Abstract
The Consensus Coding Sequence (CCDS) project (http://www.ncbi.nlm.nih.gov/CCDS/) is a collaborative effort to maintain a dataset of protein-coding regions that are identically annotated on the human and mouse reference genome assemblies by the National Center for Biotechnology Information (NCBI) and Ensembl genome annotation pipelines. Identical annotations that pass quality assurance tests are tracked with a stable identifier (CCDS ID). Members of the collaboration, who are from NCBI, the Wellcome Trust Sanger Institute and the University of California Santa Cruz, provide coordinated and continuous review of the dataset to ensure high-quality CCDS representations. We describe here the current status and recent growth in the CCDS dataset, as well as recent changes to the CCDS web and FTP sites. These changes include more explicit reporting about the NCBI and Ensembl annotation releases being compared, new search and display options, the addition of biologically descriptive information and our approach to representing genes for which support evidence is incomplete. We also present a summary of recent and future curation targets.
INTRODUCTION
Biological and biomedical research has come to rely on accurate and consistent annotation of genes and their products on genome assemblies. High-quality genome assemblies, such as the human and mouse reference genome assemblies that are maintained by the Genome Reference Consortium (GRC) (1), are particularly amenable to high-definition gene annotation. Reference annotation of these genomes is available from various sources, including the National Center for Biotechnology Information (NCBI) (2), Ensembl (3), the Vertebrate Genome Annotation database (Vega) (4) and GENCODEgenes.org. Each annotation group has independent goals and policies, which result in some annotation variation. Nevertheless, much of the annotation provided by these individual groups is identical, thus providing users with a higher degree of confidence in the accuracy of these annotations.
The Consensus Coding Sequence (CCDS) project (5) has been established to identify a gold standard set of protein-coding gene annotations that are identically annotated on the human and mouse reference genome assemblies by the participating annotation groups. The participating members are from NCBI, the European Bioinformatics Institute (EBI), the Wellcome Trust Sanger Institute (WTSI) and the University of California Santa Cruz (UCSC). Manual annotation is provided by the Reference Sequence (RefSeq) group at NCBI (2) and by the Human and Vertebrate Analysis and Annotation (HAVANA) group at WTSI (4). A combination of manual and automated genome annotations provided by NCBI and Ensembl (which incorporates manual HAVANA annotations) are compared to identify annotations with matching genomic coordinates; NCBI and Ensembl-coding region annotations must be identical at the CDS start and stop codons and at every splice site to be considered for the CCDS database. Each matching annotation is assigned a unique identifier known as a CCDS ID, which is tracked and reported in the database. Matching CCDS sequences and related metadata are available to users via a public FTP site (ftp://ftp.ncbi.nlm.nih.gov/pub/CCDS/) or a web-based interface (www.ncbi.nlm.nih.gov/CCDS/) that includes individual report pages for each CCDS ID. Information on how to access CCDS data was described previously (5), including details on CCDS report page features and how to access CCDS data tracks on various genome browsers. In addition, the README file (ftp://ftp.ncbi.nlm.nih.gov/pub/CCDS/README) on the FTP site provides full descriptions of the various download files that are available.
CCDS matching annotations indicate concordance between different annotation groups with diverse policies, and they thus represent high-quality annotations that can be used as standards. The integrity of the CCDS dataset is maintained through stringent quality assurance (QA) testing and on-going manual curation (6). QA testing includes checks for possible conflicts within the coding sequence and its annotated structure, protein-coding potential, possible problems with the genome sequence, and assessing the quality of matched NCBI and Ensembl annotation. Curatorial updates to existing CCDS IDs require unanimous agreement by all collaborators. A process flow has been established to facilitate review of CCDS IDs that have been flagged by any member of the collaboration for update or withdrawal, with the voting members for curation updates being RefSeq, HAVANA and UCSC. The CCDS dataset is an integral part of the GENCODE gene annotation project (7) and it is used as a standard for high-quality coding exon definition in various research fields, including clinical studies (8,9), large-scale epigenomic studies (10), exome projects (11) and exon array design (12). Due to the consensus annotation of CCDS exons by the independent annotation groups, exome projects in particular have regarded CCDS coding exons as reliable targets for downstream studies (e.g. for single nucleotide variant detection), and these exons have been used as coding region targets in commercially available exome kits (12).
GROWTH OF THE CCDS DATASET
The CCDS datasets for human and mouse are periodically reanalyzed and publicly distributed as CCDS releases (Table 1). Historically, CCDS releases occurred following coordinated whole-genome annotation runs by both NCBI and Ensembl. This policy has been changed to allow CCDS updates on a more regular basis following annotation updates by either NCBI or Ensembl. Human updates are expected to occur roughly every 6 months and mouse releases will be yearly. Additional CCDS updates will occur for both species following assembly updates at which time we still require both groups to have generated updated annotation on the new assembly. Reducing the time interval between releases allows for the CCDS dataset to represent more updated annotations. However, because genomic annotation is updated by NCBI and Ensembl at different times, CCDS content may not yet reflect the most recent manual annotation curation efforts. Figure 1A indicates that the number of CCDS IDs for both human and mouse continues to increase with each new CCDS analysis. Since 2011, the human and mouse CCDS dataset sizes have increased by 1279 and 906 CCDS IDs, respectively.
Table 1.
CCDS release information for human and mouse
| Species | CCDS release | NCBI annotation release | Ensembl annotation release | Assembly name | Assembly ID | CCDS release date |
|---|---|---|---|---|---|---|
| Homo sapiens | 1 | 35.1 | 23 | NCBI35 | GCF_000001405.11 | 02-03-2005 |
| 3 | 36.2 | 41 | NCBI36 | GCF_000001405.12 | 26-02-2007 | |
| 5 | 36.3 | 47 | NCBI36 | GCF_000001405.12 | 30-04-2008 | |
| 6 | 37.1 | 55 | GRCh37 | GCF_000001405.13 | 02-09-2009 | |
| 8 | 37.2 | 62 | GRCh37.p2 | GCF_000001405.14 | 20-04-2011 | |
| 9 | 37.3 | 64 | GRCh37.p5 | GCF_000001405.17 | 07-09-2011 | |
| 11 | 103 | 68 | GRCh37.p9 | GCF_000001405.21 | 25-10-2012 | |
| 12 | 104 | 71 | GRCh37.p10 | GCF_000001405.22 | 30-04-2013 | |
| Mus musculus | 2 | 36.1 | 39 | MGSCv36 | GCF_000001635.15 | 10-10-2006 |
| 4 | 37.1 | 47 | MGSCv37 | GCF_000001635.16 | 28-11-2007 | |
| 7 | 37.2 | 61 | MGSCv37 | GCF_000001635.18 | 24-01-2011 | |
| 10 | 38.1 | 68 | GRCm38 | GCF_000001635.20 | 14-08-2012 | |
| 13 | 103 | 72 | GRCm38.p1 | GCF_000001635.21 | 05-08-2013 |
Figure 1.
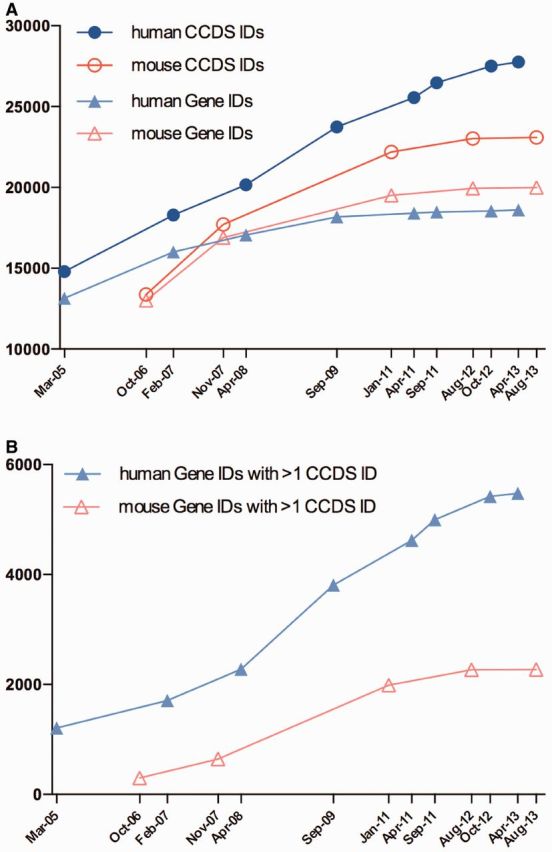
CCDS release statistics for human and mouse. The Y-axis indicates counts of CCDS IDs or Gene IDs and the X-axis shows CCDS release dates. (A) Growth in the number of CCDS IDs at each release date (Table 1) compared with the number of Gene IDs with at least one protein isoform in the CCDS dataset. (B) Growth in the number of Gene IDs with more than one protein isoform in the CCDS dataset. All data used to generate the graphs are available in the CCDS Releases and Statistics page (http://www.ncbi.nlm.nih.gov/CCDS/CcdsBrowse.cgi?REQUEST=SHOW_STATISTICS) on the website.
Most of the recent growth in the CCDS dataset comes from an increase in the number of genes with more than one splice variant obtaining a CCDS ID. Since 2011, the number of human genes in the CCDS database increased by 135, whereas the number of human genes having more than one splice variant with a CCDS ID increased by 479. A similar pattern was observed in mouse (Figure 1B), bringing the total number of genes with more than one CCDS ID to 7752, of which over 100 have more than six CCDS IDs. Therefore, the CCDS dataset is increasingly representing more alternative splicing events with each new release. The CCDS dataset is expected to continue to grow in the next few years as a result of targeted curation initiatives (see below), which will allow for an increase in the representation of protein-coding genes and more protein isoforms per gene. Also, as both curation groups begin to integrate RNAseq data available from the Illumina Human Body Map 2.0 project (NCBI GEO accession GSE30611), ENCODE (13) and other sources such as GTEx data (14) into their pipelines, the number of alternatively spliced transcripts will increase.
NEW FEATURES
The CCDS database has incorporated several new features to improve user browsing, to add useful biological information that was either previously internal or not categorized, and to increase gene representation in the dataset.
Updated reporting of CCDS and annotation release information
The CCDS web resource and FTP site now specifies which NCBI and Ensembl annotation releases were compared and which reference genome assembly was used for each CCDS release. Previously, each CCDS release was named after an ‘NCBI Build’ that included a genome assembly ‘build’ identifier followed by a ‘.version’ qualifier to indicate the NCBI annotation run of that genome assembly, e.g. human Build 37.2 was the second NCBI annotation run on the GRCh37 reference genome assembly (GRCh37.p2 assembly release). However, the ‘build’ terminology was confusing because it was suggestive of a new reference genome assembly, and it did not convey which Ensembl annotation release was used for comparison to NCBI annotation. The NCBI genome annotation pipeline (http://www.ncbi.nlm.nih.gov/genome/annotation_euk/process/) has therefore dispensed with ‘build’ terminology for NCBI annotation releases, and it now refers to them based on a three-digit identifier that completely dissociates the annotation run from the assembly name (e.g. Homo sapiens Annotation Release 104), and which will continue to be incremented for subsequent annotation releases for each species. In addition to reporting the NCBI and Ensembl release numbers, we also report a CCDS release number which increments sequentially with each human or mouse update (Table 1). The CCDS release number may be cited in publications that refer to the use of a specific time snapshot of the CCDS dataset. A tabular report of the CCDS release numbers and dates, and the corresponding NCBI and Ensembl annotation release numbers, is available on the ‘CCDS Releases and Statistics’ page (http://www.ncbi.nlm.nih.gov/CCDS/CcdsBrowse.cgi?REQUEST=SHOW_STATISTICS). In addition, the official reference genome assembly names and accession.version numbers from NCBI’s Assembly database (http://www.ncbi.nlm.nih.gov/assembly/) are provided. This information is also reported in the ‘BuildInfo’ FTP files (ftp://ftp.ncbi.nlm.nih.gov/pub/CCDS). All coordinates reported for each CCDS ID in either the FTP site or individual report pages are based on that particular assembly for each particular CCDS release. The reference genome assembly name and a link to the associated assembly accession has also been added to individual CCDS report pages in the ‘Chromosomal Locations’ section. Furthermore, all search result and CCDS report pages have been updated to reflect the updated terminology and to include both the NCBI and Ensembl annotation release information.
New attribute section on CCDS report pages
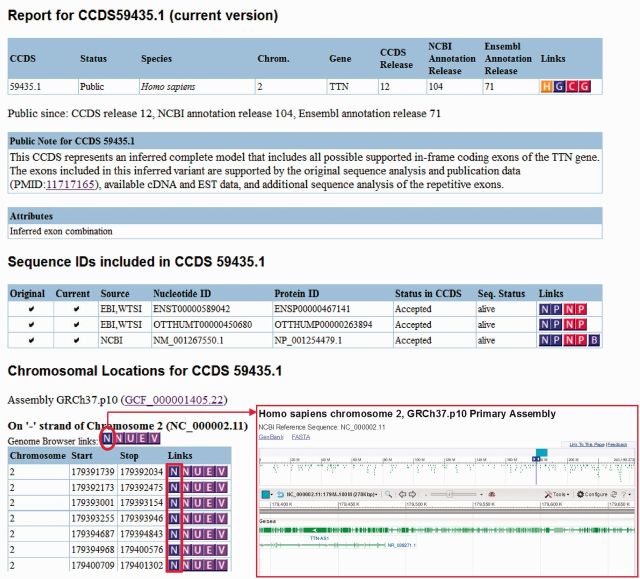
Some coding sequences have special characteristics that may represent biologically valid exceptions to general rules. For instance, the annotated start codon may not be the first one that is in-frame with the remaining coding sequence, or it may not be a canonical AUG start codon. Such phenomena are found during the process of curation or are identified by QA testing. Curators from each group review associated supporting data to determine if the particular characteristic is valid for the relevant coding sequence. Annotation decisions may be based on in-depth review of transcript and homology evidence, publication data or joint CCDS collaborator guidelines, and may also involve extensive collaborator discussion, as described previously (6). Valid coding sequences, with such special characteristics, are tracked by each annotation group with appropriately tagged attributes and the CCDS database now displays associated attribute information in relevant CCDS report pages. The attributes are displayed in a separate area between the ‘Public Note’ and ‘Sequence IDs’ sections of CCDS reports, as shown for CCDS59435 in Figure 2. The types of attributes displayed, together with a description and an example of each, are shown in Table 2. Similar attributes have also been added to the NCBI Nucleotide flat files for the RefSeq accessions associated with each tagged CCDS ID, e.g. NM_003376.5 associated with CCDS4907 has the ‘Non-AUG initiation codon’ attribute and these attributes are also flagged in the Vega database using the same or similar phrasing. The CCDS FTP site includes human and mouse supplemental files reporting attributes associated with CCDS IDs.
Figure 2.
CCDS database screenshot showing the partial report page for CCDS59435. This CCDS ID is associated with the ‘Inferred exon combination’ attribute, as explained in the Public Note. The exon locations table is partially visible in the Chromosomal Locations section. The blue ‘N’ icon circled in red links to a graphical view of the entire genomic region in NCBI’s Nucleotide database (boxed inset image). The blue ‘N’ icons boxed in red link to the sequences of each individual exon in the Nucleotide database.
Table 2.
Attribute types currently found in CCDS reports
| Attribute | Description | Counta | CCDS example |
|---|---|---|---|
| CDS uses downstream AUG | The annotated start codon is not the first AUG found in-frame with the CDS | 482 | 816.1 |
| Contains selenocysteine | A UGA codon encodes a selenocysteine residue instead of resulting in translation termination | 56 | 42 457.1 |
| Inferred exon combination | The CDS exon combination lacks full-length transcript support in INSDC databases (15) | 160 | 44 873.1 |
| NonAUG initiation codon | The annotated start codon is not AUG | 73 | 4907.2 |
| Nonsense-mediated decay (NMD) candidate | The transcript may escape NMD (24) and produce a protein | 152 | 37 108.1 |
| Ribosomal slippage (translational frameshift) | The CDS contains an experimentally verified translational frameshift due to ribosomal slippage | 5 | 58 639.1 |
aCounts reflect data available as of 30 August 2013.
Inclusion of inferred CCDS representations
A major goal of the CCDS collaboration is to represent only high quality and supported coding sequences in the dataset. Ideally, this means that every CCDS representation should include full-length transcript support for the CDS exon combination, in addition to other evidence such as conservation, functional data or predicted domain structure in the protein. In practice, however, many valid transcript variants or very long proteins lack full-length transcript support, as available in public International Nucleotide Sequence Database Collaboration (INSDC) databases (15). As a result of continuous collaborator review, some CCDS IDs were withdrawn due to a lack of full-length transcript support. Not only has this resulted in the withdrawal of alternatively spliced transcript variants for certain genes, but it has also resulted in the complete loss of CCDS representation for some genes that are otherwise known to be protein coding.
In order to fulfill the CCDS project’s goal to represent as many consistently annotated protein-coding genes as possible, the collaboration has recently revised the policy requiring full-length transcript support for the CDS region and now includes inferred exon combination representations in the dataset. The CCDS web reports of such representations are flagged with the ‘Inferred exon combination’ attribute described in Table 2. This attribute is typically used in two scenarios: (i) when a known protein-coding gene lacks full-length transcript support but a full-length protein can be inferred from partial transcript and/or homology, orthology or publication data, e.g. CCDS44873 representing the KMT2D gene; or (ii) when the gene contains cassette exons or multi-exon cassettes that are individually supported (by transcript, conservation or publication data) but where full-length transcript support for a complete complement of exons is lacking. For example, CCDS59435 represents the longest possible full-length splice variant of the TTN gene (Figure 2) with an exon combination that is not supported by any single transcript.
Updated graphical linking for the full genomic span of CCDS representations
CCDS report pages contain a ‘Chromosomal Locations’ section that includes a list of CDS exons together with the genomic coordinates for each exon, and with links to various genome browsers (purple icons; see Figure 2). The browser links within the exon table display each exon region separately, whereas the links found in the ‘Genome Browser links:’ line on top of the table link to the entire genomic span of the coding sequence. To increase user options, a link has been added to display either the full genomic span of the CDS via a graphical view from NCBI’s Nucleotide database (blue ‘N’ icon on top of the exon table; Figure 2 inset image), or to display the sequence of each exon in the Nucleotide database (blue ‘N’ icons within the table).
Updated database searching
The main search field area of the CCDS database supports various searching by CCDS ID, gene symbol, RefSeq accession (with the prefix NM_ or NP_) or gene, transcript or protein IDs provided by Ensembl (ENS) and HAVANA (OTT). A new feature is the ability to search the Nucleotide or Protein fields using an NCBI sequence GI (16) in addition to the NCBI accession number (or accession.version). NCBI provides a new GI identifier for any sequence change, and therefore, searching with a GI facilitates the rapid retrieval of a CCDS report for a specific sequence. In addition, the superseded term ‘build’, as described above, has been replaced with ‘release’. Users can now search the database by ‘All Releases’ or by ‘Current Releases’. We are planning to add support to search for CCDS attributes in the near future.
CURATION STATUS
Since the representation of all known protein-coding genes in the dataset is a priority for the CCDS project, the collaboration attempts to include at least one CCDS representation for each gene. Some genes continue to be omitted due to uncertainty about the gene type (protein-coding versus pseudogene or long noncoding RNA), insufficient supporting evidence or concerns about the genomic sequence such as assembly gaps, sequence errors, insertion or deletion polymorphisms or nonsense polymorphisms. Curation of human genes continues to be a focus for the CCDS collaboration; curation guidelines and challenges were previously reported (6). Members of the collaboration also report sequence quality concerns to the GRC (1), track protein-coding loci that lack any CCDS ID due to an assembly or sequence quality issue, review GRC sequence updates that affect those reported genes, and review and update CCDS proteins, when needed, based on assembly updates released by the GRC. Approximately 170 human protein-coding genes lacked CCDS representation due to genome problems in the GRCh37 chromosome assemblies. The GRC released sequence regions, known as FIX patches, that correct these (and other) identified genome problems for the GRCh37 assembly. However, the CCDS resource currently only tracks chromosome coordinates and thus FIX patch releases are tracked internally as anticipated gains in the CCDS dataset following a GRC assembly update. The upcoming GRCh38 assembly release incorporates FIX patch regions into the primary chromosome accessions. It is therefore expected that most of these missing genes will be correctly annotated by the NCBI and Ensembl annotation pipelines and subsequently gain a CCDS ID. CCDS analysis following an assembly update is dependent on availability of updated annotation from both NCBI and Ensembl and is expected to occur in the second quarter of 2014.
Since summer 2012, monthly web meetings and regular correspondence among the collaborators resulted in improved representation for several difficult human gene regions (e.g. the PILRB complex locus, or the DUX4 cluster on chromosome 4), as well as those genes that have less abundant or primarily partial transcript evidence (e.g. mouse Mphosph9 and human RUNX2). This coordinated curation effort encompassed standardizing annotation for several noncoding RNA genes and pseudogenes, in addition to generating consistent annotation for the protein-coding genes in the regions analyzed. This concerted review effort also resulted in planned reinstatement of some previously withdrawn CCDS IDs, either due to new data that have become available for a gene or because the collaborators can agree on a representative inferred model, e.g. CCDS3700 representing the human C4orf21 gene. The collaboration maintains an internal interface to track genes lacking CCDS representation. In addition, CCDS curators initiate and reply to incoming correspondence with external researchers; we have consulted with outside experts on some inferred models and have coordinated to meet the needs of the clinical variant reporting community by addressing requests made by the Locus Reference Genomic (LRG) project (17) to represent a specific model as a reference standard (e.g. the OBSCN and TTN genes). LRG genomic region records are annotated with mRNA and CDS features, many of which have CCDS IDs, and provide a stable coordinate system for reporting mutations for clinical studies.
The emergence of new data types has also been helpful for making some decisions. For instance, ribosome profiling (RP) data (18) can be used to determine start codon usage and is a useful approach to identify proteins that are likely annotated from the wrong start codon. The CCDS project is currently using published reports of annotation errors (or confirmations of CDS start sites) that have been identified using this new data type as additional evidence for curation decisions. In the case of CCDS2748 representing the human CCDC12 gene, the collaborators recently agreed to update this CCDS to use a downstream in-frame start codon based on RP evidence (19), which did not support the upstream start codon. It was determined that the previous representation likely represented a rarer and more extended 5′ variant. The update uses an internal downstream promoter that is supported by homology data, transcription start site scoring based on CAGE tag densities (20), and promoter-associated epigenomic modifications from the ENCODE project (13). However, while new data types can be helpful for some decisions, they have limitations. For instance, RP can indicate multiple initiation events, sometimes with nonAUG start codons or alternative reading frames scattered throughout the transcript; it is not clear whether a stable protein is produced from each initiation event. RP data are currently only available for a few cell types, and is therefore expected to miss some cell type-specific transcript variants and isoforms. In addition, the data are not yet easily accessible on common genome browser tracks. It is expected that more RP data will become available in the future. Other emerging technologies, such as more robust proteomics data (21), that confirm the existence of the specific protein, or N-terminal protein sequencing, would also provide valuable datasets for CCDS curation.
FUTURE PROSPECTS
Similar to the recent addition of certain attribute types to CCDS reports (Table 2), other attribute types could be propagated to CCDS reports and/or the FTP site in the future. For instance, we are working on expanding the CCDS report pages to include an indication of apparent retrogenes (22) and human:mouse orthologs. We are also working on adding links to the UniProt knowledgebase (23) and information about Swiss-Prot proteins (or specific isoforms) that match the CCDS protein (allowing for polymorphisms). We expect that this addition will be of general interest to users of the CCDS dataset. The manually curated Swiss-Prot subset of UniProtKB includes rich protein feature annotation that is not in scope for the CCDS database. Although the RefSeq project does propagate many features from Swiss-Prot records onto RefSeq protein records, additional information and alternative protein-oriented displays are available on the UniProt website, which may be relevant to those accessing the CCDS web pages. Adding this information to CCDS report pages will support navigation to UniProt records where more information may be available.
We anticipate a continued curation focus for both the mouse and human genomes on gene clusters, readthrough loci, and other gene regions with more complex transcript support. In the effort to provide a CCDS ID for all known or reasonably inferred protein-coding loci, we find that we have been increasingly reviewing loci of an uncertain type (e.g. genes for which the protein-coding capacity is questionable). Both the human and mouse genomes contain a number of uncharacterized genes for which the collaborators have yet to agree upon a gene type. We anticipate an increased focus on this category of data in the next year. While some of these loci may turn out to be long noncoding RNAs or pseudogenes, it is expected that at least some of them may have future CCDS representation, possibly tracked with a qualifying inference or other attribute. Coordinated curation of these uncertain loci by the CCDS collaboration will also improve NCBI and Ensembl genome annotation consistency for the loci not in scope for the CCDS database.
ACKNOWLEDGEMENTS
The authors wish to thank the programmers, database and curation staff at Ensembl, NCBI, HAVANA and UCSC for their contributions to the CCDS analysis, maintenance and continuing curation efforts. The authors also thank the UniProt Consortium, HGNC and MGI for many useful discussions.
FUNDING
Work performed at NCBI was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine; Work performed at UCSC was funded by the National Human Genome Research Institute (NHGRI) for the ENCODE project [prime award 10U41 HG007234-01 under sub-award 2186-03 from the Wellcome Trust Sanger Institute]; Work performed at WTSI was funded by the Wellcome Trust [WT077198] for HAVANA, and by the Wellcome Trust [WT062023]; National Human Genome Research Institute [5U54HG00455-04] for Ensembl. Funding for open access charge: The Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, Chen HC, Agarwala R, McLaren WM, Ritchie GR, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harte RA, Farrell CM, Loveland JE, Suner MM, Wilming L, Aken B, Barrell D, Frankish A, Wallin C, Searle S, et al. Tracking and coordinating an international curation effort for the CCDS Project. Database J. Biol. Databases Curation. 2012;2012 doi: 10.1093/database/bas008. bas008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dias C, Sincan M, Cherukuri PF, Rupps R, Huang Y, Briemberg H, Selby K, Mullikin JC, Markello TC, Adams DR, et al. An analysis of exome sequencing for diagnostic testing of the genes associated with muscle disease and spastic paraplegia. Hum. Mutat. 2012;33:614–626. doi: 10.1002/humu.22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, Gunderson KL. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 11.Meynert AM, Bicknell LS, Hurles ME, Jackson AP, Taylor MS. Quantifying single nucleotide variant detection sensitivity in exome sequencing. BMC Bioinformatics. 2013;14:195. doi: 10.1186/1471-2105-14-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parla JS, Iossifov I, Grabill I, Spector MS, Kramer M, McCombie WR. A comparative analysis of exome capture. Genome Biol. 2011;12:R97. doi: 10.1186/gb-2011-12-9-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura Y, Cochrane G, Karsch-Mizrachi I. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2013;41:D21–D24. doi: 10.1093/nar/gks1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalgleish R, Flicek P, Cunningham F, Astashyn A, Tully RE, Proctor G, Chen Y, McLaren WM, Larsson P, Vaughan BW, et al. Locus Reference Genomic sequences: an improved basis for describing human DNA variants. Genome Med. 2010;2:24. doi: 10.1186/gm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. Genome-wide annotation and quantitation of translation by ribosome profiling. Curr. Protoc. Mol. Biol. 2013 doi: 10.1002/0471142727.mb0418s103. Chapter 4, Unit 4 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Liu B, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl Acad.Sci. USA. 2012;109:E2424–E2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi H, Lassmann T, Murata M, Carninci P. 5′ end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc. 2012;7:542–561. doi: 10.1038/nprot.2012.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet. 2013;14:35–48. doi: 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]
- 22.Ding W, Lin L, Chen B, Dai J. L1 elements, processed pseudogenes and retrogenes in mammalian genomes. IUBMB Life. 2006;58:677–685. doi: 10.1080/15216540601034856. [DOI] [PubMed] [Google Scholar]
- 23.Uniprot Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]