Abstract
The BioSamples database at the EBI (http://www.ebi.ac.uk/biosamples) provides an integration point for BioSamples information between technology specific databases at the EBI, projects such as ENCODE and reference collections such as cell lines. The database delivers a unified query interface and API to query sample information across EBI’s databases and provides links back to assay databases. Sample groups are used to manage related samples, e.g. those from an experimental submission, or a single reference collection. Infrastructural improvements include a new user interface with ontological and key word queries, a new query API, a new data submission API, complete RDF data download and a supporting SPARQL endpoint, accessioning at the point of submission to the European Nucleotide Archive and European Genotype Phenotype Archives and improved query response times.
INTRODUCTION
The EBI’s BioSamples database provides a single point of entry to sample data stored in EBI assay databases and delivers a dedicated query interface and API for accessing sample data. Samples are arranged into sample groups for ease of query, submission and to allow attributes to be added at the group level rather than at the sample specific level. This supports cases where values or attributes must be expressed as binned values across samples. This happens when the information is not available or cannot be provided at the sample level for ethical reasons (1). When querying BioSamples users are offered links to assay databases, for example, to sequence information in the European Nucleotide Archive or ENA (2), gene expression microarrays in ArrayExpress (3) or proteomics data PRoteomics IDEntifications database or PRIDE (4). The EBI’s BioSamples database is developed in parallel with the NBCI’s BioSamples database (5), which fulfils a similar function at the NCBI. This article describes data growth and new features implemented since our previous publication in 2011 (6).
The EBI BioSamples database has doubled in size since January 2012 when 1 million samples were described in the BioSamples database, as of October 2013 2 846 137 samples are available as 80 232 groups. Data growth is attributed to new data sources, and increased volume of data from existing sources. New data sources include 22 288 samples from The Cancer Genome Atlas (http://cancergenome.nih.gov/), 920 441 samples from the Catalogue of Somatic Mutation in Cancer—COSMIC (7); 920 441 samples in 10 737 groups. Addition of samples from these sources provides interoperability between resources where, for example, COSMIC identifiers are included, e.g. in Ensembl (8).
INFRASTRUCTURAL IMPROVEMENTS
An updated web interface delivers new search functionality, improved tabular layout, ontology supported queries and access revised help documentation (see Figure 1). The experimental factor ontology (EFO) (9) is now for used query expansion using synonyms and child terms thus allowing more specific searches to be made. Users may select indexed key words or an EFO term for their queries in combination with Boolean Operators to refine their searches. EFO has been expanded in parallel to support BioSamples use cases, including an import of a ‘SLIM’ of the Uber Anatomy Ontology Uberon (10) and a genetic disease classification from Orphanet (11). These were selected based on analysis of common user queries and provide enhanced queries for samples based on anatomical and disease characteristics.
Figure 1.
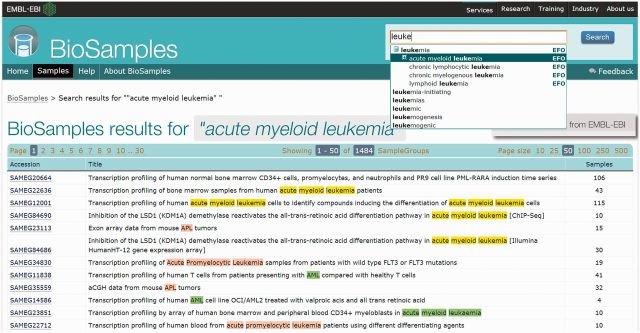
The new BioSamples web interface showing search results for a query of ‘acute myeloid leukemia’. Auto-complete on the search box suggests appropriate ontology and keyword terms as the query is entered by the user, including more specific terms from EFO such as subtypes of the disease. Highlighted results are colour coded for clarity; exact matches (yellow), synonyms (green) and more specific terms (red).
New programmatic interfaces are available for both retrieving and submitting data. The API supports queries by sample or sample group accession, and queries of samples, or sample groups by their attributes. For example, the URL http://www.ebi.ac.uk/biosamples/xml/sample/SAME42581 returns all information about a sample in XML format. The APIs are documented on the BioSamples database help pages, and example queries are provided.
Sample information can also be submitted to the BioSamples database through an API via a JSON representation of the SampleTab tabular submission format. Part of the submission API is a SampleTab validation service, including a web page interface (http://www.ebi.ac.uk/biosamples/sampletab). This service is used for pre-submission validation of both manual and programmatic submissions. To maintain quality for directly submitted sample information, use of the submission web service is restricted by API keys, which can be obtained from biosamples@ebi.ac.uk.
DATA INTEGRATION
The BioSamples database has improved interoperability with other EBI resources and with external groups. All submissions of sample information to ENA and European Genotype Phenotype Archive (http://www.ebi.ac.uk/ega) are now assigned a BioSamples Accession, which is returned to the submitter immediately as part of the submission process. Users can also pre-register sample(s) and re-use those accession(s) when submitting to resources such as ENA and EGA. By preregistering samples the BioSamples staff curates submitted data, and the BioSamples database becomes a single source of sample information across multiple experimental technologies and databases. This, in turn, encourages the use of BioSamples identifiers in other repositories to identify and link equivalent samples.
Several major research projects have established links with the BioSamples database. For example, the HipSci project (http://www.hipsci.org/) pre-register information about donors and cell lines, including the relationships between them, with BioSamples database. The ENCODE (12) data coordination centre is working with BioSamples database to ensure their existing sample records are updated and annotated with ontology terms and in specifying relationships between samples in ENCODE datasets. To date sample information from users is submitted directly to the BioSamples database through both manual and automatic processes, both of which are supported by the curatorial staff.
Other locations around the world have also established repositories of sample information, including the NCBI BioSample database. The BioSamples database at EBI is using a common accessioning scheme previously agreed with NCBI and DDBJ, and we expect that data exchange will be implemented in early 2014.
As sample data can be identified by multiple accessions assigned by EBI and external databases an identifier and URL resolution service ‘MyEquivalents’ has been deployed. It provides mapping between different, but equivalent, sample identifiers. For example, human RNA-Seq data deposited at EBI may have identifiers for ArrayExpress, ENA and BioSamples database as these resources share records. In time the BioSamples database identifier will be the only sample identifier for new submissions, but until then, and to preserve backwards compatibility for legacy data, the MyEquivalents service provides redirection URLs and web services describing mappings. More information and the source code for the MyEquivalents software is available (https://github.com/EBIBioSamples/myequivalents).
Finally, as a component of the EBI Resource Description Framework (RDF) platform (http://www.ebi.ac.uk/rdf) RDF is now available for the BioSamples database content. The schema is derived from the SampleTab format, supported by integration with existing ontologies such as the Ontology of Biomedical Investigations (13) and EFO. Data are made available as RDF and also for query via a SPARQL endpoint for which example queries are documented.
FUTURE WORK
The development of the process and tools supporting EBI-NCBI data exchange is underway in collaboration with NCBI. EBI has completed a test parse and load of the current NCBI BioSamples database content and we are examining and mapping attributes used by the NBCI’s and EBI’s databases to deliver a core set of common attributes and context for these, for example, those required by standards such as The Minimal Information about a MetaGenome (14). The core attributes list will be used to facilitate data exchange, provide improved searches across attributes and drive context specific displays to ensure like attributes are displayed together for specific experiment types, e.g. latitude, longitude and depth for ocean samples. We will further improve our API and GUI access by implementing improved support for single sample level queries by technology and assay types.
FUNDING
EMBL core budget (in part) provided by the EMBL member countries with contributions from the European Commission grants CAGEKID [HEALTH-F4-2010-241669]; ENGAGE [HEALTH-F4-2007-201413 from the European Commission FP7 program]; BioMedBridges [European Commission FP7 Capacities Specific Programme 284209]; Gen2Phen [European Commission FP7 200754]; Diachron [European Commission FP7 601043]; a grant from the Wellcome Trust and the Medical Research Council [award WT098503]. Funding for open access charge: EMBL Core.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank our NBCI colleagues particularly Tanya Barrett. We have benefitted from discussions with HIPSCI, Gramene, Encode and 1000 Genomes projects about optimal access models and data representation needs for their respective communities and species. We thank many of the EBI staff, in particular, James Malone, Ilkka Lappalainen, Lisa Skipper, Attila Csordas, Henning Hermjakob, Sarah Hunter, John Overington, Christoph Steinbeck, Paul Flicek, Ewan Birney and Rolf Apweiler for discussion on BioSamples database applications, use cases and implementation. We thank our colleagues Melissa Haendel and Chris Mungall for assistance with the Uberon SLIM generation and Segolene Ayme and Ana Rath of Orphanet.
REFERENCES
- 1.Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y. Identifying personal genomes by surname inference. Science. 2013;339:321–324. doi: 10.1126/science.1229566. [DOI] [PubMed] [Google Scholar]
- 2.Cochrane G, Alako B, Amid C, Bower L, Cerdeño-Tárraga A, Cleland I, Gibson R, Goodgame N, Jang M, Kay S, et al. Facing growth in the European Nucleotide Archive. Nucleic Acids Res. 2013;41:D30–D35. doi: 10.1093/nar/gks1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, et al. ArrayExpress update—an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, et al. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett T, Clark K, Gevorgyan R, Gorelenkov V, Gribov E, Karsch-Mizrachi I, Kimelman M, Pruitt KD, Resenchuk S, Tatusova T, et al. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012;40:D57–D63. doi: 10.1093/nar/gkr1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gostev M, Faulconbridge A, Brandizi M, Fernandez-Banet J, Sarkans U, Brazma A, Parkinson H. The BioSample Database (BioSD) at the European Bioinformatics Institute. Nucleic Acids Res. 2012;40:D64–D70. doi: 10.1093/nar/gkr937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shepherd R, Forbes SA, Beare D, Bamford S, Cole CG, Ward S, Bindal N, Gunasekaran P, Jia M, Kok CY, et al. Data mining using the Catalogue of Somatic Mutations in Cancer BioMart. Database. 2011;2011:bar018. doi: 10.1093/database/bar018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malone J, Holloway E, Adamusiak T, Kapushesky M, Zheng J, Kolesnikov N, Zhukova A, Brazma A, Parkinson H. Modeling sample variables with an Experimental Factor Ontology. Bioinformatics. 2010;26:1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, Haendel MA. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012;13:R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rath A, Olry A, Dhombres F, Miličić Brandt M, Urbero B, Ayme S. Representation of rare diseases in health information systems: the orphanet approach to serve a wide range of end users. Hum. Mutat. 2012;33:803–808. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brinkman RR, Courtot M, Derom D, Fostel JM, He Y, Lord P, Malone J, Parkinson H, Peters B, Rocca-Serra P, et al. Modeling biomedical experimental processes with OBI. J. Biomed. Semantics. 2010;1:S7. doi: 10.1186/2041-1480-1-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat. Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]


