Abstract
To understand newly sequenced genomes of closely related species, comprehensively curated reference genome databases are becoming increasingly important. We have extended CyanoBase (http://genome.microbedb.jp/cyanobase), a genome database for cyanobacteria, and newly developed RhizoBase (http://genome.microbedb.jp/rhizobase), a genome database for rhizobia, nitrogen-fixing bacteria associated with leguminous plants. Both databases focus on the representation and reusability of reference genome annotations, which are continuously updated by manual curation. Domain experts have extracted names, products and functions of each gene reported in the literature. To ensure effectiveness of this procedure, we developed the TogoAnnotation system offering a web-based user interface and a uniform storage of annotations for the curators of the CyanoBase and RhizoBase databases. The number of references investigated for CyanoBase increased from 2260 in our previous report to 5285, and for RhizoBase, we perused 1216 references. The results of these intensive annotations are displayed on the GeneView pages of each database. Advanced users can also retrieve this information through the representational state transfer-based web application programming interface in an automated manner.
INTRODUCTION
Cyanobacteria constitute a large taxonomic group within the domain of eubacteria. They are widely used as model organisms to study the fundamental aspects of photosynthesis, in basic and applied plant-related research, in biotechnology for the development of third-generation biofuels and for their evolutionary contributions for the whole biosphere. CyanoBase was originally developed as a genome database for Synechocystis sp. PCC 6803, the first cyanobacterial genome sequenced in 1996 (1). CyanoBase subsequently has been extended to include additional cyanobacteria and related species (2–4), covering 39 organisms. Rhizobia, a collective name of the genera Rhizobium, Sinorhizobium, Mesorhizobium and Bradyrhizobium, are agronomically important bacteria because they have the ability to establish nitrogen-fixing symbioses with leguminous plants. RhizoBase was initiated as a genome database for Mesorhizobium loti strain MAFF303099 sequenced in 2000 (5) and was extended to include other rhizobia and related species, encompassing 18 organisms till date.
Regarding CyanoBase and RhizoBase, we have been accumulating gene annotations by incorporating evidence from published data. To maintain the quality of annotations, the involvement of the research communities of cyanobacteria and rhizobia was essential. Therefore, to assist in the submission procedure of new annotations, we developed the TogoAnnotation system (6) and also conducted in-house curation efforts to ensure that annotations are as comprehensive as possible. New sequencing technologies and automatic genome processing pipelines [e.g., MiGAP (7) and DNA Databank of Japan (DDBJ) Pipeline (8,9)] have been certainly accelerating prokaryotic genome analyses. However, it is difficult to estimate the functions of predicted genes without the information from carefully curated reference annotations of model organisms. Thus, for this, the manually curated annotations in CyanoBase and RhizoBase provide fundamental information for the interpretation of high-throughput sequencing data.
Regarding data reusability, it is important to provide a high level of accessibility and interoperability of the reference annotations. For accessibility, CyanoBase and RhizoBase use a common database system to provide the same types of functionalities, user interfaces and application programming interfaces. For interoperability, we have introduced Semantic Web technologies (10) for representing data in a standard format and providing an advanced query interface.
DATA CURATION
Reference genomes
CyanoBase and RhizoBase integrate reference genomes from original genome projects conducted by Kazusa DNA Research Institute and from public sequence databases. By the inclusion of recent genome sequencing projects, we added 4 and 17 new genome entries in CyanoBase and RhizoBase, respectively (4,5). As a result, CyanoBase is extended to currently include 39 completely sequenced genomes, and RhizoBase contains 18 completely sequenced genomes and two partially sequenced genomic regions, such as the symbiosis island (newly incorporated genomes are listed in Supplementary Table S1). We have integrated automatic gene annotations including BLAST and the InterPro search results in the new cyanobacterial and rhizobial genomic databases before the manual curations described in the following sections.
Manual curation
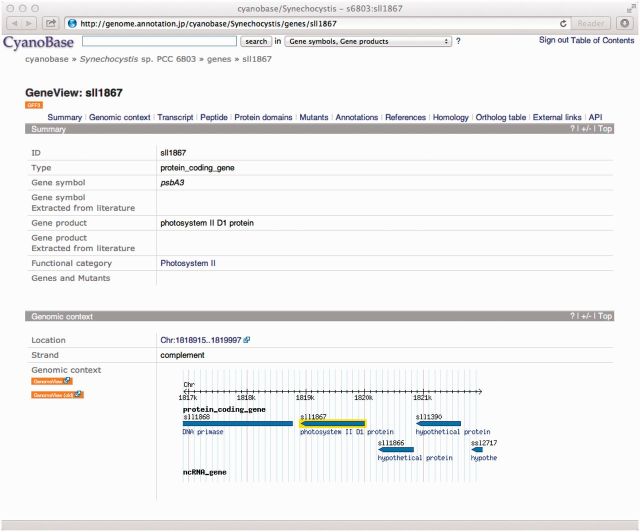
Expert curators extracted gene symbols and full names from full sections of the peer-reviewed research literature and annotated them using the Sequence Ontology (SO) terms (11) to indicate types of annotations. These annotations are immediately reflected in the ‘Extracted from literature’ fields in the ‘Summary’ section of the GeneView page of each database (Figure 1). We have been accepting community submissions to both databases including gene structure refinements, gene families, gene functions, gene symbols and links to other resources. In addition, submitted data are manually inspected by expert curators before becoming integrated.
Figure 1.
An example GeneView page for the sll1867 gene of Synechocystis sp. PCC 6803. Manually curated gene symbol(s) and gene product(s) are shown in the ‘Gene symbol Extracted from literature’ and ‘Gene symbol Extracted from literature’ fields in the ‘Summary’ section.
Curation platform
Manual curation is still one of the most important and most difficult tasks in genome projects. Therefore, methodological and technological solutions are urgently needed to reduce annotation costs. To address this issue, we have developed a web-based genome annotation tool, TogoAnnotation (http://togo.annotation.jp). This tool, which is derived from KazusaAnnotation (4), provides an easy way to access, edit and store annotation data over a flexible web interface based on social bookmarking web services architecture.
Curated genes
CyanoBase and RhizoBase have grown considerably since their introduction. The content of CyanoBase and RhizoBase and their composition are summarized in Table 1. A statistical summary of annotations conducted in August 2013 indicated that 138 896 cyanobacterial genes were curated from 5285 published references. Hence, the number of references investigated for CyanoBase increased by 3025 in comparison with our previous report in 2010 (4). For example, of the 3725 genes contained in the Synechocystis sp. PCC 6803 genome, 3067 (82.3%) have been already annotated with gene symbols, protein names and gene definitions from the literature. Users are able to access the annotation of each gene on the ‘Reference’ section of the GeneView page and to find annotated data [e.g. the photosystem II D1 protein (psbA3) currently have 386 citations http://genome.microbedb.jp/cyanobase/Synechocystis/genes/sll1867#references].
Table 1.
Number of curated publications and annotated genes for each organism of CyanoBase and RhizoBase
| Database | Organism | References | Annotations | Annotated genes | Total genes |
|---|---|---|---|---|---|
| CyanoBase | Synechocystis sp. PCC 6803 | 2346 | 80 204 | 3064 | 3725 |
| CyanoBase | Anabaena sp. PCC 7120 | 959 | 29 154 | 2754 | 6223 |
| CyanoBase | Synechococcus elongatus PCC 7942 | 815 | 17 060 | 794 | 2715 |
| CyanoBase | Thermosynechococcus elongatus BP-1 | 270 | 6768 | 2528 | 2528 |
| CyanoBase | Synechococcus sp. PCC 7002 | 264 | 3999 | 265 | 3235 |
| CyanoBase | Nostoc punctiforme ATCC 29133 | 151 | 3349 | 768 | 6794 |
| CyanoBase | Chlorobium tepidum TLS | 143 | 5532 | 751 | 2310 |
| CyanoBase | Anabaena variabilis ATCC 29413 | 119 | 1731 | 258 | 5724 |
| CyanoBase | Prochlorococcus marinus MED4 | 64 | 2155 | 390 | 1756 |
| CyanoBase | Gloeobacter violaceus PCC 7421 | 52 | 5600 | 4483 | 4484 |
| CyanoBase | Prochlorococcus marinus MIT9313 | 44 | 919 | 248 | 2326 |
| CyanoBase | Prochlorococcus marinus SS120 | 37 | 539 | 135 | 1928 |
| CyanoBase | Arthrospira platensis NIES-39 | 9 | 787 | 260 | 6676 |
| CyanoBase | Trichodesmium erythraeum IMS101 | 5 | 22 | 14 | 4498 |
| CyanoBase | Synechococcus sp. WH8102 | 5 | 38 | 22 | 2579 |
| CyanoBase | Synechococcus elongatus PCC 6301 | 2 | 5 | 2 | 2580 |
| RhizoBase | Bradyrhizobium japonicum USDA110 | 550 | 26 636 | 8366 | 8374 |
| RhizoBase | Sinorhizobium meliloti 1021 | 240 | 9801 | 1990 | 6287 |
| RhizoBase | Mesorhizobium loti MAFF303099 | 115 | 2373 | 865 | 7343 |
| RhizoBase | Rhizobium sp. pNGR234ab | 107 | 5224 | 989 | 990 |
| RhizoBase | Rhizobium leguminosarum bv. viciae 3841 | 83 | 3426 | 781 | 7342 |
| RhizoBase | Rhizobium sp. NGR234 | 8 | 46 | 17 | 6437 |
AVAILABILITY
Application programming interface
CyanoBase and RhizoBase are based on the same in-house developed genome database system offering a representational state transfer-based web application programming interface for automated retrieval of data by third-party tools and computer programs. As an output, various widely used formats are supported, including TSV, CSV, FASTA and GFF3 (4).
Semantic Web application
To improve data integration within CyanoBase, RhizoBase and other microorganism databases in the near future, we have introduced Semantic Web technologies for the standard representation and common exchange protocol of data (10). First, we developed a generic ontology for semantically describing genomic annotations in cooperation with the DDBJ and the Database Center for Life Science (DBCLS). Based on this ontology, we converted annotations stored in the CyanoBase and RhizoBase databases into the resource description framework (RDF) format. The result is accessible from our SPARQL Protocol and RDF Query Language (SPARQL) endpoint at http://genome.microbedb.jp/sparql. A list of available resources is summarized in Table 2.
Table 2.
Summary of data types and the number of items accessible from the SPARQL endpoint
Currently, databases of bacterial model organisms are maintained and distributed independently. To ensure that these data are interoperable for a large-scale genomic analysis, we collaborated with the MicrobeDB.jp (http://microbedb.jp/) and the TogoGenome (http://togogenome.org/) projects for sharing prokaryotic genome annotations as RDF data through respective SPARQL endpoints. Such standardization reduces duplicated efforts and improves reusability while allowing each database to update their own resources independently. In addition, it is beneficial for end users that they can use a variety of data sources with common software through the standard web service interface in a unified and automated manner.
Change of site URL
We have migrated the server hosting CyanoBase and RhizoBase from Kazusa DNA Research Institute to the National Institute of Genetics. Consequently, the location of these databases has changed to http://genome.microbedb.jp/.
Social media
We have been delivering timely announcements on Twitter. Users can follow @cyanobase and @rhizobase on Twitter to receive the latest information on database updates and server maintenance of the CyanoBase and RhizoBase databases.
License
All data in our database is provided under the Creative Commons CC0 public domain license (4).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Integrated Database Project, Ministry of Education, Culture, Sports, Science and Technology of Japan; National Bioscience Database Center (NBDC) of the Japan Science and Technology Agency (JST); Kazusa DNA Research Institute Foundation. Funding for Open Access: National Bioscience Database Center.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Eli Kaminuma for suggestions and comments on the database development, Yasuhiro Tanizawa for the operation of the CyanoBase and RhizoBase web services and members of Kazusa DNA Research Institute for the development of RhizoBase. They also thank members of MicrobeDB.jp and TogoGenome projects for the collaboration on the Semantic Web-based developments.
REFERENCES
- 1.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura Y, Kaneko T, Hirosawa M, Miyajima N, Tabata S. CyanoBase, a www database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803. Nucleic Acids Res. 1998;26:63–67. doi: 10.1093/nar/26.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura Y, Kaneko T, Tabata S. CyanoBase, the genome database for Synechocystis sp. strain PCC6803: status for the year 2000. Nucleic Acids Res. 2000;28:72. doi: 10.1093/nar/28.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakao M, Okamoto S, Kohara M, Fujishiro T, Fujisawa T, Sato S, Tabata S, Kaneko T, Nakamura Y. CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Res. 2010;38:D379–D381. doi: 10.1093/nar/gkp915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, et al. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000;31(7):331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- 6.Okubo T, Tsukui T, Maita H, Okamoto S, Oshima K, Fujisawa T, Saito A, Futamata H, Hattori R, Shimomura Y, et al. Complete genome sequence of Bradyrhizobium sp. S23321: insights into symbiosis evolution in soil oligotrophs. Microbes Environ. 2012;27:306–315. doi: 10.1264/jsme2.ME11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugawara H, Ohyama A, Mori H, Kurokawaw K. Microbial genome annotation pipeline (MiGAP) for diverse users. 20th Int. Conf. Genome Informatics. Kanagawa, Japan. 2009;S-001:1–2. [Google Scholar]
- 8.Kaminuma E, Mashima J, Kodama Y, Gojobori T, Ogasawara O, Okubo K, Takagi T, Nakamura Y. DDBJ launches a new archive database with analytical tools for next-generation sequence data. Nucleic Acids Res. 2010;38:D33–D38. doi: 10.1093/nar/gkp847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagasaki H, Mochizuki T, Kodama Y, Saruhashi S, Morizaki S, Sugawara H, Ohyanagi H, Kurata N, Okubo K, Takagi T, et al. DDBJ read annotation pipeline: a cloud computing-based pipeline for high-throughput analysis of next-generation sequencing data. DNA Res. 2013;20:383–390. doi: 10.1093/dnares/dst017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katayama T, Wilkinson MD, Micklem G, Kawashima S, Yamaguchi A, Nakao M, Yamamoto Y, Okamoto S, Oouchida K, Chun HW, et al. The 3rd DBCLS BioHackathon: improving life science data integration with Semantic Web technologies. J. Biomed. Semantics. 2013;4:6. doi: 10.1186/2041-1480-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato S, Shimoda Y, Muraki A, Kohara M, Nakamura Y, Tabata S. A large-scale protein protein interaction analysis in Synechocystis sp. PCC6803. DNA Res. 2007;14:207–216. doi: 10.1093/dnares/dsm021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimoda Y, Shinpo S, Kohara M, Nakamura Y, Tabata S, Sato S. A large scale analysis of protein-protein interactions in the nitrogen-fixing bacterium Mesorhizobium loti. DNA Res. 2008;29:13–23. doi: 10.1093/dnares/dsm028. [DOI] [PMC free article] [PubMed] [Google Scholar]