Abstract
PubChem’s BioAssay database (http://pubchem.ncbi.nlm.nih.gov) is a public repository for archiving biological tests of small molecules generated through high-throughput screening experiments, medicinal chemistry studies, chemical biology research and drug discovery programs. In addition, the BioAssay database contains data from high-throughput RNA interference screening aimed at identifying critical genes responsible for a biological process or disease condition. The mission of PubChem is to serve the community by providing free and easy access to all deposited data. To this end, PubChem BioAssay is integrated into the National Center for Biotechnology Information retrieval system, making them searchable by Entrez queries and cross-linked to other biomedical information archived at National Center for Biotechnology Information. Moreover, PubChem BioAssay provides web-based and programmatic tools allowing users to search, access and analyze bioassay test results and metadata. In this work, we provide an update for the PubChem BioAssay resource, such as information content growth, new developments supporting data integration and search, and the recently deployed PubChem Upload to streamline chemical structure and bioassay submissions.
INTRODUCTION
The PubChem BioAssay database (http://pubchem.ncbi.nlm.nih.gov) (1–4) is a public repository for biological activity data of small molecules and RNAi reagents, hosted by the National Center for Biotechnology Information (NCBI) (5), a division of the National Library Medicine under the National Institutes of Health since 2004. BioAssay test results are linked to the chemical structures of tested small molecules and the sequencing data of screened RNA interference (RNAi) reagents as available. In addition, the information content in the BioAssay database is linked to several biomedical and literature databases hosted at NCBI, including PubMed, Protein, Gene, Nucleotide, BioSystems, Taxonomy, OMIM and protein 3D structure associated with bioassay targets. PubChem is committed to offer biomedical researchers free access to this information. BioAssay data can be searched, accessed and analyzed by Entrez queries as well as via a suite of web-based and programmatic tools provided by PubChem, making PubChem a widely used public information system for accelerating chemical biology research and drug development. Table 1 provides a summary for BioAssay services and the corresponding URLs. Most of the web-based services can also be accessed at http://pubchem.ncbi.nlm.nih.gov/assay.
Table 1.
A list of PubChem BioAssay services
| Service | Description | URL example |
|---|---|---|
| BioAssay service home | Access a list of BioAssay services | http://pubchem.ncbi.nlm.nih.gov/assay/ |
| BioAssay search | Search BioAssay database with Entrez | http://www.ncbi.nlm.nih.gov/pcassay/ |
| BioAssay search, advanced page | An interface for searching multiple search fields | http://www.ncbi.nlm.nih.gov/pcassay/limits |
| BioAssay text search, advanced page | An interface for reviewing search history and refining search results with Boolean operation | http://www.ncbi.nlm.nih.gov/pcassay/advanced |
| BioAssay summary | Access and download a bioassay record | http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=myAID |
| BioAssay data retrieval tool | Retrieve a full data table or an active subset from a single bioassay record |
http://pubchem.ncbi.nlm.nih.gov/assay/assaydata.html?aid=myAID http://pubchem.ncbi.nlm.nih.gov/assay/assaydata.html?act=act&aid=myAID |
| BioAssay data selection tool | Select a user-defined data subset from a single bioassay record | http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?q=t&aid=myAID |
| Bioactivity data tool | Retrieve multiple-assay bioactivity data for a single substance sample (SID); chemical structure (CID); protein target (GI); or gene target (GeneID) |
http://pubchem.ncbi.nlm.nih.gov/assay.cgi?sid=mySID http://pubchem.ncbi.nlm.nih.gov/assay.cgi?sid=myCID http://pubchem.ncbi.nlm.nih.gov/assay.cgi?sid=myGI http://pubchem.ncbi.nlm.nih.gov/assay.cgi?sid=myGeneID |
| BioActivity summary (compound-centric) | Summarize and analyze bioactivity data for a set of records, presented from the compound point of view | http://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.cgi?tab=1 |
| BioActivity summary (assay-centric) | Summarize and analyze bioactivity data for a set of records, presented from the assay point of view | http://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.cgi?tab=2 |
| BioActivity summary (target-centric) | Summarize and analyze bioactivity data for a set of records, presented from the target point of view | http://pubchem.ncbi.nlm.nih.gov/assay/bioactivity.cgi?tab=3 |
| Structure-activity relationship analysis (SAR) | Analyze and visualize structure-activity relationship with clustering tools and a heatmap-style display | http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?p=heat |
| Scatter plot/histogram | Analyze bioassay test results with histogram or scatter plot | http://pubchem.ncbi.nlm.nih.gov/assay/plot.cgi?plottype=2 |
| Dose-response curve tool | Analyze bioassay test results and visualize dose-response curve | http://pubchem.ncbi.nlm.nih.gov/assay/plot.cgi?plottype=1 |
| Related BioAssay | Summarize bioassay relationship by overlap of active compounds, target sequence similarity, deposited annotation, same publication, common pathways and same assay project | http://pubchem.ncbi.nlm.nih.gov/assay/assayHeatmap.cgi? |
| PubChem PUG/SOAP | PubChem programmatic tool for data retrieval | http://pubchem.ncbi.nlm.nih.gov/pug/pughelp.html |
| PUG/REST | PubChem REST api for data retrieval | http://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST.html |
| Bioassay download tool | A flexible download interface | http://pubchem.ncbi.nlm.nih.gov/assay/assaydownload.cgi |
| BioAssay FTP | FTP for all PubChem BioAssay records and related information | ftp://ftp.ncbi.nlm.nih.gov/pubchem/Bioassay/ |
| BioAssay data standard | XML data specification for PubChem BioAssay data model | ftp://ftp.ncbi.nlm.nih.gov/pubchem/data_spec |
| PubChem upload | Substance and bioassay submission system | http://pubchem.ncbi.nlm.nih.gov/upload |
Developing and managing a public archive system for complex bioassay data has been both challenging and rewarding. In the past 9 years, PubChem has come a long way to manage the rapidly growing data and meet the increasing demand from the community. PubChem has become a leading public bioassay data repository by (i) supporting broad types of bioactivity information with an optimized bioassay data standard, (ii) maintaining steady enhancement of database infrastructure and scalability, (iii) providing and enhancing a streamlined data upload system, (iv) integrating with other biomedical information resources and (v) expanding and empowering search, retrieval, analysis and download tools. In this work, we provide an update on several aspects of the information resource, including data content growth, database infrastructure consolidation, new search indices, project-based bioassay links and newly developed web services including target-based bioactivity data tools and the recently deployed PubChem Upload system.
BioAssay DATA CONTENT GROWTH
The BioAssay database has been growing substantially during the past years (Figure 1). As of 1 September 2013, the BioAssay database has received >700 000 depositions of bioassays (Figure 1A). Counting solely the latest version of each bioassay record by accession (i.e. AID), the database contains 200 000 000 bioactivity outcome summaries (Figure 1B) and 1 200 000 000 data points representing biological properties for 2 800 000 small molecule samples, 1 900 000 chemical structures and 108 000 RNAi reagents (Figure 1C). This information represents tens of thousands of potential modulators for >8000 protein targets and 30 000 genes critical for biological process, hence providing rich information on chemical and RNAi tools for chemical and molecular biology research.
Figure 1.
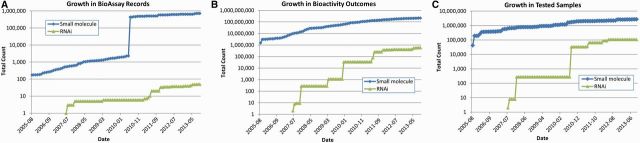
Growth in PubChem BioAssay. (A) Records, (B) bioactivity outcomes (counted by AID–SID pair) and (C) unique tested samples.
The content in the PubChem BioAssay database is contributed by >50 organizations worldwide including US government-funded institutions, pharmaceutical companies, research laboratories and collaborators hosting chemical biology databases. A summary of bioassay vendors and submission counts is provided at http://pubchem.ncbi.nlm.nih.gov/sources#assay. BioAssay datasets added during the past 2 years include (i) small molecule data from screening centers of the NIH Molecular Libraries and Imaging Program [Molecular Library Program (MLP)] (http://commonfund.nih.gov/molecularlibraries/), ICCB-Longwood/NSRB Screen Facility at the Harvard Medical School (http://iccb.med.harvard.edu/), EPA Tox21 (http://epa.gov/ncct/Tox21/) and Milwaukee Institute for Drug Discovery (http://www4.uwm.edu/drugdiscovery/); (ii) a curated dataset records from the Meiler Lab at Vanderbilt University, which derives the ultimate bioactivity outcome of a small molecule by combining multiple bioassay results in PubChem to facilitate cheminformatics studies (6); (iii) curated datasets from literature extraction by IUPHAR-DB (7) and ChEMBL (8); and (iv) small interfering RNA (siRNA) data from Drosophila RNAi Screening Center, ICCB-Longwood/NSRB Screening Facility at the Harvard Medical School (http://iccb.med.harvard.edu/), Cancer Research UK Cambridge Research Institute, Department of Molecular Cell Biology at Weizmann Institute of Science, Institut National de la Sante et de la Recherche Medicale (INSERM), Peterson Lab at Genentech and ten Dijke Lab at Leiden University Medical Center. Many of these newly added siRNA datasets are associated with recent publications in journals such as Nature Cell Biology (9–11), Genome Research (12), J Virol (13), Cancer Research (14), PNAS (15,16), Nature (17–19), Science (20,21) and Nature Genetics (22). Each of these bioassay records is linked to the corresponding abstract in PubMed, allowing PubChem users to track down the publication easily. Vice versa, users of PubMed also gain access to the corresponding bioassay datasets through this cross-link.
PubChem continues to mirror the ChEMBL database (8) hosted at the European Bioinformatics Institute. Multiple ChEMBL releases and database changes over the past 2 years have been incorporated into PubChem. Recently added annotations at ChEMBL are recorded via the Categorized Comment field of the PubChem BioAssay data model (1). Binding, surface, ligand and lipophilic ligand efficiency indices are added to a bioassay record as additional test results. As a result, many of the bioassay records in PubChem have gone through multiple updates. Annotation for bioactivity outcome (e.g. active or inactive) is largely missing in the ChEMBL datasets, hindering their integration with the rest of PubChem data and analysis tools. In such a case, PubChem now assigns bioactivity outcome using a ≤50 μM cutoff based on readouts, such as IC50, EC50 or Ki, allowing a larger portion of the ChEMBL data blended in the PubChem system.
DATABASE INFRASTRUCTURE ENHANCEMENT
A robust and scalable database system is crucial to support the rapid growth of PubChem BioAssay. A set of relational databases and tables is designed and set up on Microsoft SQL servers to (i) accept bioassay submission from depositors, (ii) archive bioassay update with version control, (iii) track embargo status, (iv) record and derive links and relationships among bioassays and other biomedical information, (v) provide search indexes, (vi) support fast data retrieval and analysis and (vii) facilitate daily update at the FTP site. Challenged by the accelerated growth of bioassay data content, great efforts have been invested in the past years to enhance the database infrastructure capacity by both hardware upgrade and revised database design. As a result, new services have been added to the PubChem resource. Furthermore, performance in bioassay data retrieval and download services have been significantly improved, thereby significantly eliminating a queuing system to minimize the user wait time.
DATA INTEGRATION AND NEW WEB SERVICES
The PubChem BioAssay database is fully integrated with other biomedical databases hosted by NCBI and provides a suite of web-based and programmatic tools to support data access, retrieval, analysis and download from PubChem or cross-linked databases (Table 1). Several new services for integrating bioassay target and bioactivity data, or grouping bioassays based on an assay project, are described later. Other developments that have focused on behind-the-scene enhancement of data retrieval without significant web interface change will not be summarized in this work.
Rapid access of bioactivity data for a protein or gene target
PubChem BioAssay closes the gap between molecular and chemical biology research by presenting and linking up information of both chemical and RNAi tools in one system supporting the study of gene function and biological pathways. The majority of small molecule screening data in PubChem are associated with protein targets, while RNAi screening data links each tested reagent to a gene. PubChem provides multiple mechanisms for cross-referencing protein and gene targets from bioactivity data (1). As a result, a protein or gene may link to many bioactivity datasets. It is critical to provide rapid access to such multi-assay bioactivity data for these protein and gene targets. Such a service provides a unique annotation service to the corresponding Entrez Protein or Gene record, which leads users to experimental data from chemical biology and RNAi research enhancing the discoverability of the NCBI Entrez system. Toward this end, two new services, the Protein Target Bioactivity Data Tool and the Gene Target Bioactivity Data Tool, were developed, respectively, to access associated bioactivity information in PubChem.
From a protein target record, such as G-protein-coupled receptor (GPCR) 35 (http://www.ncbi.nlm.nih.gov/protein/NP_005292.2), bioactivity data for this protein target can be accessed by the link ‘BioAssay by Target (Summary)’. As shown in Figure 2A, this Protein Target Bioactivity Data Tool draws and identifies each tested substance, together with its bioactivity results, assay title and a link to detailed data such as dose-response curves. The data table is sorted by bioactivity outcome and potency of the substances by default, showing first active data and potent reagents. Graphical filters are provided at the top of the page, allowing one to drill down to a data subset of one’s interest. For example, this GPCR protein has a ‘Probe’ filter highlighting three chemical probes discovered by a high-throughput screening (HTS) project for selective GPR35 antagonists.
Figure 2.
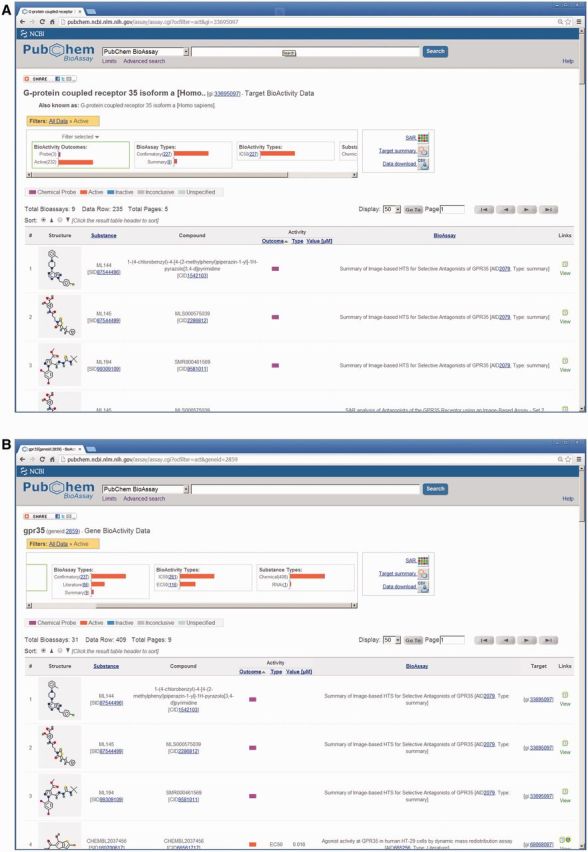
Bioactivity data for a (A) protein target and (B) gene target.
The bioactivity data for the relevant gene target record (http://www.ncbi.nlm.nih.gov/gene/2859), can be accessed by the link ‘BioAssay by Target (Summary)’. With this Gene Target Bioactivity Data Tool, a similar summary of relevant bioassay activity results is displayed as shown in Figure 2B. Note that, using a gene identifier in this case, additional data are retrieved including RNAi test results (as indicated with the filter ‘RNAi’ shown under ‘Substance Types’), which indicates that GPR35 functions as a cellular gene repressing HPV18 LCR as identified by a genome-wide siRNA screen. This example illustrates the power of aggregating bioactivity data across datasets onto a unified display. The Gene Target Bioactivity Data Tool is particularly useful for accessing datasets from multiple depositors and literature-based data from many journal articles. Moreover, it links simultaneously to findings in chemical biology research and RNAi screenings, enabling users to evaluate the biological role of a gene and to identify its small molecular regulators using data shown on the same display.
BioAssays associated with the same assay project
PubChem tracks the relationships among bioassay records as indicated by submitters. PubChem has also developed several computational methods for identifying additional bioassay linkages based on target sequence similarity, common active compounds and biological pathways as well as datasets abstracted from the same publication (1). To better support decision making, PubChem now clusters and links up bioassays based on assay projects. This feature aims to use data deposited by a network, such as the NIH MLP and the Tox21 program. MLP-funded screening laboratories are required to deposit data progressively into PubChem as an assay project continues. It usually takes months or years to finish an assay project aimed at developing chemical probe; hence, often multiple bioassay datasets are submitted to PubChem for the same project but under distinct accessions (AIDs). These datasets are highly relevant, often covering a primary HTS result, follow-ups with dose-response and toxicity testing, or counter screenings against biologically related targets, different cell lines or using different assay methods. PubChem allows submitters to specify such relationships via the cross-reference (XRef) data field. On the other hand, it is up to the submitters to provide all links as new data are made available. As a result, cross-references to related bioassay datasets unfortunately may be lacking or incomplete among many datasets, making it difficult for users to discover these key associations.
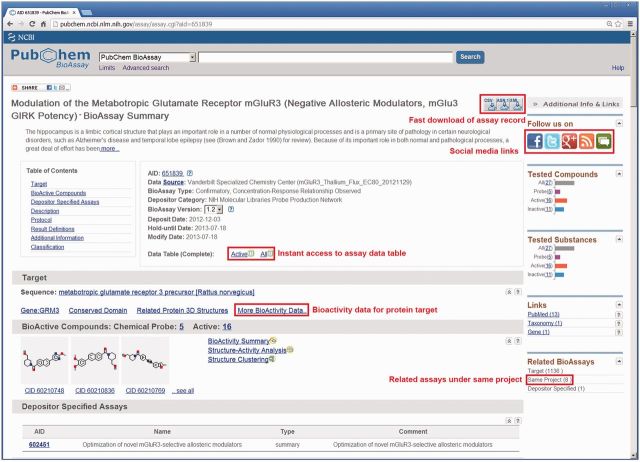
To improve this situation, it is now a common practice to create a ‘Summary’ bioassay at the outset of a multi-assay project and then link each subsequent-related assay back to that summary record. This means that the submitter only needs to specify a single link for each bioassay record to the same summary and all other links between related assays are automatically generated. As a result, assay projects are indexed on top of the individual records. Users visiting any bioassay record can access all relevant datasets of the same project, without the need for the submitter to specify all connections. As shown in Figure 3, the links to these related bioassays are labeled in the BioAssay Summary service as ‘Same Project’ under the ‘Related BioAssays’ section. The Modulation of the Metabotropic Glutamate Receptor mGluR3 (GRM3) assay (http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=651839) indicates only one ‘Depositor Specified’ assay, whereas eight bioassay records were identified as related to the same project by the new procedure. One may see details of the related bioassays by clicking the link ‘Same Project’.
Figure 3.
BioAssay Summary page for bioassay record AID: 651839. New and enhanced features are highlighted, including fast download, instant access to data table, link to additional bioactivity data targeting GRM3, link to related bioassays on the same project and links to social media account.
PUBLIC ACCESS
BioAssay record and BioAssay summary service
A PubChem BioAssay record can be accessed via the BioAssay Summary service at http://pubchem.ncbi.nlm. nih.gov/assay/assay.cgi?, where myAID is a valid BioAssay accession (AID). As shown in Figure 3 for the GRM3 assay (AID: 651839), the BioAssay Summary service provides (i) full access to submitted information, including bioassay protocol descriptions, assay data and cross-references, (ii) derived bioassay relationships and (iii) tools for evaluating tested compounds, studying SAR or researching target. For the ‘Target’ section, a link ‘More Bioactivity data’ has been recently added to gather all bioactivity data in PubChem associated with the GRM3 target. The BioAssay Summary service now provides instant access to bioassay data table and enhanced function for data download with improved database infrastructure. With the recently launched PubChem Social Media outreach, links to social media accounts are now provided on this page.
BioAssay search
Keyword search in the PubChem BioAssay database is supported by NCBI Entrez at http://www.ncbi.nlm.nih.gov/pcassay/. Textual information in PubChem BioAssay is indexed under numerous fields. An advanced interface is provided at http://www.ncbi.nlm.nih.gov/pcassay/limits (Limits page) to access multiple indices and filters (1). Based on information provided in categorized comment fields and keywords in the title of a bioassay record, new filters were added to support the identification of records containing (i) biochemical assay, (ii) cell-based assay, (iii) protein–protein interaction bioactivity and (iv) in vivo or in vitro assay. A newly added menu ‘Assay Project’ can be used to select an assay project and accessing related datasets. ChEMBL depositor information is also indexed to support sub-setting ChEMBL records. As a result, although http://www.ncbi.nlm.nih.gov/pcassay/?term=ChEMBL[sourcename] retrieves all ChEMBL bioassays in PubChem, http://www.ncbi.nlm.nih.gov/pcassay/?term=%22ChEMBL%3A%3AScientific+Literature%22%5BSourceName%5D[SourceName] retrieves literature-based records from ChEMBL, and http://www.ncbi.nlm.nih.gov/pcassay/?term=%22ChEMBL%3A%3ASt+Jude+Malaria+Screening%22%5BSourceName%5D[SourceName] retrieves ChEMBL records deposited by St Jude Malaria Screening.
PubChem BioAssay FTP AND DOWNLOAD
PubChem provides multiple services for users to download bioassay records, which have been described previously (1). This primarily includes (i) an enhanced download function at the Summary service (shown in Figure 3), (ii) a web-based BioAssay download service at http://pubchem.ncbi.nlm.nih.gov/assay/assaydownload.cgi, with a flexible interface supporting full or partial data download by specifying bioassay accessions (AIDs) and tested substance accessions (SIDs) and (iii) daily updated PubChem BioAssay FTP at ftp://ftp.ncbi.nlm.nih.gov/pubchem/Bioassay, providing open access to all bioassay datasets. While the primary FTP structure remains the same, one new FTP directory ‘Extras’ is added to offer additional information of the BioAssay resource. In this folder, the file ‘Cid2BioactivityLink’ provides a list of tested compounds and the corresponding URLs linking to associated bioactivity data. Similarly, the ‘Gi2BioactivityLink’ and ‘Geneid2BioactivityLink’ files provide the list of the corresponding bioactivity data links for protein and gene targets, respectively. The ‘Aid2GiGeneid’ contains all the bioassay (AID), protein target (GI) and gene target (Gene ID) associations in the BioAssay database. Also, a file for assay project-based related bioassays is added to the directory at ftp://ftp.ncbi.nlm.nih.gov/pubchem/Bioassay/AssayNeighbors/. Column headers for the comma-separated values (CSV) format has been modified to provide consistency among multiple download methods (ftp://ftp.ncbi.nlm.nih.gov/pubchem/Bioassay/CSV/README). Readout names are now provided in CSV files to ease data parsing and interpretation. In addition, PubChem PUG/SOAP (http://pubchem.ncbi.nlm.nih.gov/pug/pughelp.html) and PUG/REST (http://pubchem.ncbi.nlm.nih.gov/pug_rest/PUG_REST.html) facilities are being developed to support programmatic retrieval of bioassay information.
PubChem UPLOAD FOR BioAssay SUBMISSION
As a public repository handling diverse and vast amounts of chemical structure and bioassay data, it is critical for PubChem to provide an efficient and user-friendly way to upload data. The recently released PubChem Upload (http://pubchem.ncbi.nlm.nih.gov/upload/) makes use of advances in web technologies to offer streamlined support for data submissions and updates to the Substance and BioAssay databases. PubChem Upload supports all functionalities and data exchange formats of its predecessor (1). Furthermore, it provides an extensive set of wizards, inline help tips and tutorials for guiding submitters to enter assay data and descriptive information. More specifically, the new assay submission capabilities offered by PubChem Upload include (i) bioassay submission wizards to assist novice users for both small molecule and RNAi screenings, (ii) improved user interface response to complex input with newer web technology, (iii) simplified new user registration upgrades for production user accounts, (iv) improved help, including hints built into user interface and tutorial, (v) extensive PubChem bioassay templates for new submissions or for record updates, (vi) full editing and integration of assay data and description tables and (vii) expanded import/export handling of spreadsheets for assays. A detailed help document, tutorial and sample submission templates for PubChem Upload are available at: http://pubchem.ncbi.nlm.nih.gov/upload/docs/upload_help.html, http://pubchem.ncbi.nlm.nih.gov/upload/tutorial/ and http://pubchem.ncbi.nlm.nih.gov/upload/docs/upload_help.html#AssaySubmission, respectively. A detailed description of PubChem Upload will be provided in a separate article.
SUMMARY
PubChem is committed to serve as a public repository for bioactivity data of small molecules and RNAi. PubChem also provides an integrated information platform with a suite of tools allowing users to query, analyze and download all database content. PubChem will continue to improve services and tools as technology advances, and to further integrate the information it contains to third party annotations and other public biomedical data. With the support of open access to the data and the delivery of the new Upload system, PubChem welcomes the community to use the resource and to contribute data content to the repository.
ACKNOWLEDGEMENTS
The authors thank all submitters who have contributed data to PubChem and the rest of the PubChem team for their support.
FUNDING
The NIH Intramural Research program. Funding for open access charge: National Insitutes of Health, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, Han L, Karapetyan K, Dracheva S, Shoemaker BA, et al. PubChem's BioAssay database. Nucleic Acids Res. 2012;40:D400–D412. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Bolton E, Dracheva S, Karapetyan K, Shoemaker BA, Suzek TO, Wang J, Xiao J, Zhang J, Bryant SH. An overview of the PubChem BioAssay resource. Nucleic Acids Res. 2010;38:D255–D266. doi: 10.1093/nar/gkp965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: integrated platform of small molecules and biological activities. Annu. Rep. Comput. Chem. 2008;4:217–241. [Google Scholar]
- 5.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butkiewicz M, Lowe EW, Jr, Mueller R, Mendenhall JL, Teixeira PL, Weaver CD, Meiler J. Benchmarking ligand-based virtual high-throughput screening with the PubChem database. Molecules. 2013;18:735–756. doi: 10.3390/molecules18010735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharman JL, Benson HE, Pawson AJ, Lukito V, Mpamhanga CP, Bombail V, Davenport AP, Peters JA, Spedding M, Harmar AJ. IUPHAR-DB: updated database content and new features. Nucleic Acids Res. 2013;41:D1083–D1088. doi: 10.1093/nar/gks960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J, Sirokmany G, Donati G, Uribe-Lewis S, Pavlidis P, et al. Diverse epigenetic strategies interact to control epidermal differentiation. Nat. Cell Biol. 2012;14:753–763. doi: 10.1038/ncb2520. [DOI] [PubMed] [Google Scholar]
- 10.Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, Peterson AS. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol. 2012;14:61–72. doi: 10.1038/ncb2410. [DOI] [PubMed] [Google Scholar]
- 11.Prager-Khoutorsky M, Lichtenstein A, Krishnan R, Rajendran K, Mayo A, Kam Z, Geiger B, Bershadsky AD. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat. Cell Biol. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- 12.Imberg-Kazdan K, Ha S, Greenfield A, Poultney CS, Bonneau R, Logan SK, Garabedian MJ. A genome-wide RNA interference screen identifies new regulators of androgen receptor function in prostate cancer cells. Genome Res. 2013;23:581–591. doi: 10.1101/gr.144774.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powell ML, Smith JA, Sowa ME, Harper JW, Iftner T, Stubenrauch F, Howley PM. NCoR1 mediates papillomavirus E8;E2C transcriptional repression. J. Virol. 2010;84:4451–4460. doi: 10.1128/JVI.02390-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, Servant N, Paccard C, Hupe P, Robert T, et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70:1793–1803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- 15.Smith JA, White EA, Sowa ME, Powell ML, Ottinger M, Harper JW, Howley PM. Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc. Natl Acad. Sci. USA. 2010;107:3752–3757. doi: 10.1073/pnas.0914818107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang SL, Yeromin AV, Zhang XH, Yu Y, Safrina O, Penna A, Roos J, Stauderman KA, Cahalan MD. Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc. Natl Acad. Sci. USA. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444:230–234. doi: 10.1038/nature05280. [DOI] [PubMed] [Google Scholar]
- 18.Gwack Y, Sharma S, Nardone J, Tanasa B, Iuga A, Srikanth S, Okamura H, Bolton D, Feske S, Hogan PG, et al. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature. 2006;441:646–650. doi: 10.1038/nature04631. [DOI] [PubMed] [Google Scholar]
- 19.Bard F, Casano L, Mallabiabarrena A, Wallace E, Saito K, Kitayama H, Guizzunti G, Hu Y, Wendler F, Dasgupta R, et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- 20.Vig M, Peinelt C, Beck A, Koomoa DL, Rabah D, Koblan-Huberson M, Kraft S, Turner H, Fleig A, Penner R, et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DasGupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science. 2005;308:826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- 22.Nybakken K, Vokes SA, Lin TY, McMahon AP, Perrimon N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat. Genet. 2005;37:1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]