Abstract
Rice is the most important staple food for a large part of the world’s human population and also a key model organism for biological studies of crops as well as other related plants. Here we present RiceWiki (http://ricewiki.big.ac.cn), a wiki-based, publicly editable and open-content platform for community curation of rice genes. Most existing related biological databases are based on expert curation; with the exponentially exploding volume of rice knowledge and other relevant data, however, expert curation becomes increasingly laborious and time-consuming to keep knowledge up-to-date, accurate and comprehensive, struggling with the flood of data and requiring a large number of people getting involved in rice knowledge curation. Unlike extant relevant databases, RiceWiki features harnessing collective intelligence in community curation of rice genes, quantifying users' contributions in each curated gene and providing explicit authorship for each contributor in any given gene, with the aim to exploit the full potential of the scientific community for rice knowledge curation. Based on community curation, RiceWiki bears the potential to make it possible to build a rice encyclopedia by and for the scientific community that harnesses community intelligence for collaborative knowledge curation, covers all aspects of biological knowledge and keeps evolving with novel knowledge.
INTRODUCTION
Rice (Oryza sativa) is not only the most important staple food feeding a large part of the world population, but also an important model organism for biological studies of crops as well as other related plants (1,2). For this reason, rice was chosen as the first crop for whole genome sequencing. The availability of genome sequences of the two most common cultivated rice subspecies, indica 93-11 and japonica Nipponbare (3,4), along with the resequencing of cultivated and wild rice accessions (5), enables the in-depth characterization of rice genes (6), exploration of agronomically important traits (7) and investigation of rice diversity and domestication (8).
Multiple databases (9–13) have been developed for rice. However, building standardized rice reference genomes with comprehensive and accurate annotations remains a formidable challenge. Extant related databases are most based on expert curation, viz., conducted manually by dedicated experts. To perform curation, expert curators often administer raw biological data, conduct a thorough literature search, extract essential information from multiple publications, curate the information using structured and controlled vocabularies and then submit the information to a knowledge database [e.g., the Reference Sequence database (11) at the National Center for Biotechnology Information] to make it public. However, with the exponentially exploding volume of rice knowledge and other relevant data, expert curation is becoming more laborious and time-consuming. Keeping biological knowledge in expert-curated databases comprehensive, up-to-date and accurate is increasingly lagging behind knowledge creation, or worse, not being done at all in fields where insufficient funds can be allocated to curation (14). Simply put, although expert-curated databases have traditionally proven important for scientific research, they are struggling with the flood of knowledge. Explosive growth in the volume of rice data requires a large number of people getting involved in knowledge curation, viz., community curation.
Community curation exploits the whole power of the scientific community for knowledge curation (15–19). A case in point that harnesses community intelligence in knowledge integration is Wikipedia (http://www.wikipedia.org). Wikipedia is an online encyclopedia, allowing any user to create/edit any content. It features collaborative knowledge curation, up-to-date content, huge coverage and low cost for maintenance (20). Despite fears that the openness of editorial capacity could lead to incorporation of significant flawed content (21), it is reported that Wikipedia rivals the traditional encyclopedia in accuracy (22). Broad participation in Wikipedia not only increases knowledge coverage and keeps knowledge up-to-date, but also improves knowledge accuracy (‘with enough eyeballs, all bugs are shallow’; http://oreilly.com/web2/archive/what-is-web-20.html). Owing to the extraordinary success of Wikipedia, it has been advocated that biological databases go wiki (23). As a consequence, more than a dozen biological wikis (24–42) have been constructed to exploit the full potential of the scientific community for knowledge curation, such as EcoliWiki for the model bacteria Escherichia coli (27), Gene Wiki for human gene annotation (28) and PDBWiki for protein structures (31). To date, however, there is no community-curated resource for rice.
To establish a platform for community integration of rice data and to facilitate efficient management of community knowledge on rice, here we develop RiceWiki (http://ricewiki.big.ac.cn), a wiki-based, publicly editable and open-content platform for community curation of rice genes. Unlike extant relevant databases, RiceWiki features harnessing community intelligence in curation of rice genes, quantifying users’ contributions in each curated gene and providing explicit authorship for each contributor in any given gene, with the aim to attract more participation from the scientific community in collaborative and collective curation of rice genes.
IMPLEMENTATION
RiceWiki has been implemented using MediaWiki (http://www.mediawiki.org; a free and open source wiki engine; version 1.18.4), MySQL (http://www.mysql.org; a free and popular relational database management system; version 5.1.58) and PHP (http://www.php.net; a widely used general-purpose scripting language; version 5.2.17) on a Red Hat Enterprise Linux Server. The wide adoption of community intelligence in knowledge curation is primarily attributable to free wiki software such as MediaWiki that provides a collaborative framework for knowledge collection, management and dissemination (that powers Wikipedia). MediaWiki allows any user to add, modify or delete any content (with customized permission control for editing) via a web browser without any extra add-ons and thus enables web content to be edited easily, swiftly and collaboratively by multiple different users. Every page in MediaWiki has an associated page named ‘History’; every change made to a page can be stored; the user responsible for every change can be identified and every history revision can be reviewed or recovered. Built on MediaWiki, RiceWiki enables users to be involved in an ongoing process of creation and collaboration that constantly changes the contents. Thus, RiceWiki can significantly ease the process of knowledge collection, curation and sharing, befitting the exploding volume of biological data.
In addition, MediaWiki allows any user to develop customized functionalities by packaging a bunch of codes as MediaWiki extensions. We installed our newly developed extension named ‘AuthorReward’ (http://www.mediawiki.org/wiki/Extension:AuthorReward) in RiceWiki, with the aim to attract more participation from the scientific community for collaborative curation of rice genes. A wiki page can be collaboratively curated by multiple users and thus may have different edit versions. For each version that is contributed by a specific person, ‘AuthorReward’ quantifies his/her contribution by factoring edit quality as well as edit quantity; the edit quantity amounts to the edit distance in comparison with its previous version (i.e. the minimum number of edit operations required to transform one string into the other), and the edit quality corresponds to whether the edit persists in comparison with the last version, ranging from −1, when the edit is entirely reverted (short-lived), to 1, indicating that the edit is totally preserved in the last version (long-lived). Because one person may perform many discontinuous edits for a wiki page, his/her contribution score in this page is the sum of quantified contributions over all contributed edits. ‘AuthorReward’ provides RiceWiki with an authorship metric; it quantifies users’ contributions and yields explicit authorship for each wiki page according to their quantitative contributions (described later in the text). Thus, ‘AuthorReward’ bears the potential to increase community participation in RiceWiki and to achieve community curation of massive biological knowledge. All extensions as well as software installed in RiceWiki can be found at http://ricewiki.big.ac.cn/index.php/Special:Version.
In RiceWiki, users can access any content, but only registered users can perform the edits. This restriction is due to a trade-off between simplicity for users to make contributions and reliability of the edits provided. Open identity provided by registration not only improves content reliability and increases users’ collaborations and communications, but it is also supportive to reward community-curated efforts by giving explicit authorship. Thus, although the requirement of registration poses an unpleasant obstacle to community curation, it is of crucial significance for bio-wikis that would like to give credit to all contributors in reward for community-provided content. Additionally, it is beneficial to greatly avoid vandalism or spammed content in bio-wikis, albeit, of course, there are multiple solutions for vandalism detection (43).
DATABASE CONTENT
RiceWiki incorporates the two common cultivated rice subspecies (O. sativa indica 93–11 and O. sativa japonica Nipponbare) and covers >66 000 rice genes. As implemented on MediaWiki, RiceWiki inherits its features: each content page is associated with a discussion page (where users can discuss content or leave a comment), a history page (where revision as well as its contributor can be recognized) and category terms (that increase the usability for information management).
The central objects of RiceWiki are rice genes and thus each gene corresponds to a specific wiki page. To provide easy access to gene-specific pages, they are built based on gene identifiers. Although they can also be based on gene names, it is noted that only a small part of rice genes has been well studied. With the ongoing studies on the biological functions of rice genes, a gene name often comes and goes as our knowledge about that entity increases, which would bring uncertainty of designations and synonyms of rice genes. Thus, gene identifiers are relatively stable to refer to specific genes, as they do not change with the accumulation of novel information. The gene information in RiceWiki was initially seeded from NCBI RefSeq (11), Ensembl (44), RAP-DB (12) and MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu).
The content of every gene in RiceWiki is structured into multiple sections, namely, ‘Annotated Information’, ‘Structured Information’, ‘Labs Working on This Gene’ and ‘References’, as well as a one-sentence summary for gene description at the top of each page (Figure 1). ‘Annotated Information’ is organized as free text and is helpful to users who share their knowledge and contribute edits without training in the curation or wiki techniques, significantly simplifying edits’ provision and lowering technological entrance barriers for wider community participation in curation. It can also fall into several sub-sections, such as ‘Function’, ‘Evolution’ and ‘Expression’, making it convenient to direct users to the sub-section(s) of interest. Although these sub-sections are preset, new sub-sections can be easily added and irrelevant sub-section(s) can be deleted. Such arrangement with multiple sub-sections enables not only automatic entry of information via application programming interface but also facilitates users to intuitively edit the information by clicking an ‘edit’ link available to each sub-section. On the contrary, ‘Structured Information’ is organized structurally in the form of a table, including gene symbol, gene description, sequence information, expression profile, external links to other related databases and a set of images from GBrowse (45) showing the genomic context of the gene and the gene structure. Albeit the structure of this table is preset, it also allows users to provide updates and additions. ‘Labs Working on This Gene’ is a list of laboratories throughout the world working on this gene, facilitating communication and collaboration in curation of this gene. ‘References’ are publications closely related to this gene and automatically formatted using the ‘Cite’ extension (http://www.mediawiki.org/wiki/Extension:Cite).
Figure 1.
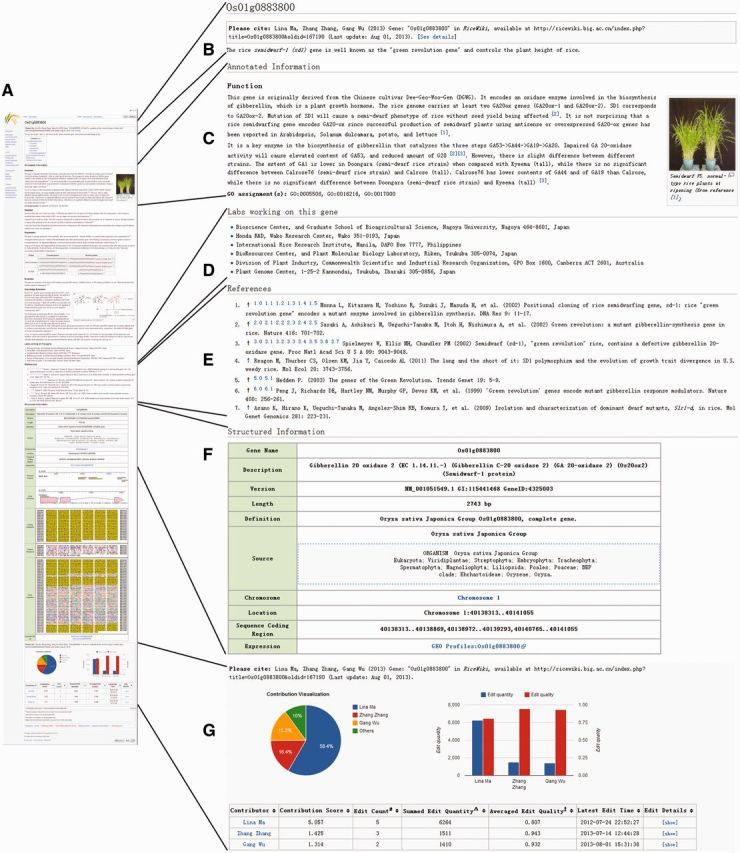
Screenshots of the RiceWiki page for the rice semidwarf-1 (sd1) gene, available at http://ricewiki.big.ac.cn/index.php/Os01g0883800. This gene was collaboratively curated by nine researchers, yielding 89 versions as of 1 August 2013. (A) Whole page, containing multiple different sections. (B) Brief authorship information in reward for community curation and a one-sentence summary for description of this gene. (C) Annotated information. It is organized as free text and contains several sub-sections. (D) Labs working on this gene. It is a list of laboratories working on this gene derived from references and provided by the community. (E) References, which are automatically generated and formatted with the help of the ‘Cite’ extension. (F) Structured information. It is organized structurally in the form of a table, including gene symbol, gene description, sequence information, gene structure, etc. (G) Authorship details generated by the ‘AuthorReward’ extension that quantifies researchers’ contributions and provides explicit authorship according to their quantitative contributions. The cutoff score for awarding authorship is configurable and set to 1 (by default) in RiceWiki.
The major focus of RiceWiki is to exploit the full potential of the scientific community in collaborative curation of rice genes. It should be noted, however, that one crucial essential to making knowledge aggregation successful in RiceWiki and also in other bio-wikis is sufficient participation (19), which requires a large number of people from the scientific community to curate biological knowledge collectively and collaboratively. Unfortunately, the current status is that, despite the presence of well-constructed bio-wikis, most researchers seldom make direct contributions. The reason is well known: most people in areas covered by bio-wikis are in academic research fields, and the currency of academic research careers is authorship, and bio-wikis do not offer attributable authorship. It is an especially severe issue for young researchers, who are most open to new technologies such as wikis, but for whom one’s publication record is the significant determinant to his/her academic professional success (41). It is inevitably time-consuming and poorly motivating for researchers to spend time performing curation without career-advancing credit (46) as opposed to writing their next paper–although, arguably, contributing to a common and open resource such as a bio-wiki may have much broader scientific impacts. There is no mechanism to reward people who perform knowledge curation in bio-wikis. It has been recognized that the major limitation deterring researchers from active participation in bio-wikis is the lack of explicit authorship and thus no credit for their contributions (17,19,41). It should be also noted, however, that not all people care about authorship, e.g., long-term Wikipedia editors. Despite this, authorship might be essential to academic-based researchers.
To attract more participation from the scientific community for RiceWiki and to make it a vivid platform for community curation of rice genes, we installed ‘AuthorReward’ (47), an extension to MediaWiki, which provides a standard practice to reward community-provided contents in bio-wikis by quantifying researchers’ contributions and providing explicit authorship according to their quantitative contributions. For each gene-centric page in RiceWiki, community curation is quantified as a contribution score for each contributor in which both edit quantity and quality are taken into account (Figure 1). Accordingly, authorship in RiceWiki is awarded only to a contributor whose contribution score is >1 (by default; this cut-off score is configurable). At the top of each page, we provide the authorship information, displaying contributor name(s), gene ID, hyperlink to this gene and last update time, which is aimed to encourage community participation and to show clearly how to cite community-curated efforts. Additionally, at the bottom of each page, we present the detailed information, including a pie chart to visualize quantified contributions of multiple contributors who were involved in curating this gene, a histogram to depict the edit quantity and quality for each contributor and a table summarizing contributor name, contribution score, edit count, edit quantity, edit quality, last edit time and edit details. As one person may perform curations for multiple different pages, we define the total contribution of this person to RiceWiki as the sum of multiple contribution scores in all participated pages. With explicit authorship as a reward for community curation, RiceWiki has the potential to attract more people to share their expertise and to provide edits on genes of their interest.
DISCUSSION AND FUTURE DIRECTIONS
The explosion of multiple different bio-wikis and RiceWiki shows the potential of ‘wikiomics’ in action (48). Considering the ever-growing volume of rice data and related literature and contrastingly the relatively small number of expert curators working on rice, it should be noted that community curation is an important complement to expert curation, and community-based bio-wikis, like RiceWiki, are not aimed for replacement of traditionally expert-driven databases. RiceWiki harnesses community intelligence in curating a wide range of rice-related topics and thus can save considerable time and efforts of expert curators. To encourage collaborative curations of rice genes in RiceWiki, expert curators can conduct quality control for community-contributed information and provide a variety of training (e.g., webinars, online tutorials and open discussion) for the community on how to perform curation in an accurate and standard manner. Meanwhile, journals can be also involved in curation by building a mechanism to regard community curation as a compulsory post-publication process and by providing obligations or incentives to authors submitting relevant information to RiceWiki. Such a mechanism has already been put into practice in the journal Plant Physiology, partnering with The Arabidopsis Information Resource to increase the curation of Arabidopsis. Wider adoption of this practice would be enormously beneficial to curatorial efficiency, accuracy and reliability (as testified by the admirable annotations for Arabidopsis). Therefore, the community as well as expert curators, authors, and journals should collaborate together to make RiceWiki more influential and to achieve community curation of massive rice knowledge.
Future directions for RiceWiki include establishment of close collaborations with laboratories in the world working on rice. ‘Nothing great is ever accomplished in isolation’—Yo-Yo Ma. Rice knowledge and related data developed by many laboratories and researchers should be added to RiceWiki and shared with the whole scientific community. We will also promote, as best as we can partnership with journals to require community curation as a compulsory post-publication. In addition, we encourage investigators/teachers to incorporate community curation of rice genes in RiceWiki as student assignments. For example, N students collaborate to curate N rice genes, where N ≥ 3, and contribution score for each student should be >1. RiceWiki will continue to integrate more types of data (e.g., mutants, repetitive elements, expression and phenotyping) from different resources and improve the connections with existing relevant databases. We have collected ∼40 000 rice-related publications from PubMed (http://www.ncbi.nlm.nih.gov/pubmed/), including title, author(s), affiliation(s), abstract and hyperlinks to the full text (if available), and currently are attempting to dig out the ‘treasure’ from the flood of literature. Therefore, RiceWiki will also integrate tools particularly for literature mining (49,50) and incorporate literature-based curated annotation, to realize automatic information retrieval and improve the credibility of community-provided contents. In response to an update of literature collection, RiceWiki will build a mechanism to send an invitation to authors with recent publications for curation of specific genes (51).
In sum, RiceWiki serves as a community-curated knowledgebase for the rice research community. It exploits the whole power of the scientific community in collaborative curation of rice genes by rewarding community-provided content through contribution quantification and explicit authorship. Such a collaborative community-contributed and contribution-rewarded resource would make it possible to build a rice encyclopedia by and for the scientific community (52) that harnesses collective intelligence for collaborative knowledge curation, covers all aspects of biological knowledge and keeps evolving with novel knowledge.
DATABASE AVAILABILITY
RiceWiki is freely available at http://ricewiki.big.ac.cn.
FUNDING
The National Natural Science Foundation of China [31200978 to L.M. and 31100915 to L.H.]; The ‘100-Talent Program’ of Chinese Academy of Sciences [Y1SLXb1365 to Z.Z.]; The National Basic Research and Development Program [973 Program; 2010CB126604 to J.X.]; National Programs for High Technology Research and Development [863 Program; 2012AA020409 to Z.Z. and J.X.], the Ministry of Science and Technology of the People’s Republic of China. Funding for open access charge: National Programs for High Technology Research and Development [863 Program; 2012AA020409], the Ministry of Science and Technology of the People’s Republic of China.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr. Jeffrey P. Townsend at Yale University for valuable discussions on this work. They also thank anonymous reviewers for constructive comments on this manuscript.
REFERENCES
- 1.Shimamoto K, Kyozuka J. Rice as a model for comparative genomics of plants. Annu. Rev. Plant. Biol. 2002;53:399–419. doi: 10.1146/annurev.arplant.53.092401.134447. [DOI] [PubMed] [Google Scholar]
- 2.Paterson AH, Freeling M, Sasaki T. Grains of knowledge: genomics of model cereals. Genome Res. 2005;15:1643–1650. doi: 10.1101/gr.3725905. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 4.Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X, Dong Y, Gutenkunst RN, Fang L, Huang L, et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2012;30:105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- 6.Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, et al. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 2010;20:646–654. doi: 10.1101/gr.100677.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 8.Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao W, Wang J, He X, Huang X, Jiao Y, Dai M, Wei S, Fu J, Chen Y, Ren X, et al. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 2004;32:D377–D382. doi: 10.1093/nar/gkh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y, Wakimoto H, Yang CC, Iwamoto M, Abe T, et al. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 2013;54:e6. doi: 10.1093/pcp/pcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, Xia Y, Li X, Hou L, Yu J. The Rice Genome Knowledgebase (RGKbase): an annotation database for rice comparative genomics and evolutionary biology. Nucleic Acids Res. 2013;41:D1199–1205. doi: 10.1093/nar/gks1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker M. Databases fight funding cuts. Nature. 2012;489:19. doi: 10.1038/489019a. [DOI] [PubMed] [Google Scholar]
- 15.Salzberg SL. Genome re-annotation: a wiki solution? Genome Biol. 2007;8:102. doi: 10.1186/gb-2007-8-1-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu JC, Aramayo R, Bolser D, Conway T, Elsik CG, Gribskov M, Kelder T, Kihara D, Knight TF, Jr, Pico AR, et al. The emerging world of wikis. Science. 2008;320:1289–1290. doi: 10.1126/science.320.5881.1289b. [DOI] [PubMed] [Google Scholar]
- 17.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, Hill DP, Kania R, Schaeffer M, St Pierre S, et al. Big data: The future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Bajic VB, Yu J, Cheung K-H, Townsend JP. In: Bioinformatics - Trends and Methodologies. Mahdavi MA, editor. Vol. 1. 2011. InTech, Rijeka, Croatia, pp. 41–56. [Google Scholar]
- 19.Finn RD, Gardner PP, Bateman A. Making your database available through Wikipedia: the pros and cons. Nucleic Acids Res. 2012;40:D9–D12. doi: 10.1093/nar/gkr1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, Cheung KH, Townsend JP. Bringing Web 2.0 to bioinformatics. Brief. Bioinformatics. 2009;10:1–10. doi: 10.1093/bib/bbn041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bidartondo MI. Preserving accuracy in GenBank. Science. 2008;319:1616. doi: 10.1126/science.319.5870.1616a. [DOI] [PubMed] [Google Scholar]
- 22.Giles J. Internet encyclopaedias go head to head. Nature. 2005;438:900–901. doi: 10.1038/438900a. [DOI] [PubMed] [Google Scholar]
- 23.Giles J. Key biology databases go wiki. Nature. 2007;445:691. doi: 10.1038/445691a. [DOI] [PubMed] [Google Scholar]
- 24.Kumar S, Schiffer PH, Blaxter M. 959 Nematode Genomes: a semantic wiki for coordinating sequencing projects. Nucleic Acids Res. 2012;40:D1295–D300. doi: 10.1093/nar/gkr826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stokes TH, Torrance JT, Li H, Wang MD. ArrayWiki: an enabling technology for sharing public microarray data repositories and meta-analyses. BMC Bioinformatics. 2008;9(Suppl. 6):S18. doi: 10.1186/1471-2105-9-S6-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoehndorf R, Bacher J, Backhaus M, Gregorio SE, Jr, Loebe F, Prufer K, Uciteli A, Visagie J, Herre H, Kelso J. BOWiki: an ontology-based wiki for annotation of data and integration of knowledge in biology. BMC Bioinformatics. 2009;10(Suppl. 5):S5. doi: 10.1186/1471-2105-10-S5-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McIntosh BK, Renfro DP, Knapp GS, Lairikyengbam CR, Liles NM, Niu L, Supak AM, Venkatraman A, Zweifel AE, Siegele DA, et al. EcoliWiki: a wiki-based community resource for Escherichia coli. Nucleic Acids Res. 2012;40:D1270–D1277. doi: 10.1093/nar/gkr880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Good BM, Clarke EL, de Alfaro L, Su AI. The Gene Wiki in 2011: community intelligence applied to human gene annotation. Nucleic Acids Res. 2012;40:D1255–D1261. doi: 10.1093/nar/gkr925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renfro DP, McIntosh BK, Venkatraman A, Siegele DA, Hu JC. GONUTS: the gene ontology normal usage tracking system. Nucleic Acids Res. 2012;40:D1262–D1269. doi: 10.1093/nar/gkr907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolser DM, Chibon PY, Palopoli N, Gong S, Jacob D, Del Angel VD, Swan D, Bassi S, Gonzalez V, Suravajhala P, et al. MetaBase—the wiki-database of biological databases. Nucleic Acids Res. 2012;40:D1250–D1254. doi: 10.1093/nar/gkr1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stehr H, Duarte JM, Lappe M, Bhak J, Bolser DM. PDBWiki: added value through community annotation of the Protein Data Bank. Database (Oxford) 2010;2010:baq009. doi: 10.1093/database/baq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodis E, Prilusky J, Martz E, Silman I, Moult J, Sussman JL. Proteopedia - a scientific ‘wiki' bridging the rift between three-dimensional structure and function of biomacromolecules. Genome Biol. 2008;9:R121. doi: 10.1186/gb-2008-9-8-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li JW, Robison K, Martin M, Sjodin A, Usadel B, Young M, Olivares EC, Bolser DM. The SEQanswers wiki: a wiki database of tools for high-throughput sequencing analysis. Nucleic Acids Res. 2012;40:D1313–D1317. doi: 10.1093/nar/gkr1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cariaso M, Lennon G. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Res. 2012;40:D1308–D1312. doi: 10.1093/nar/gkr798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mader U, Schmeisky AG, Florez LA, Stulke J. SubtiWiki–a comprehensive community resource for the model organism Bacillus subtilis. Nucleic Acids Res. 2012;40:D1278–D1287. doi: 10.1093/nar/gkr923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Csosz E, Mesko B, Fesus L. Transdab wiki: the interactive transglutaminase substrate database on web 2.0 surface. Amino Acids. 2009;36:615–617. doi: 10.1007/s00726-008-0121-y. [DOI] [PubMed] [Google Scholar]
- 38.Zhao D, Wu J, Zhou Y, Gong W, Xiao J, Yu J. WikiCell: a unified resource platform for human transcriptomics research. OMICS. 2012;16:357–362. doi: 10.1089/omi.2011.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelder T, van Iersel MP, Hanspers K, Kutmon M, Conklin BR, Evelo CT, Pico AR. WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 2012;40:D1301–D1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mons B, Ashburner M, Chichester C, van Mulligen E, Weeber M, den Dunnen J, van Ommen GJ, Musen M, Cockerill M, Hermjakob H, et al. Calling on a million minds for community annotation in WikiProteins. Genome Biol. 2008;9:R89. doi: 10.1186/gb-2008-9-5-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann R. A wiki for the life sciences where authorship matters. Nat. Genet. 2008;40:1047–1051. doi: 10.1038/ng.f.217. [DOI] [PubMed] [Google Scholar]
- 42.Dai L, Xu C, Tian M, Sang J, Zou D, Li A, Liu G, Chen F, Wu J, Xiao J, et al. Community intelligence in knowledge curation: an application to managing scientific nomenclature. PLoS One. 2013;8:e56961. doi: 10.1371/journal.pone.0056961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potthast M, Stein B, Gerling R. Automatic vandalism detection in Wikipedia. Adv. Inform. Retr. 2008;4956:663–668. [Google Scholar]
- 44.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Callaway E. No rest for the bio-wikis. Nature. 2010;468:359–360. doi: 10.1038/468359a. [DOI] [PubMed] [Google Scholar]
- 47.Dai L, Tian M, Wu J, Xiao J, Wang X, Townsend JP, Zhang Z. AuthorReward: increasing community curation in biological knowledge wikis through automated authorship quantification. Bioinformatics. 2013;29:1837–1839. doi: 10.1093/bioinformatics/btt284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waldrop M. Big data: wikiomics. Nature. 2008;455:22–25. doi: 10.1038/455022a. [DOI] [PubMed] [Google Scholar]
- 49.Muller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei CH, Kao HY, Lu Z. PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res. 2013;41:W518–W522. doi: 10.1093/nar/gkt441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bunt SM, Grumbling GB, Field HI, Marygold SJ, Brown NH, Millburn GH. Directly e-mailing authors of newly published papers encourages community curation. Database (Oxford) 2012;2012:bas024. doi: 10.1093/database/bas024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Q, Li J, Xue Y, Han B, Deng XW. Rice 2020: a call for an international coordinated effort in rice functional genomics. Mol. Plant. 2008;1:715–719. doi: 10.1093/mp/ssn043. [DOI] [PubMed] [Google Scholar]


