:14287–14294. doi: 10.1074/jbc.275.19.14287. [DOI] [PubMed] [Google Scholar]
- 10.Kaur H, Garg A, Raghava GP. PEPstr: a de novo method for tertiary structure prediction of small bioactive peptides. Protein Pept. Lett. 2007;14:626–631. doi: 10.2174/092986607781483859. [DOI] [PubMed] [Google Scholar]
- 11.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 12.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joosten RP, te Beek TA, Krieger E, Hekkelman ML, Hooft RW, Schneider R, Sander C, Vriend G. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2011;39:D411–D419. doi: 10.1093/nar/gkq1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pendley SS, Yu YB, Cheatham TE., 3rd Molecular dynamics guided study of salt bridge length dependence in both fluorinated and non-fluorinated parallel dimeric coiled-coils. Proteins. 2009;74:612–629. doi: 10.1002/prot.22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 16.Pearson WR. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- 17.Otvos L., Jr Peptide-based drug design: here and now. Methods Mol. Biol. 2008;494:1–8. doi: 10.1007/978-1-59745-419-3_1. [DOI] [PubMed] [Google Scholar]
- 18.McGregor DP. Discovering and improving novel peptide therapeutics. Curr. Opin. Pharmacol. 2008;8:616–619. doi: 10.1016/j.coph.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Gautam A, Singh H, Tyagi A, Chaudhary K, Kumar R, Kapoor P, Raghava GP. CPPsite: a curated database of cell penetrating peptides. Database. 2012;2012:bas015. doi: 10.1093/database/bas015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapoor P, Singh H, Gautam A, Chaudhary K, Kumar R, Raghava GP. TumorHoPe: a database of tumor homing peptides. PLoS One. 2012;7:e35187. doi: 10.1371/journal.pone.0035187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wynendaele E, Bronselaer A, Nielandt J, D'Hondt M, Stalmans S, Bracke N, Verbeke F, Van De Wiele C, De Tre G, De Spiegeleer B. Quorumpeps database: chemical space, microbial origin and functionality of quorum sensing peptides. Nucleic Acids Res. 2013;41:D655–D659. doi: 10.1093/nar/gks1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Dorpe S, Bronselaer A, Nielandt J, Stalmans S, Wynendaele E, Audenaert K, Van De Wiele C, Burvenich C, Peremans K, Hsuchou H, et al. Brainpeps: the blood-brain barrier peptide database. Brain Struct. Funct. 2012;217:687–718. doi: 10.1007/s00429-011-0375-0. [DOI] [PubMed] [Google Scholar]
- 23.Kondejewski LH, Jelokhani-Niaraki M, Farmer SW, Lix B, Kay CM, Sykes BD, Hancock RE, Hodges RS. Dissociation of antimicrobial and hemolytic activities in cyclic peptide diastereomers by systematic alterations in amphipathicity. J. Biol. Chem. 1999;274:13181–13192. doi: 10.1074/jbc.274.19.13181. [DOI] [PubMed] [Google Scholar]
- 24.Qin C, Zhong X, Bu X, Ng NL, Guo Z. Dissociation of antibacterial and hemolytic activities of an amphipathic peptide antibiotic. J. Med. Chem. 2003;46:4830–4833. doi: 10.1021/jm0341352. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Ding Y, Wen H, Lin Y, Hu Y, Zhang Y, Xia Q, Lin Z. QSAR modeling and design of cationic antimicrobial peptides based on structural properties of amino acids. Comb. Chem. High Throughput Screen. 2012;15:347–353. doi: 10.2174/138620712799361807. [DOI] [PubMed] [Google Scholar]
- 26.Wang P, Hu L, Liu G, Jiang N, Chen X, Xu J, Zheng W, Li L, Tan M, Chen Z, et al. Prediction of antimicrobial peptides based on sequence alignment and feature selection methods. PLoS One. 2011;6:e18476. doi: 10.1371/journal.pone.0018476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torrent M, Di Tommaso P, Pulido D, Nogues MV, Notredame C, Boix E, Andreu D. AMPA: an automated web server for prediction of protein antimicrobial regions. Bioinformatics. 2012;28:130–131. doi: 10.1093/bioinformatics/btr604. [DOI] [PubMed] [Google Scholar]
- 28.Stalmans S, Wynendaele E, Bracke N, Gevaert B, D'Hondt M, Peremans K, Burvenich C, De Spiegeleer B. Chemical-functional diversity in cell-penetrating peptides. PLoS One. 2013;8:e71752. doi: 10.1371/journal.pone.0071752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gautam A, Chaudhary K, Kumar R, Sharma A, Kapoor P, Tyagi A, Raghava GP. In silico approaches for designing highly effective cell penetrating peptides. J. Transl. Med. 2013;11:74. doi: 10.1186/1479-5876-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders WS, Johnston CI, Bridges SM, Burgess SC, Willeford KO. Prediction of cell penetrating peptides by support vector machines. PLoS Comput. Biol. 2011;7:e1002101. doi: 10.1371/journal.pcbi.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobchev DA, Mager I, Tulp I, Karelson G, Tamm T, Tamm K, Janes J, Langel U, Karelson M. Prediction of cell-penetrating peptides using artificial neural networks. Curr. Comput. Aided Drug Des. 2010;6:79–89. doi: 10.2174/157340910791202478. [DOI] [PubMed] [Google Scholar]
- 32.Li YW, Li B, He J, Qian P. Structure-activity relationship study of antioxidative peptides by QSAR modeling: the amino acid next to C-terminus affects the activity. J. Pept. Sci. 2011;17:454–462. doi: 10.1002/psc.1345. [DOI] [PubMed] [Google Scholar]
- 33.Li YW, Li B. Characterization of structure-antioxidant activity relationship of peptides in free radical systems using QSAR models: key sequence positions and their amino acid properties. J. Theor. Biol. 2013;318:29–43. doi: 10.1016/j.jtbi.2012.10.029. [DOI] [PubMed] [Google Scholar]