Abstract
NONCODE (http://www.bioinfo.org/noncode/) is an integrated knowledge database dedicated to non-coding RNAs (excluding tRNAs and rRNAs). Non-coding RNAs (ncRNAs) have been implied in diseases and identified to play important roles in various biological processes. Since NONCODE version 3.0 was released 2 years ago, discovery of novel ncRNAs has been promoted by high-throughput RNA sequencing (RNA-Seq). In this update of NONCODE, we expand the ncRNA data set by collection of newly identified ncRNAs from literature published in the last 2 years and integration of the latest version of RefSeq and Ensembl. Particularly, the number of long non-coding RNA (lncRNA) has increased sharply from 73 327 to 210 831. Owing to similar alternative splicing pattern to mRNAs, the concept of lncRNA genes was put forward to help systematic understanding of lncRNAs. The 56 018 and 46 475 lncRNA genes were generated from 95 135 and 67 628 lncRNAs for human and mouse, respectively. Additionally, we present expression profile of lncRNA genes by graphs based on public RNA-seq data for human and mouse, as well as predict functions of these lncRNA genes. The improvements brought to the database also include an incorporation of an ID conversion tool from RefSeq or Ensembl ID to NONCODE ID and a service of lncRNA identification. NONCODE is also accessible through http://www.noncode.org/.
INTRODUCTION
Non-coding RNAs (ncRNAs) constitute a significant fraction of the transcriptome (1). Widespread application of high-throughput RNA sequencing (RNA-seq), with the aid of computational methods, has revealed increasing number of ncRNAs identified from various organisms (2). Especially, long non-coding RNAs (lncRNAs), which are considered to be >200 nt in length and are often multiexonic (3), have been identified to play critical roles in various processes including embryonic development (4), dosage compensation (5) and immune response (6). Owing to their functional significance, databases that integrate comprehensive information about lncRNAs can be helpful for understanding biological processes. However, existing lncRNA resources such as lncRNAdb (7), ncRNAdb (8) and Rfam (9) fail to cover most of the newly identified lncRNAs in recent studies. Consequently, we updated the NONCODE database to version 4.0, to keep up-to-date with the latest discovery of lncRNAs. The number of lncRNA entries in NONCODE version 4.0 has increased to 210 831.
Despite a lack of protein-coding ability, lncRNAs are similar to mRNAs in many ways (10). LncRNAs are involved in alternative splicing patterns that resemble mRNAs (11). Moreover, the majority of lncRNAs are spliced with similar exon/intron lengths to protein-coding genes (11,12). Considering the increasing number of lncRNA transcripts, proposing lncRNA gene structures is now necessary to gain a systematic understanding of lncRNAs. However, existing resources merely describe gene structures of lncRNAs. Following the classical definition of ‘gene’ for protein-coding RNAs (13), NONCODE version 4.0 unites genomic sequences encoding a coherent set of overlapping long non-coding transcripts into an lncRNA gene. Because many lncRNAs reside within or overlap protein-coding loci (11), we then classified lncRNAs genes into four categories according to their genomic location in relation to protein-coding genes: antisense, intergenic, sense exonic and sense non-exonic, respectively.
The emergence of a large amount of RNA-seq data not only facilitates identification and characterization of lncRNAs but also provides clues to understanding expression patterns (14) and potential functions of lncRNAs (15). For human and mouse, NONCODE version 4.0 presents expression patterns across various tissues, as well as predicted functions of lncRNAs inferred from public RNA-seq data. Other improvements of NONCODE version 4.0 include iLncRNA, an online lncRNA identification pipeline based on user supplied data, and an ncRNA ID conversion tool allowing query of accessions from various RNA databases. An overview of updates in NONCODE version 4.0 is shown in Figure 1.
Figure 1.
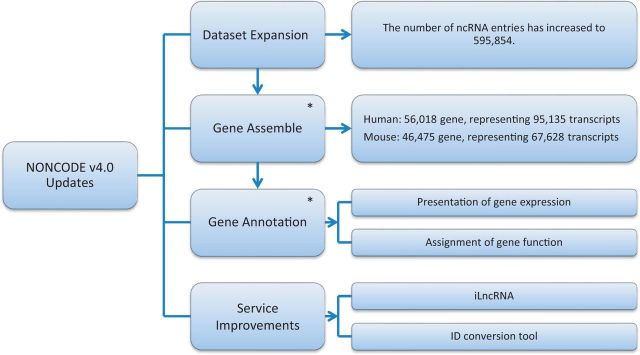
Overview of updates in NONCODE version 4.0. Through processes of data collection, redundancy elimination and filtration, the number of ncRNA entries in NONCODE version 4.0 has increased to 595 854. For lncRNAs from human and mouse, different transcripts that intersect any exon of other other and reside on the same DNA strand are considered to belong to the same gene and clustered into a single gene record. This step results in 56 018 genes from 95 135 transcripts in human and 46 475 genes from 67 628 transcripts in mouse. Using public RNA-seq data of human and mouse, presentation of expression and assignment of function is annotated for each lncRNA gene. All tools and services in NONCODE have been updated. In addition, ID conversion tool and iLncRNA is new in this version. The two fields marked with asterisk (*) are specifically for lncRNAs.
NONCODE has already proven to be an important resource in the realm of ncRNA databases, and is therefore incorporated into other ncRNA databases such as fRNAdb — a large collection of ncRNAs (16), GeneCards — the comprehensive human gene compendium (17) and DIANA-LncBase — a database for miRNAs targets on lncRNAs (18). We believe that the recent improvements in NONCODE version 4.0 will significantly contribute to the enhancement of these and potentially other ncRNA databases.
DATA COLLECTION, REDUNDANCY ELIMINATION AND FILTRATION
Based on former versions of NONCODE (19–21), new data sets from literatures and other specialized databases were collected. For literature mining, we first retrieved literature published since May 1, 2011, from PubMed, using the key words ‘ncrna', ‘noncoding', ‘non-coding', ‘no code', ‘non-code', ‘lncrna' or ‘lincrna', and found 4572 relevant articles. Then sequences, genome locations and other relevant information concerning transcripts from manually selected reports on new ncRNAs were retrieved. Next, the latest releases of Ensembl (22) and RefSeq (23) were integrated to supplement our manual curation efforts. In total, 118 148, 141 194 and 35 445 transcripts were retrieved from literature, Ensembl and RefSeq, respectively.
A process of redundancy elimination was then performed on the ncRNAs collected from literature and specialized databases mentioned earlier in text, together with existing data in NONCODE version 3.0. Cuffcompare program in Cufflinks suite (24) was used to map the whole ncRNA data set back to annotations of itself. Transcripts completely matching each other, annotated with class code ‘=’ by Cuffcompare, were considered to be redundant and grouped in a single record.
Each transcript we collected was considered non-coding in the resource it came from. However, the same transcript might be assigned mutually exclusive annotations in different resources due to respective standards of distinguishing non-coding from protein-coding RNAs.
Therefore, we used two screening criteria for all transcripts in NONCODE version 4.0 to ensure no inclusion of protein-coding transcripts. First, all transcripts kept by the redundancy elimination step were compared with a reference set containing known protein-coding RNAs from Ensembl and RefSeq by Cuffcompare. Those completely matching protein-coding transcripts, annotated with class code ‘=’, were discarded. Second, the coding potential of each transcript was evaluated by our CNCI program (25). Transcripts classified into coding sequences by CNCI were discarded. The left transcripts were kept with high confidence to be non-coding and entered into NONCODE. Finally, 595 854 ncRNAs were finally recorded. Data expansion of NONCODE mainly resulted from new collection of lncRNAs of human and mouse. In all of the 210 831 lncRNA transcripts of the final catalog of NONCODE, 95 135 and 67 628 come from human and mouse, respectively, whereas the other 48 068 come from other organisms.
DEFINITION AND CATEGORIZATION OF lncRNA GENES
LncRNAs are similar to mRNAs in regards to alternative splicing (26), thus we united genomic sequences encoding a coherent set of overlapping lncRNAs into an lncRNA gene, following the classical definition of ‘gene’ for protein-coding RNAs. Different transcripts that intersected any exons of one other and resided on the same DNA strand were considered to belong to the same gene and clustered into a single gene record. In this way, 56 018 and 46 475 lncRNA genes were generated from 95 135 and 67 628 lncRNAs for human and mouse, respectively. Both lncRNA transcripts and genes were designated systematically in NONCODE. LncRNA transcripts from a same organism were numbered subsequently, starting with ‘NON’ followed by a symbol representing the organism. For example, ‘NONMMUT000020' denotes a transcript from mouse (the beginning ‘NON’ stands for ‘noncoding’; the following ‘MMU' stands for ‘Mus musculus'; the next letter ‘T' stands for ‘transcript'). Likewise, lncRNA genes were named sequentially with the middle letter ‘T' replaced by ‘G' representing ‘gene'.
Considering that the genomic context of lncRNAs may provide suggestions about their functional role, we subsequently classified lncRNA genes into the following biotypes according to their location with respect to protein-coding genes:(i) antisense, which have transcripts that intersect protein-coding genes on the opposite strand; (ii) intergenic, which are a subset of non-coding RNA loci located between protein-coding genes; (iii) sense exonic, which have transcripts that intersect protein-coding exons on the same strand; and (iv) sense non-exonic, which overlap with protein-coding genes in respect to transcription boundaries but not overlap in respect to processed exons.
Take mouse as an example, applying this categorization automatically to the lncRNA data set of NONCODE version 4.0 results in the following distribution: antisense (6653), intergenic (19 067), sense exonic (12 111) and sense non-exonic (9312). See Figure 2 for further details on the lncRNA genes.
Figure 2.
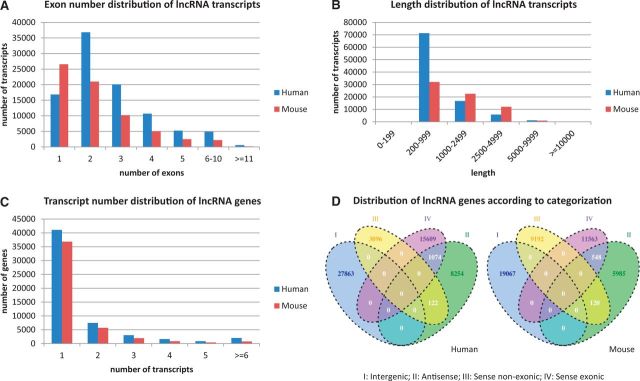
Details of lncRNA transcripts and genes. (A) Exon number distribution of human and mouse lncRNA transcripts. (B) Length distribution of human and mouse lncRNA transcripts. (C) Number of transcripts per gene for human and mouse. (D). Distribution of human and mouse lncRNA genes according to categorization.
ncRNA ANNOTATION
One significant characteristic of NONCODE is its comprehensive annotation information. Each transcript in NONCODE is annotated with the following information:(i) basic description, including the ncRNA name, alias, sequence, length, genomic location, coding potential assessment by CPC (27) and CNCI, organisms and references; (ii) biological information, concerning its function, cellular role, cellular location and process function class (PfClass); and (iii) expression indication, including independent sources of multi-tissue expression profiles and potential function predicted based on a coding–non-coding co-expression network (28,29), especially for lncRNAs.
In this update, lncRNA genes are also annotated with two important features, as follows:
Presentation of lncRNA gene expression
We made full use of public RNA-seq data of human and mouse to provide indication of lncRNA functions. Human BodyMap 2.0 data (ENA archive: ERP000546) from human across 16 tissues and another RNA-seq data set from mouse across six different tissues (ENA archive: ERP000591) were downloaded. Cufflinks assembled transfrags from these raw RNA-seq for human and mouse, respectively. Cuffcompare then compared assembled transcripts with a reference annotation set composed of lncRNA genes from NONCODE version 4.0. At the same time, Cuffdiff calculated the FPKM of each reference lncRNA gene, representing expression level of it. The expression profile of each lncRNA gene across various tissue types is presented as a bar graph.
Assignment of lncRNA gene function
Functional predictions may guide and assist future investigations of lncRNAs. We applied lnc-GFP (30), a bi-colored network-based global function predictor, to the same RNA-seq data mentioned earlier in text to predict probable functions for lncRNA genes. A total of 20 100 lncRNA genes in NONCODE version 4.0 have been annotated with potential functions with a suitable parameter setting.
SERVICE UPDATE
The NONCODE database is based on MySQL and the Web site is powered by an Apache server. NONCODE has a user-friendly interface with a number of convenient browse and search options. Several useful services are available for users to access the NONCODE data, including BLAST, UCSC Genome Browser, SOAP API, DAS and an online submission system. UCSC Genome Browser has been upgraded in the new NONCODE version, whereas all other services are new additions. Furthermore, two online services have been added, which are the ID conversion tool and iLncRNA, the lncRNA identification pipeline.
ID conversion tool
Recent advances in non-coding RNA research have led to the creation of several ncRNA resources. A given ncRNA transcript tends to be assigned different accession in different databases. In such situations, an ID conversion tool is necessary to facilitate more efficient user queries. For example, a transcript variant of the lncRNA gene termed HOTAIR (31) was assigned identifier ‘NR_003716’ in RefSeq. The ID conversion tool of NONCODE version 4.0 would recognize and convert it to ‘NONHSAT028508’. So far, NONCODE version 4.0 supports ID mapping between NONCODE identifiers and accessions from RefSeq and Ensembl.
iLncRNA
Owing to the development of next-generation sequencing technology, ncRNAs are now more easily and more accurately identified by sequencing transcriptomes (32). In this update, we provide iLncRNA an online pipeline for lncRNA identification based on assembled gtf files. As shown in Figure 3, transcript files in gff or gtf format either mapped and assembled from raw RNA-seq data or generated in other way are required as input for the identification pipeline. First, transcripts from input files <200 nt would be removed. The remaining transcripts were considered putative lncRNA transcripts, which were then subjected to the same fitration process comprises Cuffcompare and CNCI described earlier in text, to exclude potential protein-coding transcripts. Cuffcompare would be used once more to remove transcripts completely matching with pseudogenes from Ensembl. There would be an additional step to distinguish novel lncRNAs from known lncRNAs. Still by using Cuffcompare, lncRNAs not completely matching with collected lncRNAs from NONCODE would be defined as novel lncRNAs. Finally, predicted result would return to submitter of uploaded data after further validation. At the same time, this result would in turn be collected by NONCODE if authorized by the submitter.
Figure 3.
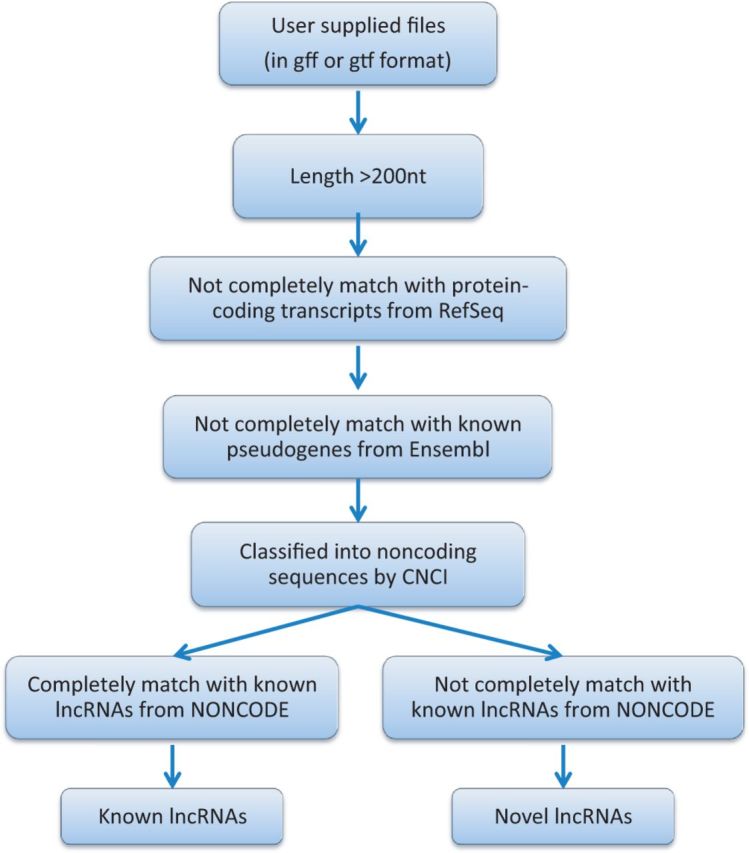
Pipeline for identification of lncRNAs for users. Refer to main text for details.
DISCUSSION
The decreasing cost and improved capability of RNA-sequencing technology has lead to numerous transcriptome data from a variety of species. As a result of this, large numbers of ncRNAs are being rapidly identified and characterized (33). Due to this situation, we updated the NONCODE database to version 4.0, to keep track of newly identified ncRNAs. Of the newly collected data, lncRNAs constitute the majority. Particularly, lncRNAs were discovered to be involved in alternative splicing patterns that resemble mRNAs. Accumulating records of lncRNA transcripts makes it both possible and necessary to establish the concept of lncRNA genes. Consequently, NONCODE version 4.0 is a step toward a more integrated knowledge database with respect to definition and categorization of lncRNA genes.
RNA-seq is also increasingly being used for gene expression profiling. Through analysis of two sets of public RNA-seq data, NONCODE version 4.0 presents expression profiles of lncRNAs across different tissues from human and mouse, respectively, as bar graphs. Moreover, potential function of lncRNAs of human and mouse is inferred from the same data by lnc-GFP, a bi-colored network-based global function predictor.
Service improvements of NONCODE include not only updating existing tools such as BLAST and UCSC Genome Browser to the latest versions but also adding an ID conversion tool, enabling queries of accessions from different resources. To help users with their own RNA-seq data, we have also provided an online pipeline for lncRNA identification, which is named iLncRNA. User supplied files in gtf or gff format assembled from raw RNA-seq using TopHat (24) or other tools can be further analyzed by this pipeline. NONCODE version 4.0 in turn would consider collecting these data into the database. Moreover, with NPInter, the ncRNA interaction databases (34,35), cooperatiting with our platform, NONCODE will stay as an informative and valuable data source for the study of lncRNAs.
FUNDING
National High-tech Research and Development Projects 863 [2012AA020402, 2012AA022501], National Key Basic Research and Development Program 973 [2009CB825401], Training Program of the Major Research plan of the National Natural Science Foundation of China [91229120], National Natural Science Foundation of China [31371320]. Funding for open access charge: Training Program of the Major Research plan of the National Natural Science Foundation of China [91229120].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
We thank Lu Yuyang for checking our annotation data. We also thank Andrew Plygawko for carefully reading our manuscript and Prof. Xiaohua Shen of Tsinghua University for help during the course of this work.
REFERENCES
- 1.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng X, Meller VH. Non-coding RNA in fly dosage compensation. Trends Biochem. Sci. 2006;31:526–532. doi: 10.1016/j.tibs.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, Bradel-Tretheway BG, Korth MJ, Castle JC, Biery MC, et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1:e00206–10. doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szymanski M, Erdmann VA, Barciszewski J. Noncoding RNAs database (ncRNAdb) Nucleic Acids Res. 2007;35:D162–D164. doi: 10.1093/nar/gkl994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karapetyan A, Buiting C, Kuiper R, Coolen M. Regulatory roles for long ncRNA and mRNA. Cancers. 2013;5:462–490. doi: 10.3390/cancers5020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, Emanuelsson O, Zhang ZD, Weissman S, Snyder M. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 14.Huang R, Jaritz M, Guenzl P, Vlatkovic I, Sommer A, Tamir IM, Marks H, Klampfl T, Kralovics R, Stunnenberg HG, et al. An RNA-Seq strategy to detect the complete coding and non-coding transcriptome including full-length imprinted macro ncRNAs. PLoS One. 2011;6:e27288. doi: 10.1371/journal.pone.0027288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma H, Hao Y, Dong X, Gong Q, Chen J, Zhang J, Tian W. Molecular mechanisms and function prediction of long noncoding RNA. Sci. World J. 2012;2012:541786. doi: 10.1100/2012/541786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mituyama T, Yamada K, Hattori E, Okida H, Ono Y, Terai G, Yoshizawa A, Komori T, Asai K. The functional RNA database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res. 2009;37:D89–D92. doi: 10.1093/nar/gkn805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belinky F, Bahir I, Stelzer G, Zimmerman S, Rosen N, Nativ N, Dalah I, Iny Stein T, Rappaport N, Mituyama T, et al. Non-redundant compendium of human ncRNA genes in GeneCards. Bioinformatics. 2013;29:255–261. doi: 10.1093/bioinformatics/bts676. [DOI] [PubMed] [Google Scholar]
- 18.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM, Hatzigeorgiou AG. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013;41:D239–D245. doi: 10.1093/nar/gks1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C, Bai B, Skogerbo G, Cai L, Deng W, Zhang Y, Bu D, Zhao Y, Chen R. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33:D112–D115. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He S, Liu C, Skogerbo G, Zhao H, Wang J, Liu T, Bai B, Zhao Y, Chen R. NONCODE v2.0: decoding the non-coding. Nucleic Acids Res. 2008;36:D170–D172. doi: 10.1093/nar/gkm1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bu D, Yu K, Sun S, Xie C, Skogerbo G, Miao R, Xiao H, Liao Q, Luo H, Zhao G, et al. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012;40:D210–D215. doi: 10.1093/nar/gkr1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41:e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 27.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao Q, Xiao H, Bu D, Xie C, Miao R, Luo H, Zhao G, Yu K, Zhao H, Skogerbo G, et al. ncFANs: a web server for functional annotation of long non-coding RNAs. Nucleic Acids Res. 2011;39:W118–W124. doi: 10.1093/nar/gkr432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo X, Gao L, Liao Q, Xiao H, Ma X, Yang X, Luo H, Zhao G, Bu D, Jiao F, et al. Long non-coding RNAs function annotation: a global prediction method based on bi-colored networks. Nucleic Acids Res. 2013;41:e35. doi: 10.1093/nar/gks967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun L, Zhang Z, Bailey TL, Perkins AC, Tallack MR, Xu Z, Liu H. Prediction of novel long non-coding RNAs based on RNA-Seq data of mouse Klf1 knockout study. BMC Bioinformatics. 2012;13:331. doi: 10.1186/1471-2105-13-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo H, Sun S, Li P, Bu D, Cao H, Zhao Y. Comprehensive characterization of 10,571 mouse large intergenic noncoding RNAs from whole transcriptome sequencing. PLoS One. 2013;8:e70835. doi: 10.1371/journal.pone.0070835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu T, Wang J, Liu C, Zhang Y, Shi B, Zhu X, Zhang Z, Skogerbo G, Chen L, Lu H, et al. NPInter: the noncoding RNAs and protein related biomacromolecules interaction database. Nucleic Acids Res. 2006;34:D150–D152. doi: 10.1093/nar/gkj025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan J, Wu W, Xie C, Zhao G, Zhao Y, Chen R. NPInter v2.0: an updated database of ncRNA interactions. Nucleic Acids Res. 2013;42:D104–D108. doi: 10.1093/nar/gkt1057. [DOI] [PMC free article] [PubMed] [Google Scholar]


