Abstract
The Global Genome Biodiversity Network (GGBN) was formed in 2011 with the principal aim of making high-quality well-documented and vouchered collections that store DNA or tissue samples of biodiversity, discoverable for research through a networked community of biodiversity repositories. This is achieved through the GGBN Data Portal (http://data.ggbn.org), which links globally distributed databases and bridges the gap between biodiversity repositories, sequence databases and research results. Advances in DNA extraction techniques combined with next-generation sequencing technologies provide new tools for genome sequencing. Many ambitious genome sequencing projects with the potential to revolutionize biodiversity research consider access to adequate samples to be a major bottleneck in their workflow. This is linked not only to accelerating biodiversity loss and demands to improve conservation efforts but also to a lack of standardized methods for providing access to genomic samples. Biodiversity biobank-holding institutions urgently need to set a standard of collaboration towards excellence in collections stewardship, information access and sharing and responsible and ethical use of such collections. GGBN meets these needs by enabling and supporting accessibility and the efficient coordinated expansion of biodiversity biobanks worldwide.
INTRODUCTION
Genome sequencing for biodiversity analysis is at the forefront of innovation and discovery due to technological advances and the sequencing of whole genomes in the last 10 years. Information generated from biodiversity genomics will revolutionize our approach to taxonomy, phylogeny, conservation, ecological monitoring, wildlife management, agriculture, drug development, zoonotic disease forecasting and even aspects of national security. Consequently, the demand is rapidly increasing for professionally preserved, managed and documented samples that yield high-molecular weight DNA and RNA from throughout the tree of life [e.g. (1,2)]. Many ambitious projects with the potential to revolutionize biodiversity research are finding access to adequate samples needed for genome sequencing to be a major bottleneck in their workflow. Examples of these projects include the Ten Thousand Vertebrate Genomes Project (Genome10 K), the Global Invertebrate Genomics Alliance (GIGA) and the International Barcode of Life Project (iBOL) (see also additional data at NAR online).
Table 1 gives an overview on certain terms used in this article.
Table 1.
Explanation of specific terms used in this article
| Term | Explanation |
|---|---|
| Biodiversity repository | A publicly accessible curated collection of biological material (in general excluding human material). Examples include museums, herbaria, botanical gardens, biobanks, seed banks, and zoos |
| Biodiversity biobank | A subset of biodiversity repositories that store DNA, RNA or tissue samples of biodiversity |
| Genome sequencing | Genome, metagenome, transcriptome, and marker sequencing |
| Genome quality | High-molecular weight DNA or RNA including the whole genome |
| Biodiversity genomics | Genome sequencing and analysis for biodiversity research |
| Genomic sample | Any biological material preserved to keep its molecular properties (in general excluding human material). Examples include DNA, RNA, and tissue |
To give an example, an analysis of data related to Corvidae, a popular and well-known bird family with ∼420 taxa, shows that a comparatively small number of sequences are discoverable and only a small proportion of these sequences are accompanied by useful voucher information. Fifty six percent (4083) of the 7250 corvid sequences found at the International Nucleotide Sequence Data Base Collaboration (INSDC, http://www.insdc.org/) (3) are related to only four taxa (Corvus corone cornix, Corvus corone corone, Pica pica and Cyanopica cyanus). Although corvids are a well-known group, they are still hiding undiscovered phylogenetic secrets [see e.g. (4)]: fifty percent of all extant and 100% of all extinct corvid taxa lack sequences.
Out of the 7250 corvid sequences, 56% (4026) lack the tag ‘specimen_voucher’ and only 21% (1548) of the sequence records are associated with a voucher number (e.g. UWBM 61 493) linking these records directly to the physical voucher specimen. Only 409 corvid voucher records associated with 860 corvid sequences (12% of all corvid sequences, 56% of vouchered corvid sequences) can be found through the Global Biodiversity Information Facility (GBIF) portal, a small number compared to the > 9 million available corvid occurence records. An adequate match for finding non-standardized voucher information is only possible through time-consuming manual searches (supporting data are available at NAR online).
A major goal of the Global Genome Biodiversity Network (GGBN) is to effectively bridge such gaps, as well as to encourage and enable scientists to complete documentation chains between vouchers, tissues, physical DNA, sequences and publications. Figure 1 illustrates the current situation described earlier in text and could be described for countless further examples.
Figure 1.
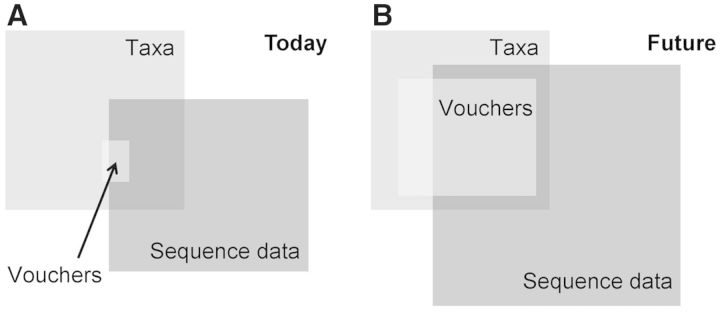
Schematic representation of (A) the current state of play regarding the proportion of sequence data available in public repositories with proper voucher specimen information, and (B) what GGBN aims to facilitate in coming years, i.e. increase both the number of taxa sequenced and the proportion of taxa sequenced for which a voucher specimen information is readily available. Box sizes are illustrative only and do not represent actual proportions.
Natural history collections play an important role in catering to the need for access to high-quality well-documented samples for genome sequencing. They have archived biodiversity specimens over centuries, often including taxa or populations that have now vanished from the wild. These collections allow researchers to access biological specimens and their data efficiently and often make further expensive field trips and subsequent time-consuming species identifications unnecessary. Biodiversity biobanks form an emerging collection type that complements classical natural history collections. While often conveniently associated with these traditional collections, biodiversity biobanks are much better suited for modern downstream molecular applications (e.g. second- and third-generation sequencing requiring high-quality DNA), as their focus lies explicitly on conserving the molecular structure of their samples. Biodiversity biobanks adhere to standard operating procedures within a sustainable collection management infrastructure to guarantee integrity, identity and ethical use of samples. They commit themselves to making samples lastingly available and accessible. By using internationally accepted standards for biodiversity information, federation and synthesis of data among collection databases is possible.
Until now, no central platform existed that would globally aggregate biodiversity biobank data. While the need for data sharing and sample access increases, information on genomic samples is still as fragmented as the geographic distribution of the repositories that maintain them. Without a central data portal that aggregates genomic sample data, biodiversity genomics studies cannot reach their full potential. The German Research Foundation (DFG)-supported DNA Bank Network was established in 2007 as a first step towards this goal (5). The GGBN aims at closing this gap on a global scale by building on the DNA Bank Network’s data model.
THE NETWORK’S MISSION
GGBN is an open network of currently 24 biodiversity repositories from across all continents (see additional data at NAR online) that came together with the aim to endorse biodiversity biobank stewardship, information sharing and ethical use of collections in compliance with national and international conventions and regulations (Convention on Biological Diversity, http://www.cbd.int/convention/text/and Nagoya Protocol on access and benefit-sharing (ABS), http://www.cbd.int/abs/doc/protocol/nagoya-protocol-en.pdf).
Key objectives of this global endeavour include establishing and adopting standards and best practices, accelerating access to specimens and data and enabling targeted collection. GGBN’s most central mission is to promote access to information on the genomic samples maintained by its members. Goals include to:
provide genome-quality DNA and tissue samples from across the tree of life for research and training;
provide free access to a global Data Portal hosting the aggregated primary specimen and sample data as well as metadata of all member institutions;
develop standards for sharing DNA and tissue information;
develop best practices related to management and stewardship of genomic samples and their derivatives, including appropriate access and benefit-sharing;
establish an infrastructure for deposition of genomic samples;
promote targeted collection and preservation of genomic samples representing a synoptic sample of life on earth;
provide a platform for biodiversity biobanking knowledge exchange;
recruit partners with different regional and taxonomic foci, to preserve global genetic diversity in an efficient, planned and concerted effort.
Global biodiversity and collections initiatives around the world such as biodiversity observatories, genome projects, monitoring and barcoding campaigns will both contribute to and rely on GGBN resources for reaching their research goals (see additional data at NAR online).
GGBN DATA PORTAL AND DATA INFRASTRUCTURE
GGBN’s Data Portal (http://data.ggbn.org) bridges the gap between biodiversity repositories, sequence databases and research results by linking globally distributed biodiversity databases of genomic samples, representing the biodiversity of life on Earth to voucher specimens, sequence data and publications. At present, >70 000 DNA and tissue samples from almost 17 000 taxa are available. These have been collected in accordance with the Convention on Biological Diversity (CBD), the Convention on International Trade in Endangered Species (CITES) and in compliance with all relevant conventions with regard to the acquisition, maintenance and use of DNA and tissue material. The Data Portal enables scientists to:
identify available resources held by GGBN members;
query, request and obtain DNA and tissue material to conduct new studies or to extend and complement previous investigations;
find collections for storing DNA and tissue samples under optimal conditions after project completion or data publication;
verify geographic locations of taxa and links to sources of genomic verification;
verify taxonomic identification through digital images;
support good scientific practice as the deposition of DNA samples and related specimens facilitates the verification of published results.
The GGBN Data Portal provides a necessary service. Natural history collections, worldwide, use different specialized collection database systems (e.g. Arctos, http://arctos.database.museum/home.cfm; Specify, http://specifysoftware.org/; KE EMu, http://www.kesoftware.com/en/ke-software-home/page-1.html; BRAHMS, http://herbaria.plants.ox.ac.uk/bol/; DiversityWorkbench (6); JACQ, http://sourceforge.net/p/jacq/_list/git?source=navbar; or in-house developments). Some collection database systems have been extended to fulfill the needs of curating a biodiversity biobank. An open-source software for the management of a biodiversity biobank has been developed in the framework of the DNA Bank Network (DNA Module, http://wiki.bgbm.org/dnabankwiki/index.php/DNA_Module).
GBIF (7), BioCASe (8) and the DNA Bank Network (5) have established an infrastructure to link biodiversity data from such systems. This is achieved by direct data integration from multiple distributed databases through wrapper software packages implementing agreed exchange protocols such as the BioCASe protocol, or DiGIR (Distributed Information Retrieval) (http://www.digir.net), and through use of publishing tools such as the IPT (Integrated Publishing Toolkit) (http://www.gbif.org/ipt). BioCASe uses ABCD (Access to Biological Collection Data) (9) and DiGIR uses Darwin Core (10) as XML data schemas. The IPT also works with Darwin Core, but the data is packaged in a Darwin Core Archive (11), in a text-based tabular format with mappings to Darwin Core terms.
Combining existing infrastructure
The GGBN Data Portal builds on GBIF’s and DNA Bank Network’s infrastructure. It allows the linkage from and to other biodiversity data information systems and provides biodiversity tissue and DNA data in a standardized way that is interoperable with all GBIF compliant data sources. The core of GGBN’s architecture is the central Data Portal and its underlying index and harvester. All recorded data belonging to a DNA or tissue sample are provided from stable disparate primary databases in each member location. As a result of a specific query, these data sets are compiled by the Network’s Data Portal into a synoptic virtual data set. The shared Data Portal facilitates and visualizes tissue and DNA data as well as specimen information of all available genomic resources and allows users to search for and request tissue and DNA in addition to allowing contributors to flag and mask sensitive data. Specimen data and digital images are rapidly imported from GBIF-compliant specimen databases.
Applying and extending the GBIF data model is advantageous for biodiversity biobanks. Local data sets are administered and updated by their providers so that redundant copies are avoided. The synoptic data set always represents the current state of knowledge. Further, GGBN’s Data Portal links to the INSDC (3), Barcode of Life Data Systems (BOLD, http://www.barcodinglife.org) (12) and Catalogue of Life (CoL, http://www.catalogueoflife.org/col/) (13), along with relevant publications. In the near future, it will furthermore link to Encyclopedia of Life (EOL, http://eol.org/) (14) and use the GBIF checklist bank. The Data Portal contains at least four interrelated types of data including vouchers, specimens, tissues, physical DNA and links to sequence data. All INSDC data portals collaborate with GBIF and recommend as best practice the submission of a voucher to a biodiversity repository and inclusion of the voucher information when submitting new sequence data. Linking to DNA repositories is an additional advantage for researchers searching for organisms and information.
Data access and sustainability
Login is only required for requesting DNA and tissue material. All members provide their data, based on the GBIF infrastructure, via web services (BioCASe) or Darwin Core Archive files (access points available via the registry at http://registry.ggbn.org). This enables third parties to retrieve data from our members. Furthermore, third parties are welcome to contact the Technical Manager of GGBN to get required data. To enable free access to genomic data via one central portal is a major goal of GGBN. The individual members may block individual samples from physical requests for certain reasons. The Botanic Garden and Botanical Museum Berlin-Dahlem (BGBM), Freie Universität Berlin is coordinating the DNA Bank Network (GGBN node with currently nine partners) and hosts the GGBN Data Portal as well as its precursor (data portal of the DNA Bank Network). The GGBN Data Portal is further developed, promoted and sustained in the framework of other projects and as a core activity of the BGBM’s Biodiversity Informatics Research Group.
GGBN use cases
Use case one
A biologist seeking the Asteraceae Cichorium intybus speeds her research by using the ‘one-stop access’ GGBN Data Portal to locate samples, specimens and data. She finds vouchered samples at the repositories A, B and C, one of which (deposited in A) has sequence data in INSDC portals. Images exist for the voucher at A. With one query, her request for access reaches all three institutions, each of which complies with GGBN guidelines on ethical uses of collections. She gains access to samples quickly and is confident that she can comply with the associated ABS requirements.
Use case two
A biologist seeks DNA sequences from Cypriniformes for cytb (cytochrome b gene) at GenBank to add to his phylogenetic tree. But some important Cyprinidae taxa are still missing to develop a well-supported tree. He finds vouchered DNA and tissue samples of missing taxa at the GGBN Data Portal and can easily order them online. After he has finished his project and has published the results, including well-cited DNA and tissue material, he sends feedback to the GGBN members from whom he received the samples. They can now update their database with new corresponding sequence accession numbers and publications. Individual scientists and the whole community gain more transparency in scientific research and a suitable infrastructure for good scientific practice.
Use case three
A Ph.D. student has finished her thesis on Agaricales at a university and no longer needs her samples. She has used the GGBN Data Portal earlier for her own study. Now she contacts one of the botanical GGBN members to deposit the DNA, tissue samples and voucher specimens from her study. After receiving a positive feedback she sends the physical material, collecting permissions and corresponding data to the GGBN member. Some of the samples are blocked for third parties, but nevertheless she can deposit them and third parties will not have access for a certain period of time. The samples and vouchers will be included in the collections of the respective GGBN member and the data will be made available via the GGBN Data Portal. Therefore, she makes use of a well-working infrastructure that allows long-term deposition for future research and verification, and she can focus on her next project.
Use case four
Members of the GIGA need access to information on specimens that could be used for future genomic studies. They require an accurate, searchable database of marine invertebrate (non-insect/non-nematode) genome-quality specimens that allows them to obtain lists of species names, literature references, current sample locations, sample origins and other vital metadata. GIGA members use the GGBN Data Portal as their main database of invertebrate genomic samples and use GGBN member institutions to contribute samples to the Network, allowing these samples to become discoverable through the GGBN Data Portal.
GGBN document library
Currently, a prototype of the GGBN document library is available at http://library.ggbn.org. Scientists can search for >4400 articles related to samples and vouchers provided via the GGBN Data Portal. Often, people visit the website after searching for a publication on the web. The articles are linked to relevant sample and voucher data.
GGBN metadata registry
The GGBN metadata registry holds collection level information of its members, and is available at http://registry.ggbn.org. Registry records currently use the Natural Collections Descriptions (NCD) data exchange standard (http://www.tdwg.org/standards/312/). This Registry helps visitors to get an overview on the GGBN members and the taxonomic and geographic scope of their collections.
Both the GGBN document library and GGBN metadata registry are hosted and supported by the BGBM and will be further developed, promoted and sustained in the framework of other projects and as a core activity of the BGBM’s Biodiversity Informatics Research Group.
FUTURE DIRECTIONS
Discoverability and coordinated expansion of biodiversity biobanks worldwide only becomes possible through a networked community of biodiversity repositories that provide trusted and transparent access to genomic samples. GGBN opens up the repositories of its member institutions to the scientific community via a shared central Data Portal that exposes collection information through a federated information infrastructure. GGBN furthermore provides a collaboration platform that helps in developing best practices for biodiversity repositories, ensures quality standards and harmonizes the exchange of samples in accordance with national and international legislation and conventions.
In times of rapid biodiversity loss and increased awareness of ABS requirements, as exemplified by the Nagoya Protocol (http://www.cbd.int/abs), the GGBN integrates widely distributed resources; it multiplies sample visibility of biodiversity repositories worldwide and unites the people who make them functional. Sharing their expertise, these people work towards the common goal of enabling more efficient and more sustainable research on biodiversity. The synergistic effect of this collaboration increases with each individual member, and GGBN welcomes and needs new members with a long-term commitment to responsible and ethical use of material deposited in biodiversity biobanks.
The GGBN Data Portal will provide web services, e.g. by using BioCASe, to enable related portals or any third party to get required data. The next version of the GGBN Data Portal will use the Harvesting and Indexing Toolkit (HIT, http://code.google.com/p/gbif-indexingtoolkit) provided by GBIF. The HIT will then be extended by the GGBN team based on the GGBN requirements, e.g. to enable multiple identifications and associations between single records.
The upcoming GGBN document library will permit users to perform full text searches for documents including publications, example use case documents on material transfer agreements (MTAs) and codes of conduct related to ABS.
The GGBN metadata registry will, in the future, use the GBIF registry via web services. Together with the GBIF developer team, the GGBN team will analyse and, if applicable, extend the GBIF registry to fulfill the requirements for biodiversity biobanks. In addition, DNA and tissue banks will be added to central registries such as Index Herbariorum (http://sweetgum.nybg.org/ih/) (15) and Registry of Biological Repositories (http://www.biorepositories.org).
Finally, GGBN intends to make the DNA and tissue data of 13 new core members discoverable between 2014 and 2016 as part of the European funded SYNTHESYS III project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation (DFG) [INST1039/1-1, INST17818/1-1, INST427/1-1, INST599/1-1]; the Smithsonian Grand Challenges Consortium for Understanding and Sustaining a Biodiverse Planet [no number assigned]; the Freie Universität Berlin [FMEx2-2013-045]; and the Danish Ministry of Science, Innovation and Higher Education [11-120379]. Funding for open access charge: Smithsonian Grand Challenges Consortium for Understanding and Sustaining a Biodiverse Planet/no number assigned.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank all reviewers for comments that improved the article. GGBN has been supported by grants from the Danish Ministry of Science, Innovation and Higher Education, the Freie Universität Berlin, the German Research Foundation and the Smithsonian Grand Challenges Consortium for Understanding and Sustaining a Biodiverse Planet. Work towards this effort has been a community effort and could not have occurred without the support from GGBN collaborator institutions and organizations.
REFERENCES
- 1.Pawlowski J, Audic S, Adl S, Bass D, Belbahri L, Berney C, Bowser S, Cepicka I, Decelle J, Dunthorn M, et al. CBOL protist working group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol. 2012;10:e1001419. doi: 10.1371/journal.pbio.1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown WY. DNA Net Earth. Global Views Policy Paper 2013-01. Washington, DC: Brookings Institution; 2013. [Google Scholar]
- 3.Nakamura Y, Cochrane G, Karsch-Mizrachi I, on behalf of the International Nucleotide Sequence Database Collaboration The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2013;41:D21–D24. doi: 10.1093/nar/gks1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.James HF, Ericson PGP, Slikas B, Lei FM, Gill FB, Olson SL, James HF, Ericson PGP, Slikas B, Lei F-M, et al. Pseudopodoces humilis, a misclassified terrestrial tit (Paridae) of the Tibetan Plateu: evolutionary consequences of shifting adaptive zones. Ibis. 2003;145:185–202. [Google Scholar]
- 5.Gemeinholzer B, Dröge G, Zetzsche H, Haszprunar G, Klenk H-P, Güntsch A, Berendsohn WG, Wägele J-W. The DNA Bank Network: the start from a German initiative. Biopreserv. Biobank. 2011;9:51–55. doi: 10.1089/bio.2010.0029. [DOI] [PubMed] [Google Scholar]
- 6.Triebel D, Neubacher D, Weiss M, Heindl-Tenhunen B, Nash TH, III, Rambold G. Integrated biodiversity data networks for lichenology–data flows and challenges. In: Nash TH III, Geiser L, McCune B, Triebel D, Tomescu AMF, Sanders WB, editors. Biology of Lichens - Symbiosis, Ecology, Environmental Monitoring, Systematics and Cyber Applications. Stuttgart: Biblioth. Lichenol; 2010. pp. 47–56. [Google Scholar]
- 7. GBIF (2011) GBIF Strategic Plan 2012-2016 - Seizing the Future, version 1.0, released March 2011. Global Biodiversity Information Facility, Copenhagen, p. 12. Accessed 2013 Aug 13. [Google Scholar]
- 8.Güntsch A, Mergen P, Berendsohn WG. The BioCASE Project - a Biological Collections Access Service for Europe. Ferrantia. 2007;51:103–108. [Google Scholar]
- 9.Holetschek J, Dröge G, Güntsch A, Berendsohn WG. The ABCD of rich data access to Natural History Collections. Plant Biosyst. 2012;146:771–779. [Google Scholar]
- 10.Wieczorek J, Bloom D, Guralnick R, Blum S, Döring M, Giovanni R, Robertson T, Vieglais D. Darwin core: an evolving community-developed biodiversity data dtandard. PLoS One. 2012;7:e29715. doi: 10.1371/journal.pone.0029715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. GBIF (2010) GBIF GNA Profile Reference Guide for Darwin Core Archives. version 1.0, released on 1 April 2011, Remsen,D.P., Döring,M., and Robertson,T. (eds). Global Biodiversity Information Facility, Copenhagen, p. 28. Accessed 2013 Aug 13. [Google Scholar]
- 12.Ratnasingham S, Hebert PDN. BOLD: the barcode of life data system. Mol. Ecol. Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roskov Y, Kunze T, Paglinawan L, Orrell T, Nicolson D, Culham A, Bailly N, Kirk P, Bourgoin T, Baillargeon G, et al. Species 2000 & ITIS Catalogue of Life. 2013. Species 2000, Reading, UK. [Google Scholar]
- 14.Wilson EO. The encyclopedia of life. Trends Ecol. Evol. 2003;18:77–80. [Google Scholar]
- 15.Thiers B. [continuously updated]. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. New York Botanical Garden's Virtual Herbarium. Accessed 2013 Aug 13. [Google Scholar]


