Abstract
This manuscript describes an update of the leaf senescence database (LSD) previously featured in the 2011 NAR Database Issue. LSD provides comprehensive information concerning senescence-associated genes (SAGs) and their corresponding mutants. We have made extensive annotations for these SAGs through both manual and computational approaches. Recently, we updated LSD to a new version LSD 2.0 (http://www.eplantsenescence.org/), which contains 5356 genes and 322 mutants from 44 species, an extension from the previous version containing 1145 genes and 154 mutants from 21 species. In the current version, we also included several new features: (i) Primer sequences retrieved based on experimental evidence or designed for high-throughput analysis were added; (ii) More than 100 images of Arabidopsis SAG mutants were added; (iii) Arabidopsis seed information obtained from The Arabidopsis Information Resource (TAIR) was integrated; (iv) Subcellular localization information of SAGs in Arabidopsis mined from literature or generated from the SUBA3 program was presented; (v) Quantitative Trait Loci information was added with links to the original database and (vi) New options such as primer and miRNA search for database query were implemented. The updated database will be a valuable and informative resource for basic research of leaf senescence and for the manipulation of traits of agronomically important plants.
INTRODUCTION
In plants, senescence occurs at the final stage of leaf development and precedes cell death (1,2). Leaf senescence is a highly coordinated process regulated by a large number of senescence-associated genes (SAGs), which are upregulated during senescence (1). Many advances in the understanding of the molecular mechanisms of leaf senescence have been achieved through the identification and characterization of hundreds of SAGs and their corresponding mutants (1,3,4). To facilitate systematical research and comparative studies of leaf senescence, we have developed a database of leaf senescence database (LSD) to collect SAGs and their mutants and phenotypes, as well as reference citations (4,5). These SAGs were retrieved based on genetic, genomic, proteomic or physiological evidence, and were classified into different categories according to their functions in leaf senescence. The first version of LSD was released in August 2010 and publicly accessible for the plant research community (5).
In the past 3 years, many advances had been made in leaf senescence studies. As a result, a large amount of new genes have been newly identified as functional SAGs. For example, SAG113, a gene that encodes protein phosphatase 2C, has been demonstrated to be a positive regulator of Arabidopsis leaf senescence (6). Transgenic plants overexpressing SAG113 exhibited an early senescence phenotype, whereas mutation in SAG113 delayed developmental senescence process (6). UGT76B1, a UDP-glycosyltransferase encoding gene, was reported to be a negative regulator of leaf senescence in Arabidopsis (7). Overexpression of UGT76B1 delayed leaf senescence, while ugt76b1 knockout lines showed a clearly early senescence phenotype (7). Moreover, genome-scale analyses have been widely used in leaf senescence studies, and thousands of SAGs have been identified and categorized by using the ATH1 Arabidopsis GeneChip microarray (8), providing a systematic view of transcriptional regulation in leaf senescence (9).
To cover the above-mentioned advances and extending the functionality of the previous LSD (5), we updated our database to a new version LSD 2.0. In this updated version, we made tremendous modifications and improvements to the original database and added several new features. First, we manually updated our collection of SAG genes and senescence-associated mutants based on recent papers published in the latest 3 years. The updated version contains 5356 genes and 322 mutants, increased by ∼4.5-fold and ∼2.8-fold, respectively (Table 1). Second, new features were included in the current version, including primer sequences, subcellular localization information, Arabidopsis seed information, images of Arabidopsis mutants and Quantitative Trait Loci (QTL) information.
Table 1.
Comparisons of gene number between the two versions of the database
| Species | Common name | LSD 1.0 | LSD 2.0 |
|---|---|---|---|
| Arabidopsis thaliana | Thale Cress | 949 | 3744 |
| Oryza sativa | Rice | 104 | 132 |
| Medicago truncatula | Barrel Clover | 31 | 31 |
| Brassica napus | Rapeseed | 15 | 8 |
| Lycopersicon esculentum | Tomato | 8 | 23 |
| Nicotiana tabacum | Tobacco | 5 | 9 |
| Brassica oleracea | Broccoli | 4 | 9 |
| Glycine max | Soybean | 4 | 12 |
| Pisum sativum | Pea | 4 | 6 |
| Sorghum bicolor | Sorghum | 4 | 26 |
| Hordeum vulgare | Barley | 3 | 14 |
| Solanum tuberosum | Potato | 3 | 3 |
| Zea mays | Maize | 3 | 94 |
| Astragalus sinicus | Chinese Milk Vetch | 1 | 1 |
| Chenopodium rubrum | Red goosefoot | 1 | 1 |
| Festuca pratensis Huds. | Fescue | 1 | 1 |
| Ipomoea nil | Japanese morning glory | 1 | 1 |
| Medicago sativa | Alfalfa | 1 | 2 |
| Rosa hybrida | Rose | 1 | 1 |
| Triticum aestivum | Wheat | 1 | 256 |
| Triticum turgidum | Wheat | 1 | 65 |
| Amaranthus hypochondriacus | Grain Amaranths | 0 | 1 |
| Arabidopsis lyrata | Thale Cress | 0 | 2 |
| Brassica campestris | Chinese cabbage | 0 | 2 |
| Brassica rapa subsp. Rapa | Turnip | 0 | 1 |
| Brassica rapa var.parachinensis | Choy sum | 0 | 5 |
| Camellia sinensis | Tea | 0 | 1 |
| Capsicum annuum | Pepper | 0 | 1 |
| Crocus sativus | Saffron | 0 | 1 |
| Cucumis melo | Muskmelon | 0 | 1 |
| Daucus carota | Carrot | 0 | 1 |
| Dianthus caryophyllus | Carnation | 0 | 1 |
| Festuca arundinacea | Tall fescue | 0 | 1 |
| Fragaria x ananassa | Strawberry | 0 | 1 |
| Ipomoea batatas | Sweet potato | 0 | 4 |
| Lolium perenne | perennial ryegrass | 0 | 4 |
| Mangifera indica | Mango | 0 | 1 |
| Musa acuminata | Banana | 0 | 882 |
| Neosinocalamus affinis | Rendle | 0 | 1 |
| Nicotiana attenuata | Solanaceae | 0 | 1 |
| Petunia hybrida | Petunia | 0 | 1 |
| Platycodon grandiflorum | Balloon flower | 0 | 1 |
| Spinacia oleracea | Spinach | 0 | 2 |
| Vigna unguiculata | cowpea | 0 | 1 |
| Total | 44 | 1145 | 5356 |
NEW FEATURES
Browsing and searching
LSD 2.0 enables users to retrieve and analyze SAGs through the Browse or Search page. Users may browse the entries by clicking the buttons of Species, Mutants, Phenotypes, QTLs and Arabidopsis Seed at the main page. A tree-like structure was designed for both species and phenotypes, and tables were also created for species, mutants, QTL and Arabidopsis Seed. In the updated version, new options such as primer and miRNA search for database query were added. Currently, the text search interface allows users to make queries with five types of data: (i) locus name, GenBank ID, alias, species and description of genes; (ii) name, type and ecotype of mutants; (iii) title, author, journal and date of literature papers; (iv) locus name, alias and keywords and (v) miRNA name.
Example of annotation
Figure 1 shows the annotation for a typical LSD entry EIN2 (AT5G03280), an essential positive regulator of ethylene signaling (10), which accelerates Arabidopsis leaf senescence through suppressing miR164 expression (11). We made detailed annotation including general information (gene ontology, sequences, protein–protein interactions and references) (Figure 1A), mutant information (Figure 1B), miRNA interaction information (Figure 1C), ortholog groups (Figure 1D), as well as cross links to several protein domain and family databases such as Pfam and Prosite (Figure 1E).
Figure 1.
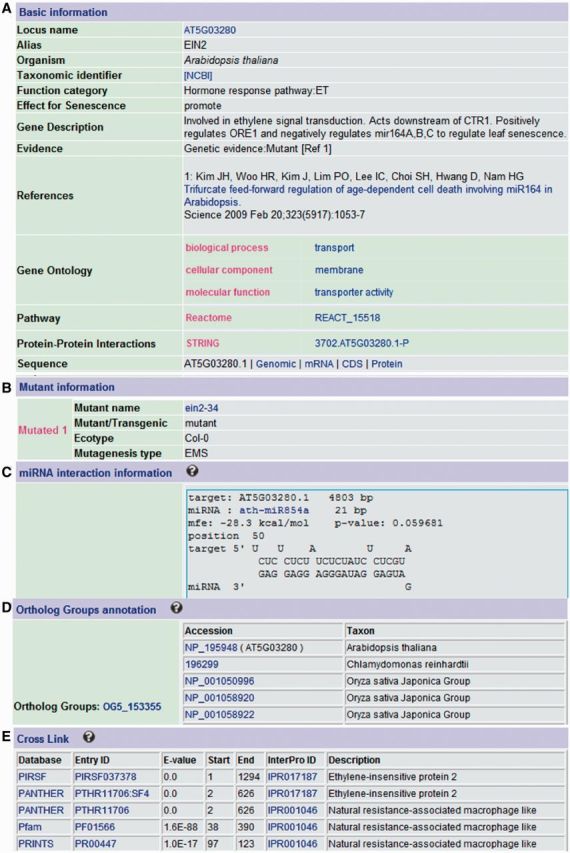
A typical entry for the Arabidopsis EIN2 gene (AT5G03280) in LSD. (A) Basic information, (B) Mutant information, (C) miRNA targets, (D) Ortholog Groups and (E) Cross Links to other databases.
Phenotype information
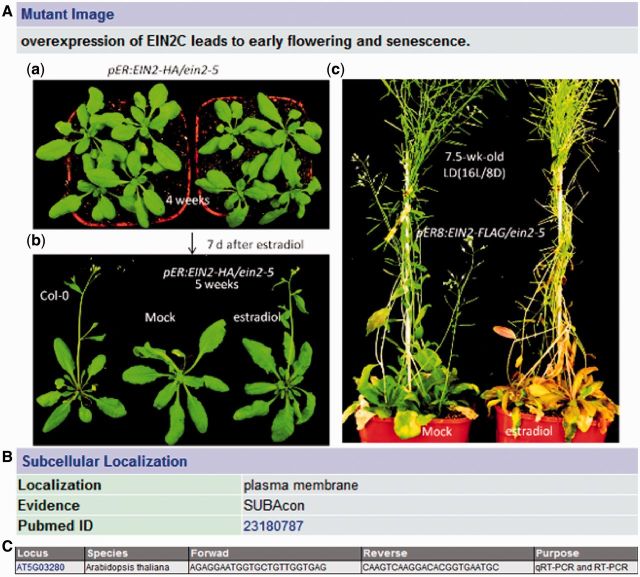
In order to determine whether SAGs collected in LSD 2.0 really affect leaf senescence process, T-DNA insertion lines were selected using the SIGnaL database (http://signal.salk.edu/) and ordered from ABRC (4). If multiple insertions were available in the same genes, the selection was based on the position of the insertions that disrupt the gene function as much as possible, such as those insertions located within exon (4). Some RNAi lines were generated by ourselves or obtained from other laboratories for further study if there was no suitable insertion line available from the Salk collections. In the previous version, phenotypes were described by text only (5). In the updated version, more than 100 pieces of phenotypic information of Arabidopsis mutants were added. As shown in Figure 2A, transgenic plants overexpressing EIN2 (12), a senescence-associated gene, exhibited early flowering and early senescence phenotype compared with ein2-5 mutant. Previous studies demonstrate that loss-of-function of EIN2 delay leaf senescence process (13), suggesting that EIN2 is a positive regulator of senescence (11), which is further supported by our results (Figure 2A).
Figure 2.
Added information for the Arabidopsis EIN2 (AT5G03280) gene in LSD 2.0. (A) Mutant image, (B) Subcellular localization and (C) primer sequences.
Newly added annotations
To help researchers easily and high-throughput study the function of SAGs, Arabidopsis seed information obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org/) was integrated into LSD. In addition, subcellular localization information of SAGs in Arabidopsis mined from literature or generated from the SUBA3 program (http://suba.plantenergy.uwa.edu.au/) (14) was added (Figure 2B). Leaf senescence is an essential developmental process that dramatically impacts on crop yields and qualities. So, in the updated version, QTL information, which was linked to the original database, was added to help scientists to study leaf senescence, one of agronomic traits, in crop, such as rice, maize and sorghum.
Design and experimental evaluation of the primers
To help researchers with high-throughput study of the expression level of SAG genes, primer sequences retrieved based on experimental evidence or designed by array designer 4 were presented (Figure 2C). To facilitate the conduction of multiple PCRs, all primers are designed to have similar properties. All primers are 19–23 nt long, with a preferred length of 21 residues. This is long enough to permit generation of gene-specific primers, while reducing the potential for cross-reactivity and allowing cost-effective generation of large primer sets. The GC contents are also similar (35–65%) to ensure uniform priming. Since PCR efficiency is decreased for very long amplicons, only short amplicons of 150–450 bp are considered during primer selection.
To evaluate the quality of the primers designed, 15 primer pairs (nine for Arabidopsis and six for rice) were tested in conventional RT–PCR experiments (Figure 3). The genes were chosen because they had been shown to be expressed specifically (SAG12) or preferentially (e.g. SAG13, WRKY75 and WRKY53) in senescent leaves as reported previously (4,15). All 15 PCRs resulted in single specific amplicons, determined by gel electrophoresis (Figure 3).
Figure 3.
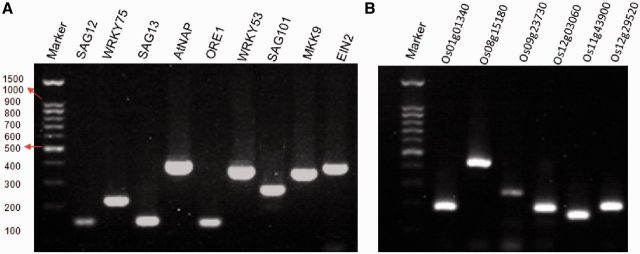
Gel electrophoresis of PCR products. (A) PCR amplifications of nine SAGs in Arabidopsis thaliana. (B) PCR amplifications of six SAGs in rice. Marker, 100-bp DNA ladder.
DICUSSION AND FUTURE PLANS
LSD is a useful resource for leaf senescence study
In recent years, genetic and genome-wide studies of leaf senescence have generated a wealth of information and led to the identification of SAGs (16). Development of the LSD with wide-spread collection and systematic annotation of SAGs provides a useful resource for the investigation of the molecular aspects of leaf senescence (4,5). After the first release in 2010, LSD has been used for comparative studies in several important crops, such as maize and wheat. For example, Sekhon et al. have annotated the maize orthologs and paralogs of the SAGs and identified 1618 genes belonging to 615 orthologous groups (17). Comparison of 815 differentially regulated genes in wheat during senescence with SAGs in LSD showed significant similarities for 181 genes (18). Functional study of some of SAGs in LSD found that transcription factors, WRKY75 and AZF2, are positive regulator of leaf senescence, while AtMKP2 delays senescence process in Arabidopsis (4).
Update and improvement of LSD in the future
In the updated version of LSD, thousands of SAGs and hundreds of mutants were added. New features including primer sequences, subcellular localization information, Arabidopsis seed information, images of Arabidopsis mutants and QTL information were also included in the current version. We will continue on this project to make further updates and improvements of LSD in the future. (i) More phenotypic information such as images and descriptions will be added. Currently, more than 2000 T-DNA homology lines of SAGs in Arabidopsis are available in our institute, and senescence phenotypic information will be collected and added into the database in the near future. In addition, we are constructing transgenic lines overexpressing SAGs in the model plant Arabidopsis thaliana and will provide phenotype information in LSD. For example, overexpression of WRKY75, a positive regulator of senescence (4), accelerates leaf senescence in Arabidopsis (Figure 4); (ii) Subcellular localization information and primer sequences of SAGs in all species will be provided; (iii) new SAGs and mutants will be added once they are identified and available and (iv) we will improve the user interface according to the suggestions and comments from the user community. We hope that LSD can be a useful platform for the research community of leaf senescence worldwide.
Figure 4.

Overexpression of WRKY75 accelerates Arabidopsis leaf senescence. Arabidopsis thaliana wild-type (WT) and WRKY75ox transgenic lines (#1 and #5) grown in soil under long-day growth conditions (16 h-light/8 h-dark) at 22°C for up to 40 days were photographed.
ACKNOWLEDGEMENTS
The authors thank members of GuoLab for kindly sharing Arabidopsis seeds. The authors would like to thank the Arabidopsis Biological Resource Center (ABRC) for propagating the T-DNA mutant lines.
FUNDING
Ministry of Science and Technology of China [2009CB119101 to H.G.]; Ministry of Agriculture of China [2010ZX08010-002 to H.G.]; Natural Science Foundation of China [31071160 to J.L.]; China Postdoctoral Science Foundation [2012M520108 and 2013T60031] and The Postdoctoral Fellowship at Peking-Tsinghua Center for Life Sciences (CLS) (to Z.L.). Funding for open access charge: The 111 Project of Peking University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu. Rev. Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 2.Guo Y, Gan S. Leaf senescence: signals, execution, and regulation. Curr. Top. Dev. Biol. 2005;71:83–112. doi: 10.1016/S0070-2153(05)71003-6. [DOI] [PubMed] [Google Scholar]
- 3.Nam HG. The molecular genetic analysis of leaf senescence. Curr. Opin. Biotechnol. 1997;8:200–207. doi: 10.1016/s0958-1669(97)80103-6. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Peng J, Wen X, Guo H. Gene network analysis and functional studies of senescence-associated genes reveal novel regulators of Arabidopsis leaf senescence. J. Integr. Plant Biol. 2012;54:526–539. doi: 10.1111/j.1744-7909.2012.01136.x. [DOI] [PubMed] [Google Scholar]
- 5.Liu X, Li Z, Jiang Z, Zhao Y, Peng J, Jin J, Guo H, Luo J. LSD: a leaf senescence database. Nucleic Acids Res. 2011;39:D1103–D1107. doi: 10.1093/nar/gkq1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang K, Gan SS. An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiol. 2012;158:961–969. doi: 10.1104/pp.111.190876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Saint Paul V, Zhang W, Kanawati B, Geist B, Faus-Kessler T, Schmitt-Kopplin P, Schaffner AR. The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell. 2011;23:4124–4145. doi: 10.1105/tpc.111.088443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell. 2011;23:873–894. doi: 10.1105/tpc.111.083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y, Gan SS. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 2012;35:644–655. doi: 10.1111/j.1365-3040.2011.02442.x. [DOI] [PubMed] [Google Scholar]
- 10.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 11.Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science. 2009;323:1053–1057. doi: 10.1126/science.1166386. [DOI] [PubMed] [Google Scholar]
- 12.Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, Jiang L, Guo H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012;22:1613–1616. doi: 10.1038/cr.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh SA, Park JH, Lee GI, Paek KH, Park SK, Nam HG. Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. 1997;12:527–535. doi: 10.1046/j.1365-313x.1997.00527.x. [DOI] [PubMed] [Google Scholar]
- 14.Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA. SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res. 2013;41:D1185–D1191. doi: 10.1093/nar/gks1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miao Y, Zentgraf U. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell. 2007;19:819–830. doi: 10.1105/tpc.106.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gepstein S. Leaf senescence–not just a ‘wear and tear’ phenomenon. Genome Biol. 2004;5:212. doi: 10.1186/gb-2004-5-3-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekhon RS, Childs KL, Santoro N, Foster CE, Buell CR, de Leon N, Kaeppler SM. Transcriptional and metabolic analysis of senescence induced by preventing pollination in maize. Plant Physiol. 2012;159:1730–1744. doi: 10.1104/pp.112.199224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantu D, Pearce SP, Distelfeld A, Christiansen MW, Uauy C, Akhunov E, Fahima T, Dubcovsky J. Effect of the down-regulation of the high Grain Protein Content (GPC) genes on the wheat transcriptome during monocarpic senescence. BMC Genomics. 2011;12:492. doi: 10.1186/1471-2164-12-492. [DOI] [PMC free article] [PubMed] [Google Scholar]