Abstract
Virus Variation (http://www.ncbi.nlm.nih.gov/genomes/VirusVariation/) is a comprehensive, web-based resource designed to support the retrieval and display of large virus sequence datasets. The resource includes a value added database, a specialized search interface and a suite of sequence data displays. Virus-specific sequence annotation and database loading pipelines produce consistent protein and gene annotation and capture sequence descriptors from sequence records then map these metadata to a controlled vocabulary. The database supports a metadata driven, web-based search interface where sequences can be selected using a variety of biological and clinical criteria. Retrieved sequences can then be downloaded in a variety of formats or analyzed using a suite of tools and displays. Over the past 2 years, the pre-existing influenza and Dengue virus resources have been combined into a single construct and West Nile virus added to the resultant resource. A number of improvements were incorporated into the sequence annotation and database loading pipelines, and the virus-specific search interfaces were updated to support more advanced functions. Several new features have also been added to the sequence download options, and a new multiple sequence alignment viewer has been incorporated into the resource tool set. Together these enhancements should support enhanced usability and the inclusion of new viruses in the future.
INTRODUCTION
‘So many sequences and yet, so little metadata’ might as well be the official slogan to the dawn of the sequencing age. Often sequence source descriptors such as host, isolation place and time, and other metadata are missing from International Nucleotide Sequence Database Collaboration (INSDC) (1) sequence records. Though metadata can sometimes be inferred from information found within the sequence record or found in the text of a research article, associating this derived metadata with the original sequence record is difficult in practice. Even when metadata are readily available, without universally accepted standards, varied but synonymous terms can hinder retrieval of relevant sequences from public database searches. Lack of data standardization extends beyond metadata and sequence annotations are often inconsistent, with the same protein annotated in different ways among different sequence records—a major impediment to sequence analysis.
Of course metadata and sequence annotation standards are but the tip of the iceberg. With so many sequences now available in public databases, under the best of circumstances, database queries often produce very large datasets, forcing the user to weed through pages of traditional text based displays. Indeed, one could argue that the explosion of sequence data now threatens to blow up traditional models of data storage, retrieval and display. This realization and the argument that such broad issues require equally broad solutions led to the development of the NCBI Virus Variation Resource (http://www.ncbi.nlm.nih.gov/genomes/VirusVariation/) (2). This comprehensive, value added web resource includes three elements—a specialized database, a unique search interface and a suite of tools and displays—all designed to support large sequence datasets.
VIRUS VARIATION 2.0
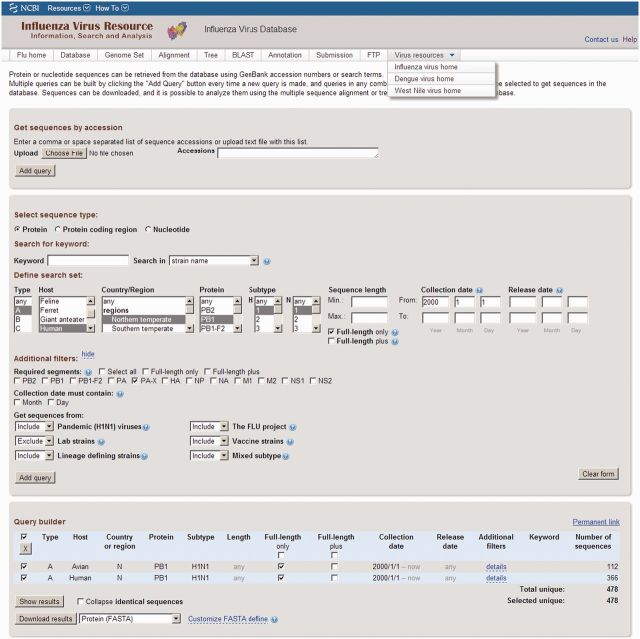
The current Virus Variation Resource is an outgrowth of the NCBI Influenza Virus Resource created in 2004 (3) in response to the National Institute of Allergy and Infectious Diseases (NIAID) Influenza Genome Sequencing Project (4). The resource was initially designed to enhance the usability of very large influenza sequence datasets, and a number of features were introduced to facilitate sequence retrieval. Among these was the development of a metadata driven search interface (Figure 1). Sequence descriptors such as country of isolation, host and protein name are parsed from GenBank (5) records during database loading using advanced strategies. These machine processes are augmented with human curation allowing data found in publications and other sources to be associated with sequences in the database. The resultant metadata are mapped to controlled vocabulary lists, and consistent terms are stored in the database, providing a single term for synonymous and misspelled ones. These metadata terms are then displayed among several menus providing users with a straightforward but comprehensive search interface through which users can retrieve nucleotide and protein sequences based on a number of biological and clinical criteria.
Figure 1.
Influenza virus database search interface. The search interface is shown with the ‘Additional filters’ selection open. A number of search criteria have been selected, and two separate searches added to the ‘Query builder’. Selected search results are indicated by the check box to the left of the ‘Query builder’ display and can be downloaded directly in a variety of formats or loaded into the Virus Variation search results interface by depressing the ‘Show results’ button.
The number of sequences in the database has grown substantially as influenza continues to be a major human pathogen and as surveillance networks and virus sequencing efforts are maintained around the world (6,7). There are now more than 292 000 individual influenza nucleotides sequences in the database, including more than 17 100 complete genome sets. The value added influenza data model was first extended to a separate Dengue virus (DENV) resource in 2009, again in response to NIAID funded genome sequencing efforts (2). DENV is mosquito borne pathogen that is thought to infect as many as 100 million people each year worldwide (8,9), and as attempts to better understand the biology of this Flavivirus have continued (10), the number of DENV sequences in the database has grown to more than 13 000 individual nucleotide sequences. Over the past 2 years, a second mosquito borne Flavivirus, West Nile virus (WNV) has been added to the Virus Variation Resource. WNV is found throughout Africa, the Middle East, southern Europe, Russia, Asia and Australia and has caused 16 196 cases of human neuroinvasive disease and 1549 deaths in the USA since 1999 (11). Moreover, WNV appears endemic to the Americas, Europe and Australia (12), and evidence supports continued WNV evolution in North America over the past decade, underscoring human health concerns (13,14). There are currently 2400 WNV nucleotide sequences in the database.
The design goal of the new Virus Variation construct is to create a resource with a single, value added data model but enough flexibility to accommodate a broad range of viruses. This approach attempts to maintain historic functionalities while leveraging shared backend support to facilitate more efficient data flow. The Virus Variation database loading pipeline is central to the new approach and is responsible for the standardized annotation of incoming nucleotide sequences, automated parsing of metadata terms from GenBank records and mapping parsed terms to a controlled vocabulary. All nucleotide sequences included in Virus Variation are processed in a similar manner. New sequences are retrieved from GenBank, and processed by a standardized set of database loading pipelines. The influenza database loading pipeline simply extracts the existent annotation from INSDC records and loads it into the database.
Influenza coding regions and other sequence features can be systematically annotated prior to INSDC database submission using the Flu Annotation Pipeline (FLAN) (15). This pipeline is publicly available from the Virus Variation web pages and first types (or genotypes) sequences by BLAST alignment to a set of virus-specific nucleotide references and then annotates protein coding regions using reference protein sequence sets specific to each virus subtype (15). Specifically, FLAN maintains a set of reference nucleotide sequences that are used to classify input influenza sequences by type (A, B or C), identify specific segments (1 through 8) and—when applicable—subtype influenza A hemagglutinin and neuraminidase segments (reference sequences available at ftp://ftp.ncbi.nih.gov/genomes/INFLUENZA/ANNOTATION/blastDB.fasta). Corresponding reference protein sets are then aligned to translated input sequences and protein coding regions predicted using the ‘Protein to nucleotide alignment tool’ (ProSplign) (15). The FLAN is continually being updated to support community needs. For example, the Influenza virus annotation tool now supports Influenza C sequences in addition to A and B, and can predict the recently discovered PA-X protein coding sequences.
The annotation pipelines for DENV and WNV are integrated into the database loading pipeline and are very similar to FLAN. The reference nucleotide records and corresponding protein sets used to annotate the two viruses are listed in Table 1. Currently, protein coding regions are extracted from INSDC records and mature peptides annotated by the pipeline and stored in the database. However, this dependency on submitted protein annotations has several shortcomings, not least of which is the inability to update protein annotations in response to evolving biological knowledge, and we are in the process of moving to a fully de novo annotation model. In the new model, all features will be annotated directly by internal pipelines using an improved version of the NCBI ‘Protein to nucleotide alignment tool’ (ProSplign 2). Annotations will also be updated on a regular basis and consistent annotation maintained irrespective of sequence submission dates or changing annotation standards.
Table 1.
DENV and WNV reference sequences
Accessions for nucleotide and protein sequences used in the Virus Variation annotation pipeline are shown. The protein names used on the Virus Variation search pages are shown within parentheses.
NEW FEATURES
A number of new features have been added to Virus Variation since the last published description of the resource. The resource web pages have been updated, including the database search interface (Figure 1). This interface now supports searches using multiple GenBank accessions as well as keyword searches for sequence patterns, strain names/definition lines and influenza drug resistance mutations. Search menus have been updated and support multiple selections, so several proteins, hosts or geographic locations can be added to a single set of search criteria. In the influenza query page there is now the option to select sequences from northern temperate, southern temperate and tropical regions in addition to the country and continent selections used throughout the resource. Searches can also be limited by both collection date and GenBank release date including year, month and day.
Several sets of virus specific filters have been added to the search interfaces to enhance usability. The ‘Full-length genomes only’ filter used in the DENV virus and WNV search interfaces limits retrieved mature protein sequences to those that are part of a complete polyprotein coding sequence (all mature proteins). On the influenza page the ‘Full-length only’ filter limits searches to protein or nucleotide sequences that include a complete coding region, from start codon to stop. A second, ‘Full-length plus’ filter restricts the search to both full-length protein or nucleotide sequences and nearly complete sequences missing only the start and/or stop codons. Complete, nearly complete and partial sequences are marked in search results. A set of ‘Additional filters’ have been added to the influenza query page, and users can now limit searches to those sequences that have a specified day and/or month in the collection date field. Users can also ‘Include’, ‘Exclude’ or ‘Only’ retrieve sequences from WHO recommended ‘Vaccine strains’, pandemic (H1N1) 2009 viruses, sequence sets with ‘Mixed subtypes’, ‘Lineage defining strains’ of well-defined lineages/clades. Currently, virus prototypes include those for the Victoria and Yamagata lineages of influenza B viruses, and the H5N1 and H9N2 subtypes of influenza A viruses. The ‘Required segments’ filter limits retrieved sequences to those where all the selected segments of the same virus isolate exist in the database.
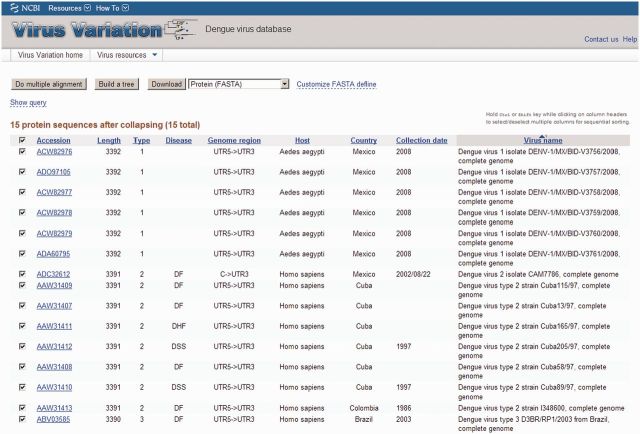
The Virus Variation search interface allows the user to build complicated datasets containing sequences retrieved using different criteria. To do this, the results from each individual database search are added to the ‘Query builder’ section at the bottom of the search interface (Figure 1), then one or more search sets selected for display on the Virus Variation search result page or direct download. The search result page displays sequences retrieved from search sets along with several sortable metadata columns and supports selection of individual sequences for download or further analysis (Figure 2). Identical sequences can be collapsed in the search results and represented by the oldest sequence in the group. Results can be downloaded as a table in XML, CSV or tab-delimited formats, or users can also download a GenBank accession list or FASTA file of selected sequences. The definition line of FASTA sequences can now be customized in the downloaded files, and users can replace original GenBank definition lines with a number of fields including host, country, date, serotype, patient age or gender, viral mutations and CDS location.
Figure 2.
Virus Variation search results interface. Results from a DENV database query are shown. The display includes a number of retrieved sequence descriptors including Accession, Length, Type, Disease, Genome region, Host, Country, Collection date and Virus name. Sequences can be selected and downloaded in a number of formats or used to construct an alignment or phylogenetic tree.
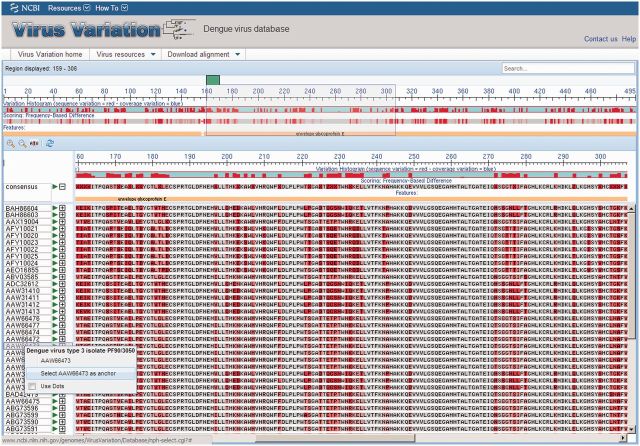
The resource sequence analysis tool set has been improved to enhance visualization of large datasets and facilitate discovery activities. A new multiple sequence alignment viewer (Figure 3) has been integrated into DENV virus and WNV resources and will soon be available for influenza virus. This tool is based on the NCBI Genome Workbench multiple sequence alignment viewer and includes a variation histogram above the alignment as well as a feature table that highlights mature protein boundaries and other important sequence features. There are a number of usability features integrated into the viewer such as selectable alignment scoring methods for individual nucleotides/amino acid residues, link outs to associated GenBank records and selectable alignment anchor sequence—either consensus or any sequence in the alignment. Alignments displayed in the viewer can also be downloaded in FASTA, Clustal, Phylip and Nexus formats for use locally or with other tools. The Virus Variation tree builder tool (16) has also been updated for all viruses, and GenBank accession numbers can be downloaded through the tree builder tool by selecting the branch of interest on the tree.
Figure 3.
Virus Variation multi-sequence alignment viewer. The results from a DENV database query were aligned and displayed in the Virus Variation multi-sequence alignment viewer. The top section of the alignment viewer includes a histogram that displays sequence and coverage variation across the alignment, a second histogram that plots the frequency of sequence differences with a shading scheme and highlights insertions and deletions with gaps and a feature table where protein names and other sequence feature identifiers are displayed. The alignment position is indicated above the histogram, and the region displayed in the lower section is highlighted by a gray box. The lower section displays the highlighted region in greater detail by default, but the magnification can be decreased or increased as desired by the user. Alignments are anchored to the consensus sequence by default, but any sequence can be selected as an anchor. Sequences identical to the consensus can be displayed as individual nucleotides or amino acids or replaced with dots—highlighting variations from the consensus.
FUTURE DIRECTIONS
The long term plan is to increase the coverage of virus sequences in the Virus Variation Resource. The flexibility of the resource should support a number of diverse viral pathogens and provide consistently annotated sequence datasets with standardized isolate descriptors. This will require continued tweaking of metadata parsing strategies and development of new virus-specific sequence annotation modules. As these annotation modules are added to our core pipeline for use by the resource, they will be made publicly available. We will also explore approaches to increase user outreach and leverage community knowledge to improve data curation, reference sequence assignment and resource usability.
FUNDING
This research was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine. Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nakamura Y, Cochrane G, Karsch-Mizrachi I. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2013;41:D21–D24. doi: 10.1093/nar/gks1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Resch W, Zaslavsky L, Kiryutin B, Rozanov M, Bao Y, Tatusova TA. Virus variation resources at the National Center for Biotechnology Information: dengue virus. BMC Microbiol. 2009;9:65. doi: 10.1186/1471-2180-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. The influenza virus resource at the National Center for Biotechnology Information. J. Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fauci AS. Race against time. Nature. 2005;435:423–424. doi: 10.1038/435423a. [DOI] [PubMed] [Google Scholar]
- 5.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briand S, Mounts A, Chamberland M. Challenges of global surveillance during an influenza pandemic. Public Health. 2011;125:247–256. doi: 10.1016/j.puhe.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Back AT, Lundkvist A. Dengue viruses—an overview. Infect. Ecol. Epidemiol. 2013;3:19839. doi: 10.3402/iee.v3i0.19839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allicock OM, Lemey P, Tatem AJ, Pybus OG, Bennett SN, Mueller BA, Suchard MA, Foster JE, Rambaut A, Carrington CV. Phylogeography and population dynamics of dengue viruses in the Americas. Mol. Biol. Evol. 2012;29:1533–1543. doi: 10.1093/molbev/msr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen LR, Brault AC, Nasci RS. West Nile virus: review of the literature. JAMA. 2013;310:308–315. doi: 10.1001/jama.2013.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pesko KN, Ebel GD. West Nile virus population genetics and evolution. Infect. Genet. Evol. 2012;12:181–190. doi: 10.1016/j.meegid.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann BR, McMullen AR, Swetnam DM, Barrett AD. Molecular epidemiology and evolution of West Nile virus in North America. Int. J. Environ. Res. Public Health. 2013;10:5111–5129. doi: 10.3390/ijerph10105111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mann BR, McMullen AR, Swetnam DM, Salvato V, Reyna M, Guzman H, Bueno R, Jr, Dennett JA, Tesh RB, Barrett AD. Continued evolution of West Nile virus, Houston, Texas, USA, 2002-2012. Emerg. Infect. Dis. 2013;19:1418–1427. doi: 10.3201/eid1909.130377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Tatusova T. FLAN: a web server for influenza virus genome annotation. Nucleic Acids Res. 2007;35:W280–W284. doi: 10.1093/nar/gkm354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaslavsky L, Bao Y, Tatusova TA. Visualization of large influenza virus sequence datasets using adaptively aggregated trees with sampling-based subscale representation. BMC Bioinformatics. 2008;9:237. doi: 10.1186/1471-2105-9-237. [DOI] [PMC free article] [PubMed] [Google Scholar]