Abstract
We have developed SMMRNA, an interactive database, available at http://www.smmrna.org, with special focus on small molecule ligands targeting RNA. Currently, SMMRNA consists of ∼770 unique ligands along with structural images of RNA molecules. Each ligand in the SMMRNA contains information such as Kd, Ki, IC50, ΔTm, molecular weight (MW), hydrogen donor and acceptor count, XlogP, number of rotatable bonds, number of aromatic rings and 2D and 3D structures. These parameters can be explored using text search, advanced search, substructure and similarity-based analysis tools that are embedded in SMMRNA. A structure editor is provided for 3D visualization of ligands. Advance analysis can be performed using substructure and OpenBabel-based chemical similarity fingerprints. Upload facility for both RNA and ligands is also provided. The physicochemical properties of the ligands were further examined using OpenBabel descriptors, hierarchical clustering, binning partition and multidimensional scaling. We have also generated a 3D conformation database of ligands to support the structure and ligand-based screening. SMMRNA provides comprehensive resource for further design, development and refinement of small molecule modulators for selective targeting of RNA molecules.
INTRODUCTION
Recently, RNAs have been unraveled as unique molecules playing critical roles in developmental and physiological processes in all living organisms (1–5). RNA is involved in the progression of diseases such as infectious diseases (6–8) (e.g. HIV, AIDS, hepatitis C), metabolic diseases (9,10) (e.g. diabetes, cancer) and triplet repeat disorders (e.g. myotonic dystrophy, Huntington’s disease) (11–21). Druggability of RNAs has been validated in high-profile targets, for instance, ribosomal RNA (rRNA) could be targeted using aminoglycosides, macrolide, tetracycline and oxazodinone (22–26). There are clinically approved antibiotics (e.g. erythromycin), which bind to RNA molecules. Structural analysis of antibiotics bound to ribosomal subunits has revealed that rRNA-small molecule recognition is mainly governed by electrostatic interaction, shape and hydrogen-bonding interactions (27–28). Recent success in crystallization of rRNA offers key structural information for RNA-based drug design (28–40). Diverse roles of RNAs provide numerous opportunities for specific targeting and modulating RNAs with small molecules.
Even though RNA adopts a favorable 3D structure, targeting of cellular RNA such as messenger RNA and microRNA for drug discovery was considered as complicated due to difficulty in crystallization, their conformational sampling and lack of RNA-specific computational tools. Excitingly, recent advances in biomedical and computational fields have provided vital information on RNA–small molecule interaction to overcome these issues to develop therapeutics for many diseases (41–44). Moreover, new developments in nuclear magnetic resonance and mass spectroscopy have made it possible to screen millions of small molecule compounds for finding selective inhibitors for specific RNA target (45–56).
Recently, number of RNA-based molecular targets have begun to grow rapidly with detailed elucidation of their structural and functional relationship (57–64). Also, small molecule inhibitors have been successfully developed for various different RNA molecules (65–75). Numerous publicly available databases are available providing information about RNA sequence, secondary structure and 3D RNA structure (76–86). To the best of our knowledge, there is no database that focuses on small molecule modulators along with their target RNA and experimentally determined binding data (Kd, Ki, IC50, ΔTm) of the corresponding RNA-inhibitor complex. The experimental data published in the literature are unstructured and difficult to navigate and as a result challenging to perform structure activity prediction. Moreover, chemical structures are depicted as ChemDraw images and are thus not searchable. The process of discovery of novel ligands to target RNA would be greatly facilitated by the availability of a small molecule database that is RNA-specific. In this regard, it would be highly valuable to collocate, organize and integrate RNA and their modulators along with experimental data in a publicly available domain that can be effectively navigated. SMMRNA is the first step in this direction. SMMRNA would facilitate the investigation of structure activity relationship between RNA and ligands, clustering of small molecules targeting RNA, provide information for fragment-based drug design and also aid in the quantitative structure activity relationship modeling of RNA.
MATERIALS AND METHODS
Data collection
The structural and experimental data were manually collected from peer-reviewed journals such as Biochemistry, Journal of American Chemical Society, Nature, Science and Journal of Molecular Biology. The relevant articles were selected from Pubmed, Web of Science and Google Scholar using keyword searches such as ‘small molecule targeting of RNA’, ‘small molecule RNA inhibitor’ and ‘targeting RNA with small molecules’. The details of RNA secondary structure, ligand structure, binding constant, IC50, binding mode and assay performed were gathered from each article. The references provided within the articles were consulted to get further information about RNA secondary structures, ligand structures, experimental binding affinities and binding mode of various ligands. RNA structures were drawn as images using Adobe Photoshop (v4). Structures of ligands were drawn using ChemBioDraw Ultra (v11.0) and saved as sdf file format. In many cases, the chemical structure of ligand molecules, drawn as scaffold and abbreviated with R-group substituent in the published articles, was redrawn in computer-readable form. Before importing these structures into the database, structures were manually checked for atom valence and correctness of representation.
Database description
The database is built using Apache HTTP (web server) along with MySQL (database server). MySQL RDBMS (relational database management system) is used for storing data (see the Supplementary File S1 for a complete description of MySQL database). The general layout of SMMRNA showing homepage and search, browse, upload, literature and help tools are shown in Figure 1.
Figure 1.
Homepage of SMMRNA database showing various analysis functionalities such as search, browse, upload, etc.
Substructure search
The command line version of VLifeMDS [VLifeMDS: Molecular Design Suite, VLife Sciences Technologies Pvt. Ltd., Pune, India, 2013 (www.vlifesciences.com)] was used for performing a substructure search. The main steps involved in substructure search (as implemented in VLifeMDS) were: (i) Inputs were user-defined, consisting of template molecules to be matched against the reference molecule. Reference molecules were already stored in the database. (ii) Details of the molecules were built that include number of atoms (of different atom type; H, O, N, Cl, F and C), number of bonds (based on their types; single, double or triple), number of rings (3-, 4-, 5- and 6-member rings) and number of aromatic rings. (iii) The first step eliminates the substructure search based on the configuration set above. If any one of the configurations of template molecule was greater than a reference molecule, then no further processing was done. For instance, if the number of F atoms in the template was 3 and the number of F atoms in reference was 2, then obviously the template substructure did not match with that of the reference. (iv) An atom from template molecules was selected (this could be the atom with maximum number of connections). Next, the graph of reference molecule was traversed to find atom with the same degree and type as the template atom. (v) Each atom of the template was traversed recursively in a similar manner. If at any stage, no matching template atom was found in the reference molecule, the search was terminated. If all the atoms of the template molecule were matched with the reference molecule, the search was deemed successful.
2D structure editor for structure search
VLife2Draw (87) was used as an editor and viewer for searching 2D molecular structures in the SMMRNA database (Figure 3A). This is a Java applet and can be run in a browser with a suitable Java plugin. VLife2Draw allows the user to draw chemical structures, and to import and export these structures in molecule file formats (mol and sdf are supported formats). VLife2Draw provides basic chemical structure editing facility to draw bonds like single-, double-, triple-, stereo-up and stereo-down bonds. Templates for drawing 3–8-membered ring structures were also provided.
Figure 3.
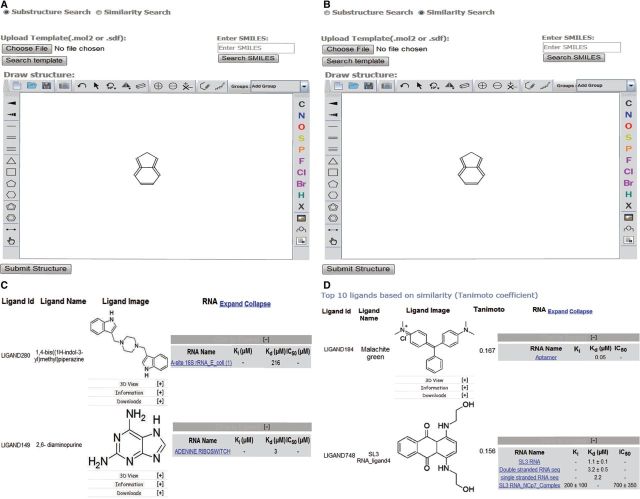
Examples of substructure and chemical similarity-based search in SMMRNA. Screenshots of (A) substructure query, (B) similarity search and (C and D) the data obtained using substructure and similarity-based search, respectively.
VLife2Draw applet runs on the client side, where a user can draw a structure that needs to be searched. After submitting the structure, applet converts these data into an sdf file format string, which is then transferred to the server. This information is captured by the server and stored in a temporary file. A Python script, written using VLifeMDS scripting framework, then uses this template molecule to search the SMMRNA database for ligand molecules that satisfy the structural similarity as indicated in the substructure search methodology.
Descriptor calculation
The command line version of VLifeMDS was used for calculating descriptors for ligands (Figure 2C). The descriptors computed by VLifeMDS are shown in Table 1.
Figure 2.
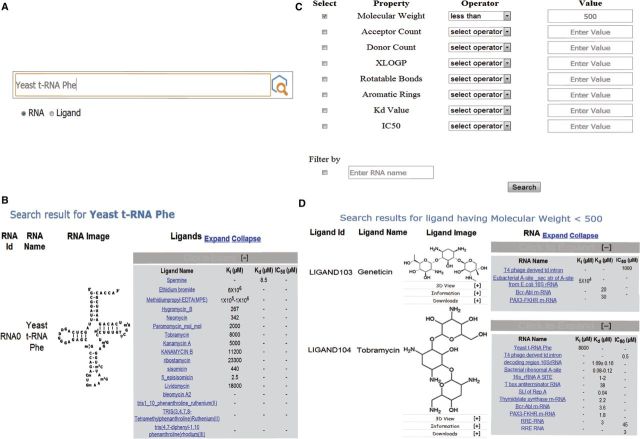
Examples of text and advanced property-based search in SMMRNA. (A) Screenshot showing dialog box for RNA and ligand text-based search. (B) Screenshot showing results based on RNA text search, e.g. using yeast t-RNA as text string, the information about the RNA and various ligands that target this particular RNA is depicted. (C) Screenshot showing dialog box for ligand search based on advanced properties such as MW, IC50, hydrogen bond donor, acceptor count, etc. (D) Screenshot showing results based on MW cutoff of <500.
Table 1.
Various descriptors along with their description and significance used for advanced search criteria
| Descriptor | Comments |
|---|---|
| Acceptor count | Number of hydrogen bond acceptors in the molecule (88). |
| Donor count | Number of hydrogen bond donors in the molecule (88). |
| Rotatable bonds | Provides information about conformation sampling of a molecule, which is useful in docking This is important in docking, where a particular conformation might show favorable binding. |
| Aromatic rings | Number of aromatic rings in a molecule. Generally, the presence of aromatic rings in a molecule result in the possibility of π–π interactions prevalent in binding sites. |
| XlogP | Distribution-coefficient of the inhibitors, drug like properties and lipophilic efficiency (89–91). |
3D structure viewing
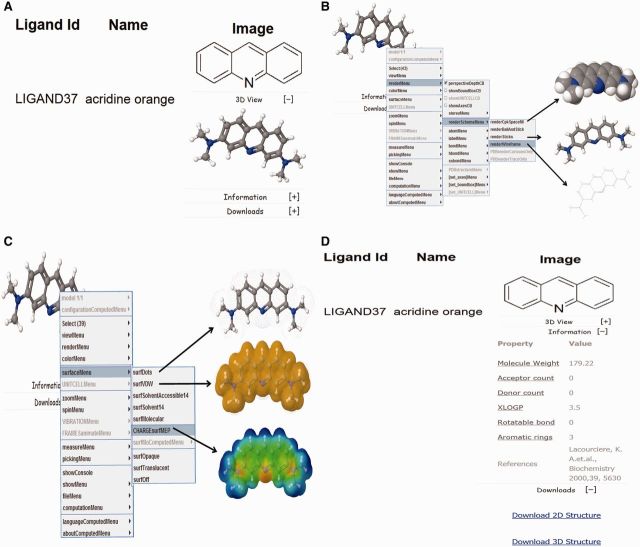
The SMMRNA database contains ligand entries as 2D coordinates. For 3D depictions, the 2D coordinates are first curated for errors followed by the addition of appropriate number of hydrogen atoms. The structures are subsequently optimized using Merck molecular force field (MMFF), using a loose gradient cutoff (1.0 kcal/mol/angstroms). The optimized structures were further processed and stored into the database in a separate field in the MOL2 file format. JMol molecular viewer was used for visualization of 3D structures of ligands (Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/). A standard ball-and-stick model is used to display the ligands (Figure 4A). Additionally, other structural displays such as Corey-Pauling-Koltun (CPK) spacefill, sticks and wireframe representation are possible by right clicking (using mouse or touchpad) and then selecting the renderMenu from the JMol dialog box (Figure 4B). It is also possible to show various molecular surfaces such as Van der Waals, solvent accessible, dots, charge, etc (Figure 4C). CPK coloring scheme of atoms is used to depict the atom types; with the prominent atom types listed as Carbon–dark gray, Hydrogen–light gray, Oxygen–red and Nitrogen–green (92,93).
Figure 4.
SMMRNA ligand screenshot showing (A) 2D and 3D structural view of the ligand, (B) various 3D structural representations using JMol dialog box, (C) various molecular surface representations such as dots, Van der Waals, charge, etc and (D) advanced property, reference and comment information and 2D and 3D structure download links.
Clustering analysis
Hierarchical, binning and multidimensional cluster analysis of the database compounds were performed using the Chemmine software (http://chemmine.ucr.edu/) (94).
3D conformer generation
High-quality 3D conformers were generated using OMEGAv2.5 from openeye (http://www.eyesopen.com/) (95). Omega uses model building and torsional search for efficient conformational sampling. In the first step of model building, the chemical structure is fragmented along sigma bonds using the makefraglib utility of OMEGAv2.5, and then the structure is reconstructed by assembly of these fragments. These fragments were refined using modified MMFF94. In the second step, an ensemble of conformers was generated using torsional angle rules as described. For each molecule, 100 conformations were generated with energy cutoff of 10 kcal/mol. The conformational searches were terminated when the energy cutoff exceeded 10 kcal/mol or when the 100 structures were built (configuration parameter file is described in the Supplementary File S2).
RESULTS AND DISCUSSION
Organization and data retrieval of SMMRNA
SMMRNA database is accessible via a user-friendly graphical user interface (GUI) at the web address www.smmrna.org. The interface allows textual search for both RNA and ligand (Figure 2), substructure and fingerprint search for ligands (Figure 3). In addition, browse facility is also provided to quickly locate ligands and RNA. The database also allows ligand-based substructure and fingerprint search (Figure 3). The ligands can be searched by their common or IUPAC names. For a molecule, this search would retrieve all the RNAs where the molecule acts as modulator together with related experimental data. Additionally for each compound, 2D and 3D structures, along with other calculated properties, appear in the drop down menu. Both 2D and 3D structures of the ligand can be downloaded (Figure 4D). For 3D representations of ligands, 2D structure was first corrected for errors, and then an appropriate number of hydrogen atoms were added. These 3D structures are energy minimized with MMFFs using gradient convergence of 1.0 kcal/(molÅ). The energy-optimized structures are further processed and stored into the database in a separate field in the MOL2 file format.
Online tools for structure and similarity search
We have implemented many web-based tools for structure-based search and the similarity analysis of various molecules reported in the SMMRNA. These tools are as described later in the text.
Advanced search
Under the search menu, an advanced tab is inserted to explore ligands using physicochemical properties such as molecular weight, XlogP and number of rotatable bonds (Figure 2C). The data generated can be filtered using a specific property or a combination of properties such as acceptor count, XlogP, etc. Each property could be controlled by an operator (greater than, less than or range) and a subsequent value for filtering. Users can further limit their search by applying a filter based on RNA names, if they are interested only in a single RNA or a class of RNA molecules.
Substructure search
A structure-based search could be easily performed by using powerful editors embedded within the web page. The user can draw a complete or partial structure of the query ligand to retrieve similar compounds (Figure 3). VLife2Draw applet runs on the client side, where a user can draw a structure that needs to be searched. After submitting the structure, applet converts these data into an sdf format string, which is then transferred to the server. The server side script captures this information and stores it into a temporary file. A Python script, written using VLifeMDS scripting framework, then uses this template molecule to search the SMMRNA database for ligand molecules that satisfy the structural similarity as indicated in the substructure search methodology. The steps implemented in VLifeMDS for performing a substructure search are as follows: (i) user-defined input to be matched against the database molecule; (ii) molecule generation using atoms (H, O, N and C), bonds (single, double and triple) and carbocyclic, heterocyclic and aromatic rings of different sizes (3-, 4-, 5-, 6-membered); (iii) atom template matching between the input and the database reference molecules; and (iv) graph matching between input and reference molecule to establish connectivity. If there is a match between atom template and graph connectivity between input and reference molecule, the search would return with the matching list of structures. Graph matching is based on graph theory, which involves modeling the pairwise similarity between two molecules. In VLifeMDS, molecules are represented by graphs, which consist of vertices (atom) and edges (bond connecting atoms). Vertices are pictorially represented as ‘dots’ or ‘circles’, whereas the edges are represented by drawing an arc. Mathematically, a graph can be represented by a matrix of atoms and bonds, and each entry (atom, bonds) contains the bond endpoint data. For substructure search matching, all it involves is to determine which graphs in a database are subgraph isomorphic to a query graph (see the Supplementary File S3 for illustration of graph matching).
Similarity search
Ligand similarity search of SMMRNA is quite versatile, as it has the capability to identify as well as rank the structures based on the query. A fingerprint represents a predefined fragment or feature found in a molecule. The fingerprint search uses a binary fingerprint for the chemical structure. A fingerprint is a series of binary (1/0) bits that are arranged in sequence. Each bit position corresponds to certain information such as the presence or absence of an atom, type of ring, element count and a substructure. During a substructure similarity search, a fingerprint of the query is computed initially. Tanimoto coefficient is based on the fingerprints, which are binary vectors, with a value of 1 and 0, with 1 indicating the ‘presence of’ and 0 indicating the ‘absence of’ fingerprints. Tanimoto coefficient quantifies the similarity between query and database molecules and has a similarity score range from 0.0 to 1.0. For computing Tanimoto coefficient, each ligand in the SMMRNA has a pre-computed fingerprint incorporated. The generated fingerprint is then matched with every entry in the database, and a similarity score, which lies between 0.0 and 1.0, is computed. The structures are sorted in descending order relative to similarity score, with the top hits representing the most similar structure to the search query (Figure 3D). SMMRNA uses OpenBabel fingerprints (http://openbabel.org/) search for similarity analysis (96).
Significance of the database
We have reviewed >900 original research articles and collected all the relevant information. In SMMRNA, we have not included the PubChem Bioassay data to avoid duplication and problems about the credibility and validation of high-throughput screening (HTS) data. The SMMRNA database contains unique molecules and includes only those molecules for which the experimental data are reported in the peer-reviewed literature. In few articles, experimental parameters were not reported, although the binding to RNA molecules was confirmed and validated with various spectroscopic and other binding techniques such as gel shift and radioactivity assay. We have included these molecules and annotated comments and references to highlight these observations so that the user can easily verify and cross validate.
The SMMRNA database is very diverse and includes molecular classes such as antibiotics, peptides, amino acids, modified amino acids, dyes, nucleotides, polycations, intercalators, inorganic and organometallic compounds, which are unique to this database (see the Supplementary File S4). One particular observation is that many antibiotics act as ligands for RNA. Literature analysis reveals (97) that there are eight important clinically approved antibiotics, whose mechanism of action involves RNA binding, and the research and development efforts on antibiotics in recent years have severely declined. The data provided here are appealing for antibiotics research and would aid in the design of improved or completely new antibiotics (using various computational methods).
Chemical space analysis of database molecules
RNA is a highly charged molecule and possesses a high degree of conformational flexibility. Thus, it is important in the early drug discovery stages to predict and optimize the favorable physicochemical properties of ligands to facilitate their efficient binding to RNA molecules. To understand whether various filters that are commonly applied to other biological targets are effective, we analyzed the molecules in the SMMRNA database using standard filters such as Lipinski Rule of Five, ZINC and PAINS filter (98–100). ZINC filter is less restricted compared with other filters and uses relaxed cutoff value of various physicochemical parameters such as molecular weight, hydrogen bond donor and acceptor count, logP and number of rotatable bonds (see the Supplementary File S5 for ZINC filters used). About two-third of the molecules failed to pass these filters. These results are important in understanding the chemical space, which we should focus in developing ligands for targeting RNA. For targeting RNA, specific physicochemical parameters such as more hydrophilicity, lower lipophilicity (especially for developing new antibiotics), high molecular weight and larger polar surface area are required. Most of the ligands that bind to RNA are highly charged, e.g. polycation derivatives and aminoglycosides. However, there are small drug-like molecules such as DAPI, etc, which can be optimized to bind RNA with varying binding affinity. It is thus possible to develop lead-like molecules to target RNA by modification of these molecules using organic synthesis. From the analysis of the molecules in SMMRNA, it is clear that we need to develop separate filter parameters for analyzing ligands targeting RNA. In particular, we need to be flexible in terms of hydrogen bond donor, hydrogen bond acceptor, number of rotatable bonds, ring size to design and synthesize diverse and optimized ligands of higher selectivity and sensitivity. Also, majority of RNA targeting molecules have significantly higher topological polar surface area as compared with the drug-like molecules, where the value is around <140 Å2. These results are highly important, as recent reviews (97) have highlighted the need to develop entirely different physicochemical parameters for targeting RNA.
Clustering analysis
Clustering is an important tool for medicinal or computational chemist and is widely used in application ranging from drug discovery to lead optimization (101,102). Clustering techniques are used to identify molecule scaffolds that share common structural properties (102). It is widely accepted that structurally similar molecules will show similar biological activity profile (103). ChemMine tools for clustering analysis include hierarchical clustering, multidimensional scaling (MDS) and binning clustering. We have used these clustering tools to identify common scaffolds based on structural and molecular descriptors. This will lead to the identification of potential molecular entities for structure activity relationship. We have performed hierarchical, binning and MDS to identify and visualize the chemical space of ligand molecules.
We have performed agglomerative hierarchical clustering method, which involves a progressive combination of the most similar molecules into related groups, and the results are shown as a dendrogram, highlighting the relationship between clusters. It has the advantage that any number of clusters can be selected. However, it is difficult to visualize the chemical space covered by database molecules. For visualization of chemical space, we performed binning partitioning clustering and MDS. Bin-based partitioning methods are useful for the identification of underrepresented chemical space, which can be fulfilled by synthesizing more compounds. In bin-based partitioning method, we subdivided multidimensional chemical space into various bins using a Tanimoto similarity cutoff of 0.4. Compounds falling into the same bin volumes are deemed to have similar chemical properties. Bin-based partitioning of database compounds, which passed the ZINC filter resulted in the creation of bins with variable bin sizes. We observed many bin clusters with sizable number of similar molecules. For example, there is an interesting cluster of antibiotics that target RNA molecules. We observed a number of bin clusters represented by just single molecular scaffold. We believe that greater medicinal chemistry effort should be made to expand the compounds representing these particular scaffolds.
3D conformation
For high-throughput virtual screening, such as ligand and structure-based drug design, we generated a multiconformer database of 647 compounds using MMFF94. For each compound, 100 conformers were generated specifying the energy cutoff of 10 kcal/mol and root-mean-square deviation of 0.5. For some compounds, the number of conformers generated was less than the specified as a result of energy and root-mean-square deviation criteria. Around 128 compounds failed as a result of the criteria specified. We believe this multiconformer database (with 33 866 conformations, see the Supplementary File S6) will be very useful, as it provides the structural information about the conformational states of all the compounds, which is vital for in silico drug designing.
CONCLUSIONS AND FUTURE PROSPECTS
We have developed SMMRNA (www.smmrna.org), a chemoinformatics platform to facilitate interactive exploring of RNA molecules and their modulators. The database comprises structural images of RNA, chemical structure of ligands, related experimental data (Kd, Ki, IC50, ΔTm values), 3D conformers and chemoinformatic information. The database can be browsed using RNA names, ligand structures, substructures and fingerprint-based chemical similarity search (see the Supplementary File S7, which shows the video tutorial for navigating SMMRNA database). SMMRNA would facilitate structure activity relationship studies, statistical analysis, fragment-based drug design, virtual screening and molecular docking studies to assist in developing RNA-based small molecule modulators (for comparison with other databases see the Supplementary File S8). In general, this publicly available database would be beneficial for the design and discovery of modulators targeting RNA-mediated diseases such as diabetes, neurodegenerative disorders and cardiovascular disorders. We are continually reviewing the older literature, patents, etc, so as to include additional experimentally validated ligand entries. We have also provided the upload facility for both ligand and RNA entries for other researchers so that SMMRNA will continue to grow and thus provide comprehensive RNA-ligand data. In future versions, we would include RNA structure drawing tool, RNA alignment tool, fragment generation capability, 3D structure comparison, automatic docking protocol and principal component analysis. The main aim of the database is to provide computational and medicinal chemistry tools for research community who are interested in medicinal chemistry, biology and biochemistry of RNA. We believe that the set of tools provided in SMMRNA would invite scientists from diverse backgrounds to initiate drug design and development of small molecule modulators for targeting various RNA molecules.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Institute of Microbial Technology [grant Infra 62 to R.P.]; Institute of Microbial Technology Research Internship (to A.M. and S.S.) and CSIR JRF (to S.L.); Recipient of Ramalingaswami fellowship (to D.K.S.). Funding for open access charge: Intramural research funds of Institute of Microbial Technology, Chandigarh.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Rethi Madathil for help in preparing the manuscript, Ms Isha for drawing additional structures and clustering analysis and Dr Kundan, Dr Ganesh and Mr Kiran for technical help.
REFERENCES
- 1.Moore PB, Steitz TA. The roles of RNA in the synthesis of protein. Cold Spring Harb. Perspect. Biol. 2011;3:a003780. doi: 10.1101/cshperspect.a003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 3.Hermann T, Westhof E. RNA as a drug target: chemical, modelling, and evolutionary tools. Curr. Opin. Biotechnol. 1998;9:66–73. doi: 10.1016/s0958-1669(98)80086-4. [DOI] [PubMed] [Google Scholar]
- 4.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 5.Noller HF. Drugs and the RNA world. Nature. 1991;353:302–303. doi: 10.1038/353302a0. [DOI] [PubMed] [Google Scholar]
- 6.Zapp ML, Stern S, Green MR. Small molecules that selectively block RNA binding of HIV-1 rev protein inhibit rev function and viral production. Cell. 1993;74:969–978. doi: 10.1016/0092-8674(93)90720-b. [DOI] [PubMed] [Google Scholar]
- 7.Malim MH, Cullen BR. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 8.Jensen KB, Green L, MacDougal-Waugh S, Tuerk C. Characterization of an in vitro-selected RNA ligand to the HIV-1 rev protein. J. Mol. Biol. 1994;235:237–247. doi: 10.1016/s0022-2836(05)80030-0. [DOI] [PubMed] [Google Scholar]
- 9.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pérez B, Ugarte M, Desviat L. In: From Nucleic Acids Sequences to Molecular Medicine. Erdmann VA, Barciszewski J, editors. Berlin/Heidelberg, Germany: Springer Verlag; 2012. pp. 357–370. [Google Scholar]
- 11.Fiszer A, Mykowska A, Krzyzosiak WJ. Inhibition of mutant huntingtin expression by RNA duplex targeting expanded CAG repeats. Nucleic Acids Res. 2011;39:5578–5585. doi: 10.1093/nar/gkr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galka-Marciniak P, Urbanek MO, Krzyzosiak WJ. Triplet repeats in transcripts: structural insights into RNA toxicity. Biol. Chem. 2012;393:1299–1315. doi: 10.1515/hsz-2012-0218. [DOI] [PubMed] [Google Scholar]
- 13.Hu J, Liu J, Yu D, Chu Y, Corey DR. Mechanism of allele-selective inhibition of huntingtin expression by duplex RNAs that target CAG repeats: function through the RNAi pathway. Nucleic Acids Res. 2012;40:11270–11280. doi: 10.1093/nar/gks907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome. Nucleic Acids Res. 2011;39:7308–7315. doi: 10.1093/nar/gkr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper TA. Chemical reversal of the RNA gain of function in myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2009;106:18433–18434. doi: 10.1073/pnas.0910643106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sicot G, Gourdon G, Gomes-Pereira M. Myotonic dystrophy, when simple repeats reveal complex pathogenic entities: new findings and future challenges. Hum. Mol. Genet. 2011;20:5. doi: 10.1093/hmg/ddr343. [DOI] [PubMed] [Google Scholar]
- 19.Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, Wang T, Hoskins J, Tran T, Housman D, Thornton CA, et al. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. J. Am. Chem. Soc. 2012;134:4731–4742. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson LM, Wood MJ. RNA therapy for polyglutamine neurodegenerative diseases. Expert Rev. Mol. Med. 2012;31:1. doi: 10.1017/erm.2011.1. [DOI] [PubMed] [Google Scholar]
- 21.Nakamori M, Gourdon G, Thornton CA. Stabilization of expanded (CTG)*(CAG) repeats by antisense oligonucleotides. Mol. Ther. 2011;19:2222–2227. doi: 10.1038/mt.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandi L, Fabbretti A, Milon P, Carotti M, Pon CL, Gualerzi CO. Methods for identifying compounds that specifically target translation. Methods Enzymol. 2007;431:229–267. doi: 10.1016/S0076-6879(07)31012-4. [DOI] [PubMed] [Google Scholar]
- 24.Lange RP, Locher HH, Wyss PC, Then RL. The targets of currently used antibacterial agents: lessons for drug discovery. Curr. Pharm. Des. 2007;13:3140–3154. doi: 10.2174/138161207782110408. [DOI] [PubMed] [Google Scholar]
- 25.Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCoy LS, Xie Y, Tor Y. Antibiotics that target protein synthesis. Wiley Interdiscip. Rev. RNA. 2011;2:209–232. doi: 10.1002/wrna.60. [DOI] [PubMed] [Google Scholar]
- 27.Harvey I, Garneau P, Pelletier J. Inhibition of translation by RNA–small molecule interactions. RNA. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 29.Clemons WM, Jr, May JL, Wimberly BT, McCutcheon JP, Capel MS, Ramakrishnan V. Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution. Nature. 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 30.Brodersen DE, Clemons WM, Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 31.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 32.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 33.Clemons WM, Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 34.Yonath A. High-resolution structures of large ribosomal subunits from mesophilic eubacteria and halophilic archaea at various functional States. Curr. Protein. Pept. Sci. 2002;3:67–78. doi: 10.2174/1389203023380828. [DOI] [PubMed] [Google Scholar]
- 35.Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 36.Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 37.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 38.Moore PB, Steitz TA. The structural basis of large ribosomal subunit function. Annu. Rev. Biochem. 2003;72:813–850. doi: 10.1146/annurev.biochem.72.110601.135450. [DOI] [PubMed] [Google Scholar]
- 39.Steitz TA, Moore PB. RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem. Sci. 2003;28:411–418. doi: 10.1016/S0968-0004(03)00169-5. [DOI] [PubMed] [Google Scholar]
- 40.Steitz TA. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 41.Dieterich C, Stadler PF. Computational biology of RNA interactions. Wiley Interdiscip. Rev. RNA. 2013;4:107–120. doi: 10.1002/wrna.1147. [DOI] [PubMed] [Google Scholar]
- 42.Chou CH, Lin FM, Chou MT, Hsu SD, Chang TH, Weng SL, Shrestha S, Hsiao CC, Hung JH, Huang HD. A computational approach for identifying microRNA-target interactions using high-throughput CLIP and PAR-CLIP sequencing. BMC Genomics. 2013;14(Suppl.1):S2. doi: 10.1186/1471-2164-14-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuzu G, Keskin O, Gursoy A, Nussinov R. Expanding the conformational selection paradigm in protein-ligand docking. Methods Mol. Biol. 2012;819:59–74. doi: 10.1007/978-1-61779-465-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sim AY, Minary P, Levitt M. Modeling nucleic acids. Curr. Opin. Struct. Biol. 2012;22:273–278. doi: 10.1016/j.sbi.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moumne R, Catala M, Larue V, Micouin L, Tisne C. Fragment-based design of small RNA binders: promising developments and contribution of NMR. Biochimie. 2012;94:1607–1619. doi: 10.1016/j.biochi.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Furtig B, Richter C, Wohnert J, Schwalbe H. NMR spectroscopy of RNA. Chembiochem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 47.Qin PZ, Dieckmann T. Application of NMR and EPR methods to the study of RNA. Curr. Opin. Struct. Biol. 2004;14:350–359. doi: 10.1016/j.sbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Furtig B, Buck J, Manoharan V, Bermel W, Jaschke A, Wenter P, Pitsch S, Schwalbe H. Time-resolved NMR studies of RNA folding. Biopolymers. 2007;86:360–383. doi: 10.1002/bip.20761. [DOI] [PubMed] [Google Scholar]
- 49.Huber W. A new strategy for improved secondary screening and lead optimization using high-resolution SPR characterization of compound-target interactions. J. Mol. Recognit. 2005;18:273–281. doi: 10.1002/jmr.744. [DOI] [PubMed] [Google Scholar]
- 50.Zhu Y, Valdes R, Jr, Jortani SA. Application of bioaffinity mass spectrometry for analysis of ligands. Ther Drug Monit. 2005;27:694–699. doi: 10.1097/01.ftd.0000179851.32093.dc. [DOI] [PubMed] [Google Scholar]
- 51.Schug KA. Solution phase enantioselective recognition and discrimination by electrospray ionization—mass spectrometry: state-of-the-art, methods, and an eye towards increased throughput measurements. Comb. Chem. High Throughput Screen. 2007;10:301–316. doi: 10.2174/138620707781662790. [DOI] [PubMed] [Google Scholar]
- 52.Oeffinger M. Two steps forward—one step back: advances in affinity purification mass spectrometry of macromolecular complexes. Proteomics. 2012;12:1591–1608. doi: 10.1002/pmic.201100509. [DOI] [PubMed] [Google Scholar]
- 53.Tuzimski T. Application of different modes of thin-layer chromatography and mass spectrometry for the separation and detection of large and small biomolecules. J. Chromatogr. A. 2011;1218:8799–8812. doi: 10.1016/j.chroma.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 54.Nakayama H, Takahashi N, Isobe T. Informatics for mass spectrometry-based RNA analysis. Mass Spectrom. Rev. 2011;30:1000–1012. doi: 10.1002/mas.20325. [DOI] [PubMed] [Google Scholar]
- 55.Krug K, Nahnsen S, Macek B. Mass spectrometry at the interface of proteomics and genomics. Mol. Biosyst. 2011;7:284–291. doi: 10.1039/c0mb00168f. [DOI] [PubMed] [Google Scholar]
- 56.Giessing AM, Kirpekar F. Mass spectrometry in the biology of RNA and its modifications. J. Proteomics. 2012;75:3434–3449. doi: 10.1016/j.jprot.2012.01.032. [DOI] [PubMed] [Google Scholar]
- 57.DeJong ES, Luy B, Marino JP. RNA and RNA-protein complexes as targets for therapeutic intervention. Curr. Top. Med. Chem. 2002;2:289–302. doi: 10.2174/1568026023394245. [DOI] [PubMed] [Google Scholar]
- 58.Franceschi F, Duffy EM. Structure-based drug design meets the ribosome. Biochem. Pharmacol. 2006;71:1016–1025. doi: 10.1016/j.bcp.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 59.Maxwell MM. RNAi applications in therapy development for neurodegenerative disease. Curr. Pharm. Des. 2009;15:3977–3991. doi: 10.2174/138161209789649295. [DOI] [PubMed] [Google Scholar]
- 60.Coyne AG, Scott DE, Abell C. Drugging challenging targets using fragment-based approaches. Curr. Opin. Chem. Biol. 2010;14:299–307. doi: 10.1016/j.cbpa.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 61.Wu W. MicroRNA: potential targets for the development of novel drugs? Drugs R D. 2010;10:1–8. doi: 10.2165/11537800-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Collie GW, Parkinson GN. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011;40:5867–5892. doi: 10.1039/c1cs15067g. [DOI] [PubMed] [Google Scholar]
- 63.Czech MP, Aouadi M, Tesz GJ. RNAi-based therapeutic strategies for metabolic disease. Nat. Rev. Endocrinol. 2011;7:473–484. doi: 10.1038/nrendo.2011.57. [DOI] [PubMed] [Google Scholar]
- 64.Deigan KE, Ferre-D'Amare AR. Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem. Res. 2011;44:1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson WD, Li K. Targeting RNA with small molecules. Curr. Med. Chem. 2000;7:73–98. doi: 10.2174/0929867003375434. [DOI] [PubMed] [Google Scholar]
- 66.Tor Y. Targeting RNA with small molecules. Chembiochem. 2003;4:998–1007. doi: 10.1002/cbic.200300680. [DOI] [PubMed] [Google Scholar]
- 67.Hermann T. Drugs targeting the ribosome. Curr. Opin. Struct. Biol. 2005;15:355–366. doi: 10.1016/j.sbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 68.Wu L, Pan J, Thoroddsen V, Wysong DR, Blackman RK, Bulawa CE, Gould AE, Ocain TD, Dick LR, Errada P, et al. Novel small-molecule inhibitors of RNA polymerase III. Eukaryot Cell. 2003;2:256–264. doi: 10.1128/EC.2.2.256-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richter SN, Palu G. Inhibitors of HIV-1 Tat-mediated transactivation. Curr. Med. Chem. 2006;13:1305–1315. doi: 10.2174/092986706776872989. [DOI] [PubMed] [Google Scholar]
- 70.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 71.Giri P, Kumar GS. Molecular aspects of small molecules-poly(A) interaction: an approach to RNA based drug design. Curr. Med. Chem. 2009;16:965–987. doi: 10.2174/092986709787581932. [DOI] [PubMed] [Google Scholar]
- 72.Aboul-ela F. Strategies for the design of RNA-binding small molecules. Future Med. Chem. 2010;2:93–119. doi: 10.4155/fmc.09.149. [DOI] [PubMed] [Google Scholar]
- 73.Neelakandan K, Babu P, Nair S. Emerging roles for modulation of microRNA signatures in cancer chemoprevention. Curr. Cancer Drug Targets. 2012;12:716–740. doi: 10.2174/156800912801784875. [DOI] [PubMed] [Google Scholar]
- 74.Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS Chem. Biol. 2012;7:73–86. doi: 10.1021/cb200447r. [DOI] [PubMed] [Google Scholar]
- 75.Kumar GS. RNA targeting by small molecules: binding of protoberberine, benzophenanthridine and aristolochia alkaloids to various RNA structures. J. Biosci. 2012;37:539–552. doi: 10.1007/s12038-012-9217-3. [DOI] [PubMed] [Google Scholar]
- 76.Liu X, Liu L, Xu Q, Wu P, Zuo X, Ji A. MicroRNA as a novel drug target for cancer therapy. Expert Opin. Biol. Ther. 2012;12:573–580. doi: 10.1517/14712598.2012.671293. [DOI] [PubMed] [Google Scholar]
- 77.Mackowiak G, Jens SD, Maaskola J, Kuntzagk A, Rajewsky N, Landthaler M, Dieterich C. doRiNA: a database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res. 2012;40:D180–D186. doi: 10.1093/nar/gkr1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Augustin R, Endres K, Reinhardt S, Kuhn PH, Lichtenthaler SF, Hansen J, Wurst W, Trumbach D. Computational identification and experimental validation of microRNAs binding to the Alzheimer-related gene ADAM10. BMC Med. Genet. 2012;13:35. doi: 10.1186/1471-2350-13-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cao Y, Wu J, Liu Q, Zhao Y, Ying X, Cha L, Wang L, Li W. sRNATarBase: a comprehensive database of bacterial sRNA targets verified by experiments. RNA. 2010;16:2051–2057. doi: 10.1261/rna.2193110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen X, Liu MX, Yan GY. RWRMDA: predicting novel human microRNA-disease associations. Mol. Biosyst. 2012;8:2792–2798. doi: 10.1039/c2mb25180a. [DOI] [PubMed] [Google Scholar]
- 82.Cho S, Jang I, Jun Y, Yoon S, Ko M, Kwon Y, Choi I, Chang H, Ryu D, Lee B, et al. MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2013;41:D252–D257. doi: 10.1093/nar/gks1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cvitkovic I, Jurica MS. Spliceosome database: a tool for tracking components of the spliceosome. Nucleic Acids Res. 2013;41:D132–D141. doi: 10.1093/nar/gks999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.George AD, Tenenbaum SA. Web-based tools for studying RNA structure and function. Methods Mol. Biol. 2011;703:67–86. doi: 10.1007/978-1-59745-248-9_6. [DOI] [PubMed] [Google Scholar]
- 86.Pang KC, Stephen S, Dinger ME, Engstrom PG, Lenhard B, Mattick JS. RNAdb 2.0—an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35:D178–D182. doi: 10.1093/nar/gkl926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Supekar K. Masters Thesis. University of Pune; 2010. 2D Chemical Structure Editor. [Google Scholar]
- 88.Wang R, Fu Y, Lai L. A new atom—additive method for calculating partition coefficients. J. Chem. Inf. Comput. Sci. 1997;37:615–621. [Google Scholar]
- 89.Cheng T, Zhao Y, Li X, Lin F, Xu Y, Zhang X, Li Y, Wang R, Lai L. Computation of octanol-water partition coefficients by guiding an additive model with knowledge. J. Chem. Inf. Model. 2007;47:2140–2148. doi: 10.1021/ci700257y. [DOI] [PubMed] [Google Scholar]
- 90.Edwards MP, Price DA. Role of physicochemical properties and ligand lipophilicity efficiency in addressing drug safety risks. Annu. Rep. Med. Chem. 2010;45:381–391. [Google Scholar]
- 91.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliver Rev. 2012;64:4–17. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 92.Corey RB, Pauling L. Molecular models of amino acids, peptides, and proteins. Rev. Sci. Instrum. 1953;24:621–627. [Google Scholar]
- 93.Koltun WL. 1965. Space filling atomic units and connectors for molecular models. Washington, DC, USA, U. S. Patent 3170246. [DOI] [PubMed] [Google Scholar]
- 94.Backman TW, Cao Y, Girke T. ChemMine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res. 2011;39:W486–W491. doi: 10.1093/nar/gkr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hawkins PC, Skillman AG, Warren GL, Ellingson BA, Stahl MT. Conformer generation with OMEGA: algorithm and validation using high quality structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010;50:572–584. doi: 10.1021/ci100031x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J. Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lewis K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 98.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 99.Lagorce D, Maupetit J, Baell J, Sperandio O, Tuffery P, Miteva MA, Galons H, Villoutreix BO. The FAF-Drugs2 server: a multistep engine to prepare electronic chemical compound collections. Bioinformatics. 2011;27:2018–2020. doi: 10.1093/bioinformatics/btr333. [DOI] [PubMed] [Google Scholar]
- 100.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 101.Downs GM, Barnard JM. Clustering methods and their uses in computational chemistry. In: Lipkowitz BK, Boyd BD, editors. Reviews in Computational Chemistry, Vol 18. New Jersey, USA: John Wiley and Sons, Wiley-VCH; 2002. pp. 1–40. [Google Scholar]
- 102.Wild DJ, Blankley CJ. VisualiSAR: a web-based application for clustering, structure browsing, and structure-activity relationship study. J. Mol. Graphics Model. 1999;17:85–89. doi: 10.1016/s1093-3263(99)00026-1. [DOI] [PubMed] [Google Scholar]
- 103.Nicholls A, McGaughey GB, Sheridan RP, Good AC, Warren G, Mathieu M, Muchmore SW, Brown SP, Grant JA, Haigh JA, et al. Molecular shape and medicinal chemistry: a perspective. J. Med. Chem. 2010;53:3862–3886. doi: 10.1021/jm900818s. [DOI] [PMC free article] [PubMed] [Google Scholar]