Abstract
PortEco (http://porteco.org) aims to collect, curate and provide data and analysis tools to support basic biological research in Escherichia coli (and eventually other bacterial systems). PortEco is implemented as a ‘virtual’ model organism database that provides a single unified interface to the user, while integrating information from a variety of sources. The main focus of PortEco is to enable broad use of the growing number of high-throughput experiments available for E. coli, and to leverage community annotation through the EcoliWiki and GONUTS systems. Currently, PortEco includes curated data from hundreds of genome-wide RNA expression studies, from high-throughput phenotyping of single-gene knockouts under hundreds of annotated conditions, from chromatin immunoprecipitation experiments for tens of different DNA-binding factors and from ribosome profiling experiments that yield insights into protein expression. Conditions have been annotated with a consistent vocabulary, and data have been consistently normalized to enable users to find, compare and interpret relevant experiments. PortEco includes tools for data analysis, including clustering, enrichment analysis and exploration via genome browsers. PortEco search and data analysis tools are extensively linked to the curated gene, metabolic pathway and regulation content at its sister site, EcoCyc.
INTRODUCTION
The central role of Escherichia coli research in the history of molecular genetics, systems biology and synthetic biology make the data generated from E. coli important not only for this model organism, but also for bacteria in general, including environmental sequencing and human microbiome studies. High-throughput molecular biology technologies are transforming biological research, making it possible to probe the detailed systems responses of organisms to perturbations in their genetics or environment. A large number of such data sets have been, and continue to be, collected for E. coli, one of the best-studied bacterial model organisms. PortEco (http://porteco.org) is a data resource that provides access to data and tools to allow users to efficiently find and integrate information from more than half a century of basic research on laboratory E. coli, its phages, plasmids and mobile genetic elements.
PortEco’s mission is to support bacterial research, by facilitating access to the massive (and continually growing) volume of experimental data for E. coli, and eventually other bacterial model systems. Making these data truly accessible requires both data handling—collection, consistent and updated processing, curation (e.g. creation of accurate data descriptions)—and databases and intuitive software for users to find and analyze existing data to help pose or answer novel research questions. PortEco is designed to be a ‘central point of access’ for such data, but it does not seek to reinvent the wheel. EcoCyc (1) already provides curated, review-level data for E. coli genes, metabolic pathways and, in collaboration with RegulonDB (2), operons and gene regulatory interactions. PortEco, by contrast, focuses on high-throughput experimental data and analysis tools, described in detail below, as well as covering genetics data and information about E. coli plasmids and phage. PortEco, through EcoliWiki (3) and GONUTS (4), also provides community input for a variety of areas. As researchers need to quickly navigate between all of these data sources, PortEco and EcoCyc have extensive reciprocal links, and the PortEco integrated search simultaneously searches PortEco and EcoCyc, as well as other, more specialized, data resources. Together, these resources create a more complete and powerful solution for the needs of researchers using E. coli as a model system for microbiology and molecular biology, for biotechnology, or as a platform for systems and synthetic biology.
INTEGRATED SEARCH
The PortEco search is designed to be a ‘one-stop’ search for information about E. coli. Searches are ‘comprehensive’, including not only PortEco data sources, but also other databases with E. coli information. On the technical side, the searches of all different data sources are carried out simultaneously via web services, and the results page is continually updated as search results arrive from each source using AJAX (asynchronous Javascript and XML). Currently, PortEco searches 16 different data sources (Table 1), and new resources that support web services-based queries can easily be added. By default, the search displays all results from each resource that are associated with the search term. However, the PortEco search is also ‘context-sensitive’: it automatically detects if the user has entered a gene name or synonym, and filters and formats the results into a ‘gene view.’ The gene view displays only those results obtained for the specific gene, and performs additional queries for more detailed information about that gene (Table 2). Users can still view the ‘full results’ even for a gene query, by clicking on the ‘view full results’ button at the top of the gene view.
Table 1.
Resources integrated into PortEco search
| Resource | Data | Maintained by PortEco |
|---|---|---|
| BioModels (5) | Quantitative models for simulating metabolic and regulatory systems | No |
| EcoCyc (1) | Genes, pathways, operons | No (sister site) |
| EcoGene (6) | Genes | No |
| EcoliWiki (3) | Genes, strains, alleles | Yes |
| GenExpDB (http://genexpdb.ou.edu) | Gene expression profiles | No |
| NCBI (7) | Genes | No |
| PANTHER (8) | Gene families and orthologs of E. coli genes | Yes |
| Pathway Commons (9) | Pathways and protein interactions | No |
| Protein Data Bank (10) | Protein 3D structures (experimental) | No |
| Protein Model Portal (11) | Protein 3D structures (both experimental and models) | No |
| PortEco GBrowse | Gene location, ChIP, ribosome profiling, RNA-seq | Yes |
| PortEco Gene Expression | Gene expression profiles; expression conditions | Yes |
| PortEco Phenotype | Knockout phenotype data | Yes |
| PortEco Textpresso | Publications (full text search) | Yes |
| STRING (12) | Predicted interacting genes | No |
| UniProt (13) | Proteins | No |
Table 2.
Additional information retrieved for PortEco search results for genes
| Additional information | Source |
|---|---|
| Thumbnail image of genomic location, links to ChIP, ribosome profiling, RNA-seq | PortEco GBrowse |
| Gene summary information | EcoCyc |
| Thumbnail image of most significant differential expression conditions | PortEco Gene Expression |
| Thumbnail image of most significant growth phenotype conditions | PortEco Phenotype |
| Knockout phenotypes, protein localization, gene essentiality | GenoBase (14) |
| Mutant alleles and phenotypes | EcoliWiki |
| Available mutant strains | EcoliWiki |
| Comprehensive lists of subfamily and family members | InterPro (15) |
PORTECO DATA: COLLECTION, PROCESSING AND CURATION
PortEco is collecting, processing and curating data from experiments in E. coli. These data types currently include the following:
Genome-scale mRNA expression data
Alleles and phenotype data for E. coli mutant strains from both curated articles and genome-scale growth experiments
Genomic features of E. coli plasmids and phage
Genome-scale protein–DNA interactions from chromatin immunoprecipitation (ChIP) experiments and genomic SELEX experiments
Genome-scale ribosome profiling data
An interactive, community-editable E. coli strain genealogy
Gene Ontology (GO) annotations of gene functions (in collaboration with EcoCyc)
Gene family trees and orthologs of E. coli genes in representative species
A corpus of E. coli scientific literature
mRNA expression data
At PortEco, we collect publicly available microarray data, with the vast majority of these data being taken from ArrayExpress (16) or the Gene Expression Omnibus [GEO, (17)]. To allow results from different laboratories and different experiment sets to be compared with one another, raw data are processed and normalized using a standard procedure before being made available at PortEco. The processing pipeline includes associating each probe on a particular microarray platform with the correct genomic coordinates [by remapping probes to the current genome sequence, many probes were designed before the current version of the sequence (18)], and associating those coordinates with the correct gene name, and an extensive list of synonyms where synonyms exist. This allows data to be retrieved regardless of what gene identifier might have been used when the data were first deposited. Each experiment (microarray) is manually curated: information about growth and treatment conditions is collected, along with names and genotypes of the strains used. The descriptions of experimental conditions accompanying publicly deposited microarray data are often abbreviated and sometimes incomplete. In such cases, we turn to the associated publications and citations therein to track down experimental details. When necessary, we contact authors for further information. In a similar fashion, we try to obtain complete genotypes for strain(s) used and, whenever possible, determine strain lineages. Details about strain constructions and lineages are entered on strain pages at EcoliWiki (for example, <http://ecoliwiki.net/colipedia/index.php/Category:Strain:BW25113> contains information about BW25113, the strain background for the Keio knockout collection). Another part of the curation process assigns each experiment (microarray) to an experimental condition category; this allows users to search for microarrays that may be related using those categories as queries. We are collaborating with the RegulonDB (2) and COLOMBOS (19) groups to establish a common set of condition terms.
At PortEco, we currently have data associated with 193 publications that have been published over the past 12 years. These data are normalized, converted to log ratios if necessary (for example, single channel Affymetrix data are converted to ratio style measurements by using either a control array as the denominator, or by using a probe’s average intensity in the data set as the denominator), and then clustered. We note that GenExpDB (http://genexpdb.ou.edu/) has some similar functionality to our expression site, also containing expression data for E. coli imported from GEO. For a given gene or genes entered into the GenExpDB search box, a heatmap can be retrieved for those genes’ expression across all conditions for which they have data available. However, GenExpDB does not provide the ability to cluster data for arbitrary genes across an arbitrary set of conditions, nor does it annotate the conditions with a consistent set of controlled vocabulary terms, instead relying on the meta-data imported from GEO. In addition, it does not provide a means by which to select the most significantly expressed genes from any given condition or set of conditions. These functionalities are all currently available from PortEco (see below).
With the advent of high-throughput sequencing, researchers now have the ability to not only determine with unprecedented detail which parts of the genome are actually transcribed, but in addition, can quantify at what level they are transcribed over a linear range spanning 5 orders of magnitude, at least two more orders than possible with microarrays (20). While there are few RNA-Seq data sets currently available for E. coli, these data sets are expected to be generated with increasing frequency, as fewer experiments are performed using microarray technology. At PortEco, we are developing standard pipelines to take the raw read data (in fastq format), and to map these data to the latest version of the genome, and to then determine expression values for each gene (in rpkm) using the latest genome annotation. As the genome sequence is updated, and as the primary annotation of the genome changes (for example, with newly described transcripts), we will be able to reprocess all data sets using the same pipeline, to provide consistent and comparable results across all RNA-Seq data sets.
Alleles and phenotypes
EcoliWiki gene pages contain >16 000 entries for alleles or phenotypes for E. coli genes. These alleles and phenotypes are a combination of alleles imported from the records of the E. coli Genetic Stock Center (21) and information from manual curation of the E. coli genetics literature. As part of EcoliWiki, these pages are available for community curation.
Nichols et al. performed large-scale determination of growth phenotypes for 3979 mutants under 324 conditions representing 114 distinct stresses (22). This data set provides a rich source of functional insights from comparison of phenotypic profiles between genes and conditions. PortEco provides two systems for browsing data from this study. The original data browser allows users to query and browse the fitness data and correlations from the authors between strains or conditions. This phenotypic profiles data browser, which was linked in the article, is one of the most heavily accessed components of PortEco. Integration with EcoliWiki allows the search to recognize records by the current gene names and synonyms. In a second-generation data browser, we have adapted the GeneXplorer system (23) used for expression data to allow users to recluster and analyze subsets of the large-scale growth phenotypes. This system provides the significant phenotypes section displayed by the PortEco search.
Genome scale protein–nucleic acid interaction data
Visualizing the locations of protein–nucleic acid interactions in the context of genes and the genome provides valuable insights about central dogma processes (replication, transcription and translation) and their regulation. Experiments of this kind include ChIP-chip (24–31), Chip-Seq (29,32) and ribosome profiling (33–35). PortEco identifies studies of these kinds in the literature and by contact with authors for studies in preparation. Data are obtained from repositories or supplemental data or, in some cases, by contacting the authors. If necessary, data are background corrected, renormalized and converted to standard file formats for display in the EcoCyc genome browser, our own GBrowse (36,37) instance, and our JBrowse (38) test site. The converted data are downloadable in common file formats (gff, wiggle, bam) for viewing in other browsers.
Information about each data set is associated with an EcoliWiki page for the relevant publication. Publications associated with browser tracks are placed in EcoliWiki categories to help users find data sets of interest among the growing corpus of experiments, and tracks from each publication are annotated in a table that allows the complete set of tracks or arbitrary subsets of tracks in EcoliWiki to be displayed in other wiki pages.
The PortEco GBrowse and JBrowse genome browsers allow users to view these data in the context of curated genomic features. We provide browsers for multiple E. coli strains, plasmids and bacteriophage. Default tracks are generated from RefSeq and Genbank records, but alternative tracks provide alternative annotations, such as operons from RegulonDB, locations of cloned inserts, known deletions and other manually curated content.
E. coli strain genealogies
In EcoliWiki, PortEco provides community-editable information about >280 strains. Stain information includes genotypes, references, construction details and sources for obtaining the strain. Strains are arranged in genealogies based on their construction. We currently include all of the strains described in the genealogies for E. coli K-12 and E. coli B described by Bachmann [in (39) and Daegelen et al. (40), respectively]. PortEco also supports pathway-genome databases for several E. coli strains, allowing comparison of these strains using BioCyc tools.
GO annotations of gene function
PortEco and EcoCyc collaborate to maintain and update the annotation of E. coli gene function for the GO consortium (41,42). We regularly aggregate and deposit an up-to-date gene annotation file that is downloadable from either PortEco or the GO consortium Web site. This file is constructed from combining annotations from UniProt with the professionally curated GO annotations from EcoCyc and community annotations from EcoliWiki and GONUTS, which provides a community GO annotation system for any protein in UniProt.
Orthologs and gene family trees
E. coli gene families and phylogenetic trees are generated and curated in collaboration with the PANTHER database (8). Currently, 2657 genes (64% of protein-coding genes) have been placed in phylogenetic trees. ‘Strict’ orthologs (i.e. genes related by vertical descent from a common ancestor) are computed from these trees in 81 other organisms (listed at http://pantherdb.org/panther/summaryStats.jsp). Hidden Markov models are created for both families and subfamilies, to allow searching for related genes in other genomes. These Hidden Markov models are run regularly on the UniProt database as part of the InterPro project (15), so users can navigate to comprehensive lists of related genes.
E. coli literature
EcoliWiki contains wiki pages for >25 000 publications. These pages allow community-editable addition of notes and discussion, links to other PortEco content and data tables for data mining, such as the track information tables described above. Articles covered in EcoliWiki are used to automatically update the literature corpus for full-text indexing by the PortEco instance of Textpresso (43), which has been modified to provide a more user-friendly interface and to provide a web service to provide relevant articles to the integrated PortEco search.
EcoliHouse: database access to gene information
EcoliHouse is a database warehouse containing multiple E. coli databases. EcoliHouse serves two purposes within PortEco. First, it is a publicly queryable MySQL database that allows scientists to issue SQL queries across multiple E. coli databases. Second, it is the database to which the PortEco web-based multigene query system sends queries to access the EcoCyc and EcoGene databases. The databases currently present within EcoliHouse are EcoCyc, EcoGene, Eco2Dbase, the UniProt complete proteome for E. coli K-12, the RefSeq E. coli K-12 MG1655 genome entry, and the Genbank E. coli K-12 MG1655 genome entry and several E. coli ChIP-chip data sets. See http://biowarehouse.ai.sri.com/EcoliHouseOverview.html for a listing of the current databases within EcoliHouse, EcoliHouse access instructions and example queries.
HIGH-THROUGHPUT DATA ANALYSIS WORKFLOWS
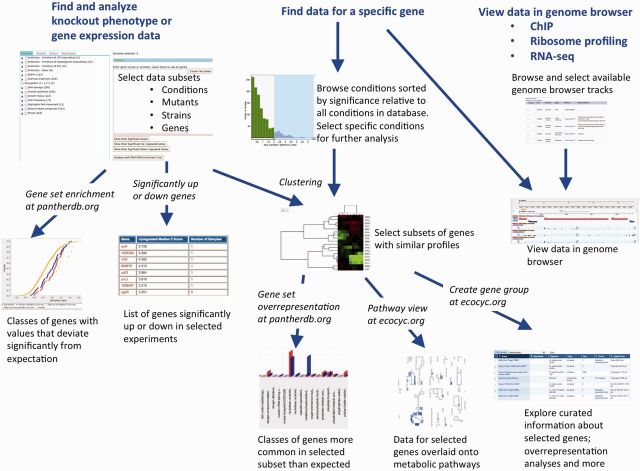
PortEco is designed to facilitate retrieval and analysis of high-throughput data sets that have been generated for E. coli (Figure 1). There are three starting points for accessing E. coli data in PortEco: (i) search for a specific gene, (ii) search for a specific set of experimental conditions (for either gene expression or growth phenotype data) and (iii) search for a specific set of experiments to view in a genome browser. PortEco uses the GeneXplorer tool (23) for display of gene expression and knockout growth phenotype data, which in PortEco is now seamlessly integrated with analysis tools from the PANTHER and EcoCyc Web sites. PortEco currently uses GBrowse (36) as a genome browser, though Jbrowse (44) is currently available on a testing site and will be fully released in the near future.
Figure 1.
Main workflows supported for retrieving and analyzing high-throughput data sets at PortEco. There are three entry points (blue text at top), and at each intermediate step the user can choose between several different paths for further analysis and exploration.
Search for a specific gene
Searching for a gene name, synonym or accession launches the PortEco gene search results view (see ‘Integrated Search’ above). From here, users can click on the genome browser link to view the genomic context and select ChIP, ribosome profiling and RNA-seq tracks to add to the view. Users will see a thumbnail of conditions where mRNA expression of that gene is up- or downregulated, and another thumbnail of conditions where the knockout of that gene has increased or decreased growth rate. Clicking on the link to analyze all data (for either expression or growth phenotype) will launch the Samples and Conditions view of the GeneXplorer tool, allowing the user to (i) browse the conditions that have the most significantly increased or decreased expression or growth, and (ii) select subsets of conditions for clustering. This allows users to find genes that are correlated with the gene of interest specifically under those conditions where the gene of interest shows a significant expression change or phenotype. Focusing on specific conditions helps to avoid spurious correlations driven by the majority of conditions where there is little or no effect on the expression or knockout phenotype of most genes. Note that because this point of entry provides the ability to retrieve data from many unrelated experiments, the notion of using log ratio data is not necessarily applicable as it is when analyzing a coherent data set from a single publication. Thus, all data are transformed into Z-scores, which indicate, in that experiment, how many standard deviations above or below the mean was a particular gene’s expression or phenotype value.
The Samples and Conditions view displays a histogram with the Z-scores for that gene’s expression or phenotype data and a list of the experiments where the Z-score for the gene is above a user-selected threshold. Once conditions of interest have been selected, the data for all genes in those conditions can be clustered, and a GeneXplorer window then shows global and zoomed ‘heatmap’ views for the clustered data. Within the zoomed view, users can see gene names, product descriptions and links to resources for more information. At this point, users have a number of options. For any particular gene they can get a list of other genes with the most highly correlated and anti-correlated expression patterns or phenotypic profiles across the selected conditions. Subclusters of genes and data can be selected and further analyzed in a number of ways, including finding overrepresented pathways/processes, viewing in the EcoCyc ‘cellular overview’ tool or sending to the EcoCyc ‘groups’ tool (1).
Search for a specific set of experimental conditions
Using the ‘cluster my genes’ tool, users can browse the available experiments for selection. As described above, the experiments have been classified manually by the type of experimental conditions, the strain(s) used, the specific mutant (if applicable) and the publication. Users can select data sets by any of these criteria, and optionally enter a subset of genes (all genes are considered by default). They can then (i) retrieve a list of genes that are significantly up or down in the selected experiments (based on Z-scores relative to all experiments in the database, as described above), (ii) analyze those conditions for enriched biological pathways/processes or (iii) cluster the patterns for different genes under the selected conditions. Selected genes and data sets are then retrieved and clustered, and displayed using GeneXplorer. Clusters can be further analyzed as described above.
Search for a specific set of experiments to view in a genome browser
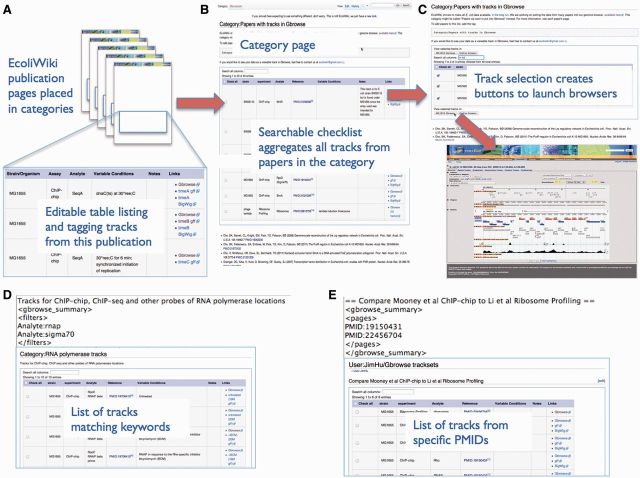
Figure 2 illustrates the use of EcoliWiki to manage and personalize views of track collections for high-throughput data. The curation of track data in EcoliWiki publication pages allows us to generate interactive tables of available data sets. These list the author and publication, the type of experiment, a brief description and the strains used. Entering a search term will dynamically filter the table to include only those entries matching the term (e.g. by entering ‘ribosome profiling’ the table will be reduced to only those types of experiments). The user can then select the data sets to launch in a genome browser. In addition to the global listing of data tracks, users can create their own custom views of subsets of tracks based on querying the global set of browser tracks from high-throughput data.
Figure 2.
(A) EcoliWiki pages for publications related to track data include a user-editable table listing relevant track data, with links to genome browsers and data files. These pages can be tagged to place them into EcoliWiki Categories. (B) A tag extension on the appropriate Category page creates a summary table aggregating all available high-throughput tracks from Category members. (C) The table can be sorted and searched for keywords and used to launch GBrowse views with the selected tracks enabled. Custom track lists can be created based on (D) keyword matches or (E) lists of PubMed IDs.
PREPUBLICATION SERVICES
In addition to allowing users to compare their data sets with publicly available data sets, users can use Porteco tools to create password-protected private views of their data. Private views of data that can be visualized as genome browser tracks, such as genome-scale protein–DNA interactions, ribosome profiling or alternative genome annotations, can be created using the custom tracks capabilities of GBrowse (36,45). This allows users to view their data in the context of other work and existing annotations. GBrowse allows users to do this without even having to tell PortEco about it. Reviewers can be provided with access to these private before publication. However, working with PortEco, we can move these temporary custom tracks into the permanent collection so that stable URLs can be included in manuscripts and the data can be opened to the public on publication. For example, Myers et al. (29) was able to provide links for ChIP-chIP and ChIP-seq data sets, while Liu et al. (34) used the PortEco browser for ribosome profiling data mapped against both the E. coli K-12 and bacteriophage lambda genomes.
In other cases, the data of interest is a set of tabular data where we can create custom web-based tools to analyze and then provide public access. We have constructed a framework to quickly construct access-controlled custom views of tabular data. Unlike tabular data in Excel or Google Spreadsheets, we can easily leverage PortEco so that tables can be searched using synonyms for accessions or gene names in the user data sets, and links from the tables to PortEco or EcoliWiki can be built in more easily than if authors built and maintained their own web interfaces for supplemental data. This approach was used to provide data browsers for the Nichols et al. (22) phenotypic profile data and the analysis of the stress-induced mutagenesis network by Al-Mamun et al. (46). As with browser tracks, we can provide URLs to the public view of the data to be included in publications. These capabilities allow a greater subset of the research community to use published data in ways that will increase the citation of the articles including these links.
CONCLUSION
PortEco has been designed to leverage and integrate with the wealth of bioinformatics data resources that include information related to E. coli. Leverage and integration are also key to how PortEco combines and extends available open-source software. Our two wiki projects leverage the broader expertise of the research community and illustrate how MediaWiki can be used to quickly build community resources for different kinds of information. In this way PortEco provides important content for use by researchers using E. coli as a model system, and illustrates a virtual model organism database approach to building a data resource.
FUNDING
National Institutes of Health (NIH) [1U24GM088849]. Funding for open access charge: NIH [1U24GM088849].
DEDICATION
The authors dedicate this article to the memory of Monica Riley (1926–2013), a pioneer in the genome biology of E. coli.
Conflict of interest statement. None declared.
REFERENCES
- 1.Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martinez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salgado H, Peralta-Gil M, Gama-Castro S, Santos-Zavaleta A, Muniz-Rascado L, Garcia-Sotelo JS, Weiss V, Solano-Lira H, Martinez-Flores I, Medina-Rivera A, et al. RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res. 2013;41:D203–D213. doi: 10.1093/nar/gks1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIntosh BK, Renfro DP, Knapp GS, Lairikyengbam CR, Liles NM, Niu L, Supak AM, Venkatraman A, Zweifel AE, Siegele DA, et al. EcoliWiki: a wiki-based community resource for Escherichia coli. Nucleic Acids Res. 2012;40:D1270–D1277. doi: 10.1093/nar/gkr880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renfro DP, McIntosh BK, Venkatraman A, Siegele DA, Hu JC. GONUTS: the gene ontology normal usage tracking system. Nucleic Acids Res. 2012;40:D1262–D1269. doi: 10.1093/nar/gkr907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chelliah V, Laibe C, Le Novere N. BioModels Database: a repository of mathematical models of biological processes. Methods Mol. Biol. 2013;1021:189–199. doi: 10.1007/978-1-62703-450-0_10. [DOI] [PubMed] [Google Scholar]
- 6.Zhou J, Rudd KE. EcoGene 3.0. Nucleic Acids Res. 2013;41:D613–D624. doi: 10.1093/nar/gks1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013;41:D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas J, Roth S, Arnold K, Kiefer F, Schmidt T, Bordoli L, Schwede T. The Protein Model Portal–a comprehensive resource for protein structure and model information. Database (Oxford) 2013;2013:bat031. doi: 10.1093/database/bat031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.UniProt Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rustici G, Kolesnikov N, Brandizi M, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Ison J, Keays M, et al. ArrayExpress update—trends in database growth and links to data analysis tools. Nucleic Acids Res. 2013;41:D987–D990. doi: 10.1093/nar/gks1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, et al. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100049. 2006.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelen K, Fu Q, Meysman P, Sanchez-Rodriguez A, De Smet R, Lemmens K, Fierro AC, Marchal K. COLOMBOS: access port for cross-platform bacterial expression compendia. PLoS One. 2011;6:e20938. doi: 10.1371/journal.pone.0020938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 21.Berlyn MB, Letovsky S. Genome-related datasets within the E. coli Genetic Stock Center database. Nucleic Acids Res. 1992;20:6143–6151. doi: 10.1093/nar/20.23.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rees CA, Demeter J, Matese JC, Botstein D, Sherlock G. GeneXplorer: an interactive web application for microarray data visualization and analysis. BMC Bioinformatics. 2004;5:141. doi: 10.1186/1471-2105-5-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho BK, Barrett CL, Knight EM, Park YS, Palsson BO. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl Acad. Sci. USA. 2008;105:19462–19467. doi: 10.1073/pnas.0807227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grainger DC, Aiba H, Hurd D, Browning DF, Busby SJ. Transcription factor distribution in Escherichia coli: studies with FNR protein. Nucleic Acids Res. 2007;35:269–278. doi: 10.1093/nar/gkl1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grainger DC, Hurd D, Goldberg MD, Busby SJ. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34:4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herring CD, Raffaelle M, Allen TE, Kanin EI, Landick R, Ansari AZ, Palsson BO. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J. Bacteriol. 2005;187:6166–6174. doi: 10.1128/JB.187.17.6166-6174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers KS, Yan H, Ong IM, Chung D, Liang K, Tran F, Keles S, Landick R, Kiley PJ. Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet. 2013;9:e1003565. doi: 10.1371/journal.pgen.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R. Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl Acad. Sci. USA. 2009;106:15406–15411. doi: 10.1073/pnas.0903846106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez-Romero MA, Busby SJ, Dyer NP, Ott S, Millard AD, Grainger DC. Dynamic distribution of SeqA protein across the chromosome of Escherichia coli K-12. MBio. 2010;1:e00012. doi: 10.1128/mBio.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, Benes V, Fraser GM, Luscombe NM. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 2011;39:2073–2091. doi: 10.1093/nar/gkq934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Jiang H, Gu Z, Roberts JW. High-resolution view of bacteriophage lambda gene expression by ribosome profiling. Proc. Natl Acad. Sci. USA. 2013;110:11928–11933. doi: 10.1073/pnas.1309739110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto K, Ishihama A, Busby SJ, Grainger DC. The Escherichia coli K-12 MntR miniregulon includes dps, which encodes the major stationary-phase DNA-binding protein. J. Bacteriol. 2011;193:1477–1480. doi: 10.1128/JB.01230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein LD. Using GBrowse 2.0 to visualize and share next-generation sequence data. Brief. Bioinform. 2013;14:162–171. doi: 10.1093/bib/bbt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachmann BJ. Derivatives and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt FC, editor. Escherichia coli and Salmonella Cellular and Molecular Biology. 2nd edn. Washington, DC: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 40.Daegelen P, Studier FW, Lenski RE, Cure S, Kim JF. Tracing ancestors and relatives of Escherichia coli B, and the derivation of B strains REL606 and BL21(DE3) J. Mol. Biol. 2009;394:634–643. doi: 10.1016/j.jmb.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 41.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu JC, Karp PD, Keseler IM, Krummenacker M, Siegele DA. What we can learn about Escherichia coli through application of Gene Ontology. Trends Microbiol. 2009;17:269–278. doi: 10.1016/j.tim.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westesson O, Skinner M, Holmes I. Visualizing next-generation sequencing data with JBrowse. Brief. Bioinform. 2013;14:172–177. doi: 10.1093/bib/bbr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stein LD, Thierry-Mieg J. Scriptable access to the Caenorhabditis elegans genome sequence and other ACEDB databases. Genome Res. 1998;8:1308–1315. doi: 10.1101/gr.8.12.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al Mamun AA, Lombardo MJ, Shee C, Lisewski AM, Gonzalez C, Lin D, Nehring RB, Saint-Ruf C, Gibson JL, Frisch RL, et al. Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science. 2012;338:1344–1348. doi: 10.1126/science.1226683. [DOI] [PMC free article] [PubMed] [Google Scholar]