Abstract
HbVar (http://globin.bx.psu.edu/hbvar) is one of the oldest and most appreciated locus-specific databases launched in 2001 by a multi-center academic effort to provide timely information on the genomic alterations leading to hemoglobin variants and all types of thalassemia and hemoglobinopathies. Database records include extensive phenotypic descriptions, biochemical and hematological effects, associated pathology and ethnic occurrence, accompanied by mutation frequencies and references. Here, we report updates to >600 HbVar entries, inclusion of population-specific data for 28 populations and 27 ethnic groups for α-, and β-thalassemias and additional querying options in the HbVar query page. HbVar content was also inter-connected with two other established genetic databases, namely FINDbase (http://www.findbase.org) and Leiden Open-Access Variation database (http://www.lovd.nl), which allows comparative data querying and analysis. HbVar data content has contributed to the realization of two collaborative projects to identify genomic variants that lie on different globin paralogs. Most importantly, HbVar data content has contributed to demonstrate the microattribution concept in practice. These updates significantly enriched the database content and querying potential, enhanced the database profile and data quality and broadened the inter-relation of HbVar with other databases, which should increase the already high impact of this resource to the globin and genetic database community.
INTRODUCTION
Hemoglobinopathies are the commonest single-gene genetic disorders in humans, resulting from pathogenic genome variants in the human α-like and β-like globin gene clusters (reviewed in 1). The human α-globin gene cluster is composed of the HBZ (OMIM number 142310), HBA2 (OMIM number 141850), HBA1 (OMIM number 141800), HBM (OMIM number 609639) and HBQ1 (OMIM number 142240) genes, which encode the ζ-, α2-, α1- and possibly μ- and θ-globin polypeptides, respectively. The human β-globin gene cluster is composed of the HBE1 (OMIM number 142100), HBG2 (OMIM number 142250), HBG1 (OMIM number 142200), HBD (OMIM number 142000) and HBB (OMIM number 141900) genes, which encode the ε-, Gγ, Aγ-, δ- and β-globin polypeptides, respectively. Single nucleotide substitutions or indels can lead to several hemoglobin variants owing to amino acid replacements, while molecular defects in either regulatory or coding regions of the human HBA2, HBA1, HBB or HBD genes can minimally or drastically reduce their expression, leading to α-, β- or δ-thalassemia, respectively.
HbVar database of hemoglobin variants and thalassemia mutations is one of the oldest and the most-appreciated locus-specific databases (LSDBs), not only from the globin but also from the wider genetic database community. HbVar was launched in 2001 and derived from previous compilations (2,3), as a publicly available LSDB, to provide timely information to interested users, e.g. the globin research community, patients and their parents and providers of genetic services and counseling. HbVar is developed such as to accommodate the need for regular data entry updates and corrections, as new hemoglobin variants and thalassemias continue to be discovered. In addition, HbVar has a comprehensive query interface that allows easy access to this information, particularly for the research community and also for physicians as an aid in diagnosis. Since its launch, HbVar has rapidly become an important data resource for the globin research community and is considered to be one of the premier LSDBs available to date (4). We report here several new updates in HbVar structure and contents, aiming at increasing the quality of the database, the accuracy and breadth of data coverage and, above all, its impact to the scientific community. Also, the various synergies with other data resources, namely LSDBs and National/Ethnic Genetic databases, are discussed.
UPDATES TO EXISTING DATA
Since the launch of HbVar (5) and the previous database updates in 2004 (6) and 2007 (7), HbVar information has been expanded by >600 additional entries and data corrections, made continually by the database curators. Also, to cope with the increased need of regular data updates and corrections, Dr Joseph Borg (University of Malta, MT), Dr Kamran Moradkhani (Nantes, FR) and Dr Philippe Joly (Lyon, FR) have joined the HbVar team as data curators for thalassemia mutations and hemoglobin variants, respectively. To identify new hemoglobin variants and thalassemia mutations not previously documented in the database, we continued to manually scan articles from the specialized journal ‘Hemoglobin’, which frequently publishes new hemoglobin variants and thalassemia mutations, and where applicable, previously undocumented variants have been entered into HbVar.
QUERY PAGE UPGRADES AND NEW FUNCTIONALITIES
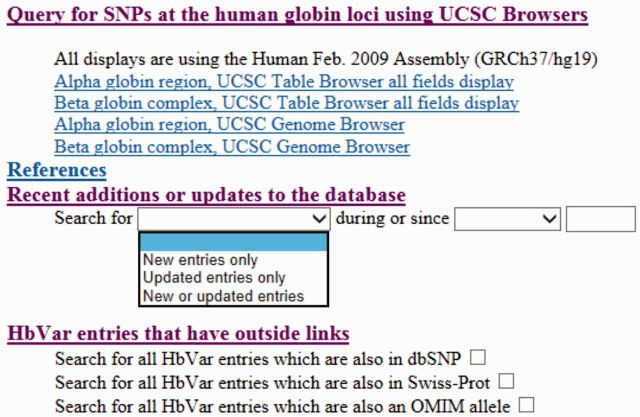
The HbVar query page has undergone a major refit in 2006 (7) to improve the clarity of display. We have now added two additional querying options in the database allowing the user to query for the most recent updates, referring to either new entries or updates of existing entries (or both; Figure 1). The user can also specify the date of the new or updated entry in the adjacent drop-down menu. In addition, we have included query options to list all HbVar entries that are also listed in other resources, such as dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP), Swiss-Prot (8) and OMIM (9; Figure 1).
Figure 1.
The additions in the updated HbVar query page. Hyperlinks allowing the user to query for genomic variants using the UCSC and PSU Genome Browsers (see also Figure 2), recent additions and/or updates of the HbVar data content and HbVar entries that have links to other databases, such as dbSNP, Swiss-Prot and OMIM (see text for details).
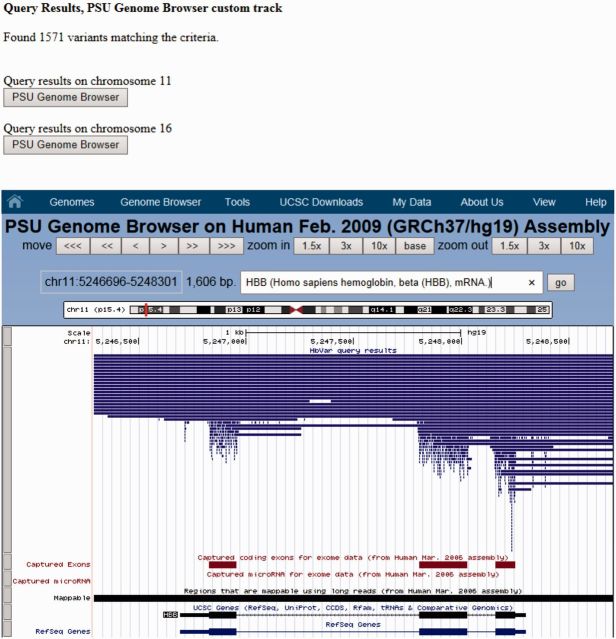
In addition to the data querying option changes, we have added new data visualization features. In addition to querying HbVar contents by gene location and globin chain, the user can also pre-select the output of the query results, in different genome browsers, namely the University of California at Santa Cruz (UCSC) Genome Browser (10) and the Pennsylvania State University Genome Browsers, using the dedicated HbVar custom tracks (Figure 2). All these upgrades have significantly enriched the content of the data output of all HbVar records, as shown in Supplementary Figure S1.
Figure 2.
Overview of the new HbVar visual display feature that couples with the PSU Genome Browser. A user can select a particular gene or genomic region in the human α- or β-globin cluster in the HbVar query page (HBB in this example) and the query returns all genomic variants (base changes, small indels and gross deletions) documented in HbVar that are present within the selected region. Note that the majority of single base changes are nicely clustered within the HBB gene exons. Similar displays can be obtained by using the UCSC Genome Browser using the HbVar custom tracks (not shown). The PSU Genome Browser toolbar is shown on top.
INTER-RELATION WITH OTHER DATABASES AND SCIENTIFIC JOURNALS
To maximize querying options and comparative data analysis, we have decided to share HbVar data content with that of other established and internationally renowned genetic data resources.
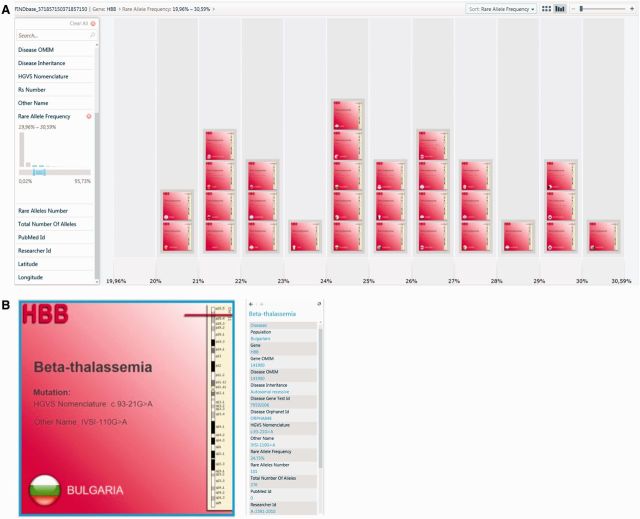
FINDbase (http://www.findbase.org; 11) is a global database documenting allele frequencies of clinically relevant genomic variations, namely, causative mutations and pharmacogenomic markers, in various populations and ethnic groups worldwide. HbVar allele frequency data were initially shared with FINDbase developers in 2006 and made available as an integral part of its data collection. Since then, data collection has been significantly updated with new α- and β-thalassemia mutation frequencies data from 28 populations and 27 ethnic groups and/or geographical regions that were extracted from the published literature (Figure 3).
Figure 3.
HBB allele frequency data visualization in FINDbase database. (A) The present query involves all HBB gene variants with an allele frequency between 19.96 and 30.59%. Query returns 30 records sorted by frequency. (B) The user can zoom in to each record (red box) for details [this record involves the HBB:c.93-21G>A allele frequency in the Bulgarian population (24.75%), documented in the data entry displayed in the list on the right (see also 11)].
Also, the entire HbVar database content has been copied to the Leiden Open-Access Variation database (LOVD; http://www.lovd.nl; 12) V2.0 platform not only to fulfill the need of comparative analyses with data from other LSDBs that have been developed in the same platform but, most importantly, to allow documentation of hemoglobinopathies-related genome variations in other genomic loci, namely, those leading to α-thalassemia, variations in genes modifying thalassemia disease severity, quantitative trait loci, etc (see later in the text).
Finally, HbVar database is inter-related with the scientific journal Hemoglobin such that each variant published in the journal should be first documented in the HbVar database.
IMPLEMENTING MICROATTRIBUTION TO ENCOURAGE DATA SUBMISSION
In 2008, the scientific journal Nature Genetics introduced the concept of microattribution in an attempt to establish an alternative reward system for scientific contributions, such as database entries and records and possibly database curation efforts. The principle of microattribution is ‘… to produce a publication workflow that is open to all journals and that draws on the expertise of all those with a stake in understanding variation at a particular region in the human genome’ (13). In this way, microattribution introduces the concept of individual genome variants curation, so that data submitters and database curators obtain all due credit for their effort and/or contribution. A prerequisite for this is that genomic variants should be deposited in stable, publicly available and well-maintained central repositories that would run independent microattribution services, based on an individual researcher’s unique identity, defined by the Open Researcher and Contributor ID consortium (http://orcid.org), Researcher ID (http://www.researcherid.com), OpenID (http://openid.net) and so on (14).
HbVar data content was used to first demonstrate the use of microattribution in practice (15). In particular, all causative globin gene variations in the α-like and β-like globin genes, as well as those genomic loci that when mutated lead to (ATRX, KLF1, BCL11A, etc) or modify a hemoglobinopathy-related phenotype (MYB, MAP3K5, PDE7B, etc), their associated phenotypes and allele frequencies, where applicable, were comprehensively documented in the 37 LOVD-based interrelated LSDBs (Table 1) and assigned a unique LSDB accession number and IDs of the contributor(s) of the variant. Each genomic variant in these LSDBs included either published variants stored against the PubMed IDs or unpublished variants contributed by individual researchers or research groups involved in hemoglobin research. Subsequently, the microattribution tables were deposited in the NCBI (http://www.ncbi.nlm.nih.gov) public repository in an effort to measure microcitations for every data contributor or data unit centrally. The microattribution article itself, comprising 51 authors from 35 institutions, was published in ‘Nature Genetics’ in 2011 (15).
Table 1.
Total number of variants deposited into the LOVD-related LSDBs (http://lovd.bx.psu.edu). The first part of the Table corresponds to the globin genes and the second part with genes not linked with the human α- and β-globin gene clusters but related to (or modifying) the resulting hemoglobinopathies phenotype and clinical picture
| Gene | Chromosome | Genomic RefSeq ID | Number of variants |
|---|---|---|---|
| Globin genes | |||
| HBA1 | 16p13.3 | NG_000002.1 | 320 |
| HBA2 | 16p13.3 | NG_000006.1 | 380 |
| HBB | 11p15.4 | NG_000007.3 | 826 |
| HBD | 11p15.4 | NG_000007.3 | 91 |
| HBG1 | 11p15.4 | NG_000007.3 | 51 |
| HBG2 | 11p15.4 | NG_000007.3 | 65 |
| Genes not linked with the human globin gene cluster | |||
| ALOX5AP | 13q12 | NC_000013.10 | 3 |
| AQP9 | 15q22.1–q22.2 | NC_000015.9 | 1 |
| ARG2 | 14q24.1–q24.3 | NC_000014.8 | 2 |
| ASS1 | 9q34.1 | NC_000009.11 | 4 |
| ATRX | Xq13.1–q21.1 | NC_000023.10 | 161 |
| BCL11A | 2p16.1 | NC_000002.11 | 9 |
| CNTNAP2 | 7q35–q36 | NC_000002.11 | 1 |
| CSNK2A1 | 20p13 | NC_000020.10 | 1 |
| EPAS1 | 2p21–p16 | NG_016000.1 | 1 |
| ERCC2 | 19q13.3 | NC_000019.9 | 6 |
| FTL1 | 13q12 | NC_000013.10 | 12 |
| GATA1 | Xp11.23 | NC_000023.10 | 1 |
| GPM6B | Xp22.2 | NC_000023.10 | 7 |
| HAO2 | 1p13.3–p13.1 | NC_000001.10 | 1 |
| HBS1L | 6q23.3 | NC_000006.11 | 12 |
| KDR | 4q11–q12 | NC_000004.11 | 4 |
| KL | 13q12 | NC_000012.10 | 5 |
| KLF1 | 19p13.13–p13.12 | NC_000019.9 | 27 |
| MAP2K1 | 15q22.1–q22.33 | NG_008305.1 | 1 |
| MAP3K5 | 6q22.33 | NG_011965.1 | 2 |
| MAP3K7 | 6q16.1–q16.3 | NC_000006.11 | 2 |
| MYB | 6q23.3 | NC_000006.11 | 12 |
| NOS1 | 12q24.2–q24.31 | NC_000012.11 | 6 |
| NOS2 | 17q11.2–q12 | NC_000017.10 | 2 |
| NOS3 | 7q36 | NC_000007.13 | 3 |
| NOX3 | 6q25.1–q26 | NC_000006.11 | 4 |
| NUP133 | 1q42.13 | NC_000001.10 | 1 |
| PDE7B | 6q23–q24 | NC_000006.11 | 4 |
| SMAD3 | 15q22.33 | NC_000015.9 | 2 |
| SMAD6 | 15q21–q22 | NC_000015.9 | 4 |
| TOX | 8q12.1 | NC_000008.10 | 21 |
According to HbVar data accesses and contribution, microattribution has been found to contribute significantly to the increase in data submission rate, showing an up to 8.2-fold increase in data submission rate as compared with previous years in which HbVar was active (15), even compared with the official HbVar launch year (2001), emphasizing the potential impact of microattribution to genome variation data submission. Also, apart from the increase in data contribution to HbVar, a number of useful conclusions were drawn from this approach, mostly derived from the clustering of HBG2/HBG1 gene promoter variants and the ATRX and KLF1 gene coding variants, from where new mutation patterns have emerged (15). Such conclusions would not have been possible without such an approach, further demonstrating the value of the immediate contribution of novel genome variants to databases even though they would not warrant classical narrative publication on their own.
DATA CONTRIBUTION IN COLLABORATIVE PROJECTS
As previously shown with microattribution, the existence of comprehensive data repositories allows comparative data analysis and reciprocally drawing conclusions that would have otherwise not been possible. HbVar data content was exploited in two such collaborative projects to study, from an evolutionary and functional perspective: (i) hemoglobin variants that may be due to the same mutation but lie on a different α-globin (HBA1 or HBA2) gene and (ii) hemoglobin variants and mutations leading to hereditary persistence of fetal hemoglobin that results from the same mutation but on either the HBG1 or HBG2 (fetal globin) gene.
In the first case, the study was performed within the context of the European Commission-funded ITHANET collaborative project (http://www.ithanet.eu), in which we were able to identify 14 different hemoglobin variants resulting from identical mutations on either one of the two human α-globin (HBA1 or HBA2) paralogous genes (16). Also, in the second project, we managed to identify 11 different hemoglobin variants resulting from identical mutations on either one of the two human γ-globin paralogous genes, while seven other promoter variants either result in non-deletional hereditary persistence of fetal hemoglobin or are benign polymorphisms (17).
PhenCode: CONNECTING GENOTYPE WITH PHENOTYPE
LSDBs are key connections between the abundance of genomic information and clinically important issues. In 2006, HbVar and GenPhen databases have been adapted to complete a path from genome sequence to functional analysis, via Encyclopedia of DNA element (ENCODE, http://www.genome.gov/10005107; 18), to human mutations (HbVar) and to clinical phenotypes of groups of patients (GenPhen). The display of clinically relevant locus-specific mutation data on the UCSC Genome Browser makes it readily available to a wide audience, and facilitates viewing of data from many sources in one context. On the other hand, links back to the original databases allow detailed queries within individual loci, which can then be combined and further analyzed accordingly. This combination of LSDBs and powerful genome browsers paves the way for drawing useful genotype–phenotype connections, which can be expanded to other loci of clinical importance, such as modifier genes. Examples of HbVar-related use of PhenCode are available at http://globin.bx.psu.edu/phencode/examples (see also 19).
DATABASE ACCESS
Since their launch in January 2001, the HbVar database and associated resources at the Globin Gene Server [http://globin.bx.psu.edu), such as the online Syllabi, are regularly used worldwide. Also, HbVar is very frequently accessed by Facebook and mobile devices. Users frequently contact the curators and the rest of the HbVar team members to submit new hemoglobin variants and/or thalassemia mutations, report missing information for existing mutants, identify inconsistencies and/or erroneous entries and even propose collaborative projects related to HbVar data records.
This fact not only shows how important users’ input is to improve data quality and accuracy but also demonstrate the impact the HbVar has in the entire globin research community.
FUTURE PROSPECTS
HbVar has become, since the year of its launch, a key resource for information about sequence variation leading to hemoglobinopathies and is still considered an exemplary LSDB from the various existing ones. Key factors that have contributed to its success are (i) its constant data update and improvements, mostly driven by the devotion of those researchers involved in this project, (ii) its dynamic data querying and visualization tools, in conjunction with the UCSC and PSU Genome Browsers and (iii) its partnership with other stable and well-respected international databases. The positive impact that HbVar has on the research community is also illustrated by the fact that funding, dedicated or related to other projects, has always been available for keeping this resource alive, in an environment where dedicated funding opportunities for database development and curation are often limited and very hard to find (20), frequently resulting in the discontinuation of many useful databases. The international recognition of HbVar also comes from the fact that although couple of other globin-related databases, such as Deniz [content migrated to the Catalog of Transmission Genetics in Arabs (CTGA) database (http://www.cags.org.ae/ctga_search.html), 21] or the Ithanet databases (http://www.ithanet.eu/index.php/db), have been developed, HbVar is still considered as the key LSDB for hemoglobin research professionals.
To ensure continuous HbVar data enrichment, we plan to implement a broader search strategy that combines manual and electronic search procedures and also tighter links to the scientific journal ‘Hemoglobin’. Also, we plan to expand the inter-relation of HbVar with other relevant high quality databases, following the successful example of HbVar and GALA databases (6), the UCSC and PSU Genome Browsers, PhenCode (19), FINDbase (11) and LOVD (12).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
United States Public Health Service grants [HG02238 to W.M. and DK065806 to R.C.H.]; European Commission grants [ITHANET 026539, GEN2PHEN 200754 to G.P.P.] and financial support from Tobacco Settlement Funds of the Commonwealth of Pennsylvania, the Huck Institute of the Life Sciences at Penn State University and the Golden Helix Foundation (London, UK). Funding for open access charge: Penn State University, Philadelphia, PA, USA.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank all the HbVar users worldwide for their valuable comments and suggestions, which help us to keep the information as updated and complete as possible and also contribute to the continuous improvement of the database profile and contents. We will always be indebted to the late Prof. Titus H.J. Huisman and his colleagues for their detailed compilations of hemoglobin variants and thalassemia mutations. The authors wish to dedicate this work to the memory of Renzo Galanello who has much contributed in the field of hemoglobin research.
REFERENCES
- 1.Weatherall DJ, Clegg JB, editors. The Thalassaemia Syndromes. 4th edn. Wiley-Blackwell; [Google Scholar]
- 2.Huisman THJ, Carver MF, Baysal E. A Syllabus of Thalassemia Mutations. Augusta, GA: The Sickle Cell Anemia Foundation; 1997. [Google Scholar]
- 3.Huisman THJ, Carver MF, Efremov GD. A Syllabus of Human Hemoglobin Variants. 2nd edn. Augusta, GA: The Sickle Cell Anemia Foundation; 1998. [Google Scholar]
- 4.Mitropoulou C, Webb AJ, Mitropoulos K, Brookes AJ, Patrinos GP. Locus-specific databases domain and data content analysis: evolution and content maturation towards clinical use. Hum. Mutat. 2010;31:1109–1116. doi: 10.1002/humu.21332. [DOI] [PubMed] [Google Scholar]
- 5.Hardison RC, Chui DH, Giardine B, Riemer C, Patrinos GP, Anagnou N, Miller W, Wajcman H. HbVar: a relational database of human hemoglobin variants and thalassemia mutations at the globin gene server. Hum. Mutat. 2002;19:225–233. doi: 10.1002/humu.10044. [DOI] [PubMed] [Google Scholar]
- 6.Patrinos GP, Giardine B, Riemer C, Miller W, Chui DH, Anagnou NP, Wajcman H, Hardison RC. Improvements in the HbVar human hemoglobin variants and thalassemia mutations for population and sequence variation studies. Nucleic Acids Res. 2004;32:D537–D541. doi: 10.1093/nar/gkh006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giardine Β, van Baal S, Kaimakis P, Riemer C, Miller W, Samara M, Kollia P, Anagnou NP, Chui DH, Wajcman H, et al. HbVar database of human hemoglobin variants and thalassemia mutations: 2007 update. Hum. Mutat. 2007;28:206. doi: 10.1002/humu.9479. [DOI] [PubMed] [Google Scholar]
- 8.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM®) Hum. Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 10.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. The UCSC genome browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgitsi M, Viennas E, Gkantouna V, van Baal S, Petricoin EF, Poulas K, Tzimas G, Patrinos GP. FINDbase: a worldwide database for genetic variation allele frequencies updated. Nucleic Acids Res. 2011;39:D926–D932. doi: 10.1093/nar/gkq1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum. Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 13.Anonymous. Human variome microattribution reviews. Nat. Genet. 2008;40:1. [Google Scholar]
- 14.Patrinos GP, Cooper DN, van Mulligen E, Gkantouna V, Tzimas G, Tatum Z, Schultes E, Roos M, Mons B. Microattribution and nanopublication as means to incentivize the placement of human genome variation data into the public domain. Hum. Mutat. 2012;33:1503–1512. doi: 10.1002/humu.22144. [DOI] [PubMed] [Google Scholar]
- 15.Giardine B, Borg J, Higgs DR, Peterson KR, Philipsen S, Maglott D, Singleton BK, Anstee DJ, Basak AN, Clark B, et al. Systematic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat. Genet. 2011;43:295–301. doi: 10.1038/ng.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moradkhani K, Préhu C, Old J, Henderson S, Balamitsa V, Luo HY, Poon MC, Chui DH, Wajcman H, Patrinos GP. Mutations in the paralogous human α-globin genes yielding identical hemoglobin variants. Ann. Hematol. 2009;88:535–543. doi: 10.1007/s00277-008-0624-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papachatzopoulou A, Patrinos GP. Functional and evolutionary implications of identical mutations in the paralogous gamma-globin genes leading to hemoglobin variants or non-deletional hereditary persistence of fetal hemoglobin. Hemoglobin. 2011;35:135–141. doi: 10.3109/03630269.2011.553019. [DOI] [PubMed] [Google Scholar]
- 18.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. ENCODE data in the UCSC genome browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giardine B, Riemer C, Hefferon T, Thomas D, Hsu F, Zielenski J, Sang Y, Elnitski L, Cutting G, Trumbower H, et al. PhenCode: connecting ENCODE data with mutations and phenotype. Hum. Mutat. 2007;28:554–562. doi: 10.1002/humu.20484. [DOI] [PubMed] [Google Scholar]
- 20.Patrinos GP, Brookes AJ. DNA, disease and databases: disastrously deficient. Trends Genet. 2005;21:333–338. doi: 10.1016/j.tig.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Tadmouri GO, Al Ali MT, Al-Haj Ali S, Al Khaja N. CTGA: the database for genetic disorders in Arab populations. Nucleic Acids Res. 2006;34:D602–D606. doi: 10.1093/nar/gkj015. [DOI] [PMC free article] [PubMed] [Google Scholar]