Abstract
As the number of prescribed drugs is constantly rising, drug–drug interactions are an important issue. The simultaneous administration of several drugs can cause severe adverse effects based on interactions with the same metabolizing enzyme(s). The Transformer database (http://bioinformatics.charite.de/transformer) contains integrated information on the three phases of biotransformation (modification, conjugation and excretion) of 3000 drugs and >350 relevant food ingredients (e.g. grapefruit juice) and herbs, which are catalyzed by 400 proteins. A total of 100 000 interactions were found through text mining and manual validation. The 3D structures of 200 relevant proteins are included. The database enables users to search for drugs with a visual display of known interactions with phase I (Cytochrome P450) and phase II enzymes, transporters, food and herbs. For each interaction, PubMed references are given. To detect mutual impairments of drugs, the drug-cocktail tool displays interactions between selected drugs. By choosing the indication for a drug, the tool offers suggestions for alternative medications to avoid metabolic conflicts. Drug interactions can also be visualized in an interactive network view. Additionally, prodrugs, including their mechanisms of activation, and further information on enzymes of biotransformation, including 3D models, can be viewed.
INTRODUCTION
The number of prescribed drugs is rising (1). A study revealed that 87.1% of people >50 years of age take at least one drug per day, and 43.3% take >5 (2). Polypharmacy, which is defined as the regular use of five or more drugs, leads to an increased risk of adverse drug reactions (ADRs). The frequency of ADR is associated with the number of drugs prescribed (3). Among hospitalized patients, ADRs have an incidence of 6.7% and are the fifth commonest cause of death (4). One possible cause for ADR might be the individual variance of drug metabolism (5), and age-related changes make elderly patients more sensitive to ADRs (6). The information is widely scattered over the scientific literature. A knowledge base of xenobiotic metabolism and the effect of polymorphisms could prevent ADR and cases of death.
Xenobiotic metabolism and detoxification (especially for drugs) are separated into three different phases of reaction. Only a few xenobiotics are excreted unchanged in urine or feces without any metabolic degradation.
Phase I and phase II reactions convert compounds to more water-soluble and often less active derivatives to increase excretion. Thereby, phase I reflects the production of reactive groups through oxidation and is primarily managed by the Cytochrome P450 family (CYP) of enzymes (7). Subsequently, the reactive groups are used to conjugate small polar molecules (phase II) to increase the polarity. Six enzyme families that provide the detoxification and excretion of xenobiotics mainly realize the conjugation (8).
Transporters (phase III) play a crucial role in pharmacokinetics by enabling the migration of hydrophilic molecules, which cannot penetrate cellular membranes. Kell et al. showed that the majority of drugs enter cells through at least one transporter (9). Those proteins form a transmembrane channel lined with hydrophilic amino acid side chains spanning the lipid bilayer (10). Two major protein superfamilies are known: 49 ATP-binding cassette transporters (ABC) (11) and 362 solute carriers (SLC) (12). These are important for absorption, distribution and excretion of drugs (13) and are involved in a broad range of physiological processes (10).
Eukaryotic ABC transporters are predominantly exporters, which require energy released by ATP hydrolysis. One problem is multidrug resistance, which is caused by active transporters. Unfortunately, 40% of human tumors develop resistance to chemotherapeutics by overexpressing ABC proteins (14). The SLC transporters facilitate passive diffusion along the concentration gradient or use concentration gradients from other substrates as a symporter or antiporter (12).
Another issue related to drug metabolism and ADR is prodrugs. Prodrugs have to be converted to active drugs by metabolic conversion (15). In general, prodrugs are non-toxic and need to have their chemical structure changed to enable their inherent medical capability. However, problems in conversion can also lead to undesired side effects. For example, the antihistamine terfenadine is a potent hERG blocker as a prodrug and a slow conversion can cause cardiac toxicity (16).
Prodrugs can be activated by photo irradiation (17), a change in pH (18) and enzymes, such as esterases or CYPs (19,20). Many prodrugs are activated by hydrolysis with the aid of esterases or phosphatases. Thereby, gastric intestinal tolerance and pharmacokinetics can be improved, but the targeting of drugs to specific cells or tissues cannot. The activation of prodrugs by CYPs might be a better approach (21).
Not only can drugs participate in the alteration of drug metabolism but food and herbs also have a proven influence; e.g. furanocoumarins in grapefruit inhibit intestinal CYP3A4 and organic anion-transporting polypeptides 1A2 (22,23).
More than 350 ingredients in food and drink, such as broccoli, alcohol and char-grilled meat, as well as herbal medicine, such as St John’s wort, are known to alter drug responses.
A comprehensive resource that combines scientific information on phase I and phase II enzymes, transporter enzymes, prodrugs, food and herbs could help to improve research in this field and prevent ADR.
MATERIALS AND METHODS
Text mining
We created a text mining approach using semantic web standards. To develop a specialized text mining pipeline, we first downloaded Medline/PubMed data from the NCBI FTP site in xml-format. Using the search engine library Apache Lucene (http://lucene.apache.org) and a tool kit for processing text with computational linguistics (http://alias-i.com/lingpipe), the data was indexed. The search engine comprises comprehensive lists of chemical compounds and drug names (24), metabolic enzymes (25) and transporters (26), including their various synonyms. Additionally, we added a list of common interaction terms, such as ‘activate’, ‘inhibit’, and ‘metabolize’. The search engine, written in Java, dynamically queries the indexed data and produces a structured query language (SQL) file containing the text mining hits. A query example is:
(DrugSynonym [TI] AND TransformerSynonym [TI]) OR
(DrugSynonym [abstract] AND TransformerSynonym [abstract]) OR
(DrugSynonym [abstract] AND InteractionTerm AND TransformerSynonym [abstract])
The positional distance between the different terms had to be restricted to reduce false-positive hits, when terms occurred far from each other in the abstract. The 22 500 records found were scored as rule-based. Duplicates were removed and a team of scientists manually processed 12 427 articles found in PubMed. Further details about the text mining approach can be found on the Web site in the frequently asked questions (FAQs) section.
Database
The database was designed as a relational database on a MySQL server. To allow chemical functionality, such as handling chemical data within MySQL, the MyChem package was included.
Information about ∼3000 CYP drug interactions and 2000 polymorphisms were extracted from the SuperCYP database (27). SuperCYP is a database with a focus on human CYPs. However, there are many other important enzymes in the metabolism of xenobiotics, such as transporters or phase II enzymes.
DATABASE FEATURES
Over 100 000 interactions were revealed. In the 12 427 articles found in PubMed, 769 drugs were attributed to those phase II enzymes that are involved in drug metabolism. Text mining was also performed for prodrugs, transporters and food. We found 125 prodrugs described in 890 PubMed articles together with their mechanism of activation, accompanying enzymes, chemical structure and identification numbers. Furthermore, ∼500 drug-transporters and 150 food interactions were identified.
Additionally, ∼200 3D structures were collected for transporters, CYPs and phase II enzymes.
The database includes four main functionalities. To provide an overview of these, a comprehensive FAQs section was created, which is helpful for first-time users. Depending on the user’s interest or needs (e.g. clinicians, researchers), different ways to browse the data were enabled.
Prodrugs
Prodrugs can be identified directly by entering the name, PubChemID, CAS number or ATC code, as well as by choosing a mechanism of activation, such as ring opening or carboxylation.
Drugs
To view the metabolism of particular drugs, users can search directly by entering the name, PubChemID or CAS number. Based on the WHO classification system, which classifies drugs into different groups according to Anatomical site of action, Therapeutical effect and Chemical structure (ATC), a tree with all of the drugs contained in the database can be viewed in their ATC group.
Cocktail
This tool enables users to see drug interactions of an individually composed drug cocktail (Figure 1A). If >1 drug interacts with the same enzyme, lines of the interaction table are shown in yellow, orange, red and dark red (Figure 1B). In the header of each column, the indication for the drugs can be chosen and the database will provide alternative drugs that are metabolized by different enzymes (Figure 1C). PubMed references are available by clicking on the interaction. Additionally, food interactions, as well as elimination half-life (EHL) times and Q0 values are displayed. A Q0 value (extrarenal excretion) of <0.3 is shown in green because those drugs are, to a large extent, excreted in the unchanged form. Clicking ‘Display network’ presents these interactions in a network view based on Cobweb (28).
Figure 1.
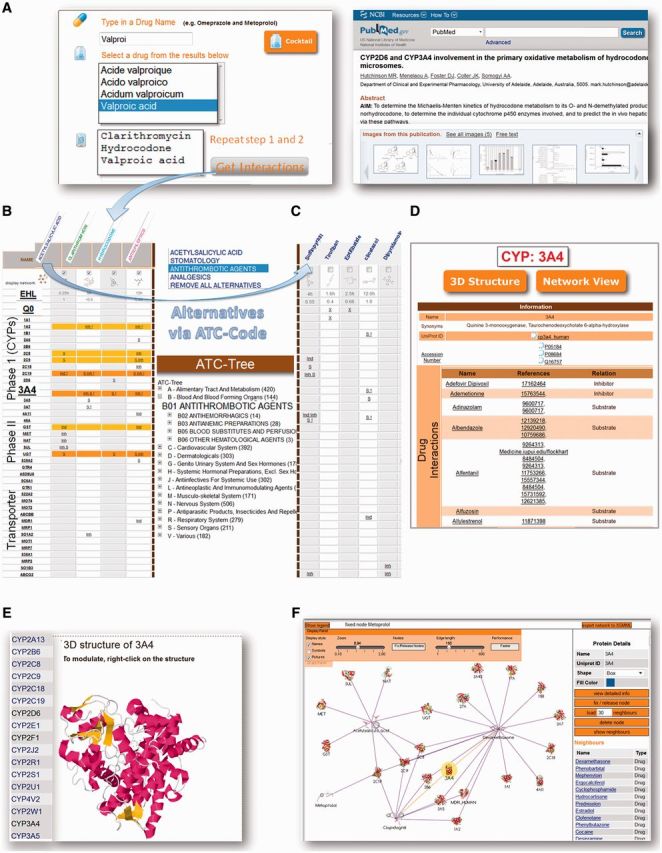
Functionalities of the ‘Transformer database’. (A) Composition of a drug-cocktail. An example of a PubMed reference is shown. PubMed references can be viewed by clicking on ‘S’, ‘Inh’ or ‘Ind’ in the result table. (B) Clicking on ‘Get Interactions’ leads to a result table, which shows the interactions between the drugs. (C) By choosing the indications of the drugs via ATC-code the user receives specific alternatives. (D) By clicking on an enzyme (e.g. CYP3A4), detailed information on the enzyme, including drug interactions, are shown. (E) The 3D structures of all enzymes can be viewed (e.g. CYP3A4). (F) Network views are provided for each enzyme and compound.
Biotransformation
To find drugs that are metabolized by specific phase I, phase II or transporter enzymes, users can perform a search by clicking on ‘Biotransformation’. This page provides (homology modeled) 3D structures of all enzymes (Figure 1E). Furthermore, a list of interacting drugs can be viewed in a table (Figure 1D) or in a network view (Figure 1F).
DATABASE USAGE
The following case illustrates the need to detect interactions with the help of the Transformer database. A five-year-old child died from a fatal opioid toxicity. She was inadvertently administered a high dose of hydrocodone (an antitussive drug) while suffering from a cold. Additionally, she was administered clarithromycin for an ear infection and valproic acid for seizures. The postmortem blood screen revealed an excessively high-hydrocodone level and, in contrast, barely measurable hydromorphone (biotransformation metabolite of hydrocodone) concentration (29). Hydrocodone is metabolized by CYP2D6, CYP3A4 and afterwards by UGT. There were three reasons for the low metabolism rate:
CYP-polymorphism: the child was found to be a CYP2D6 poor metabolizer,
inhibition of CYP3A4 by clarithromycin and
inhibition of UGT by valproic acid.
Figure 1B shows the Transformer database results for this drug combination. All interactions described in this case are displayed and colored because of enzyme overload. Nevertheless, parts of the available information of drug–enzyme interactions are experimental data and offer no evidence for drug interactions in humans and clinical work, although Q0 and elimination half-life times could be relevant. The Transformer database, however, provides a platform for detecting mutual drug impairments and could help to appraise the drug response. The database is a comprehensive resource on drug enzyme/transporter interactions and could be a sound starting-point for further research.
The database will be updated yearly to add new drugs/compounds and interactions.
AVAILABILITY
The Transformer database is publicly available via http://bioinformatics.charite.de/transformer and should be used under the terms of the Creative Commons Attribution-Noncommercial-Share Alike 3.0 License.
FUNDING
Funding for open access charge: SynSys (EU Framework 7) and DKTK (BMBF), BMBF Immunotox.
Conflict of interest statement. None declared.
REFERENCES
- 1.Banerjee A, Mbamalu D, Ebrahimi S, Khan AA, Chan TF. The prevalence of polypharmacy in elderly attenders to an emergency department—a problem with a need for an effective solution. Int. J. Emerg. Med. 2011;4:22. doi: 10.1186/1865-1380-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan TK, Williamson M, Pirotta M, Stewart K, Myers SP, Barnes J. A national census of medicines use: a 24-hour snapshot of Australians aged 50 years and older. Med. J. Aust. 2012;196:50–53. doi: 10.5694/mja11.10698. [DOI] [PubMed] [Google Scholar]
- 3.Sato I, Akazawa M. Polypharmacy and adverse drug reactions in Japanese elderly taking antihypertensives: a retrospective database study. Drug Healthc. Patient Saf. 2013;5:143–150. doi: 10.2147/DHPS.S45347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998;279:1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 5.Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA. 2001;286:2270–2279. doi: 10.1001/jama.286.18.2270. [DOI] [PubMed] [Google Scholar]
- 6.Marusic S, Bacic-Vrca V, Obreli Neto PR, Franic M, Erdeljic V, Gojo-Tomic N. Actual drug-drug interactions in elderly patients discharged from internal medicine clinic: a prospective observational study. Eur. J. Clin. Pharmacol. 2013;69:1717–1724. doi: 10.1007/s00228-013-1531-7. [DOI] [PubMed] [Google Scholar]
- 7.Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem. Res. Toxicol. 2001;14:611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 8.Jakoby WB, Ziegler DM. The enzymes of detoxication. J. Biol. Chem. 1990;265:20715–20718. [PubMed] [Google Scholar]
- 9.Kell DB, Dobson PD, Bilsland E, Oliver SG. The promiscuous binding of pharmaceutical drugs and their transporter-mediated uptake into cells: what we (need to) know and how we can do so. Drug Discov. Today. 2013;18:218–239. doi: 10.1016/j.drudis.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Yan Q. Pharmacogenomics of membrane transporters an overview. Methods Mol. Biol. 2003;227:1–20. doi: 10.1385/1-59259-387-9:1. [DOI] [PubMed] [Google Scholar]
- 11.Velamakanni S, Wei SL, Janvilisri T, van Veen HW. ABCG transporters: structure, substrate specificities and physiological roles:a brief overview. J. Bioenerg. Biomembr. 2007;39:465–471. doi: 10.1007/s10863-007-9122-x. [DOI] [PubMed] [Google Scholar]
- 12.He L, Vasiliou K, Nebert DW. Analysis and update of the human solute carrier (SLC) gene superfamily. Hum. Genomics. 2009;3:195–206. doi: 10.1186/1479-7364-3-2-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa T, Tamura A, Saito H, Wakabayashi K, Nakagawa H. Pharmacogenomics of the human ABC transporter ABCG2: from functional evaluation to drug molecular design. Naturwissenschaften. 2005;92:451–463. doi: 10.1007/s00114-005-0019-4. [DOI] [PubMed] [Google Scholar]
- 14.Landry Y, Gies JP. Drugs and their molecular targets: an updated overview. Fundam. Clin. Pharmacol. 2008;22:1–18. doi: 10.1111/j.1472-8206.2007.00548.x. [DOI] [PubMed] [Google Scholar]
- 15.Albert A. Chemical aspects of selective toxicity. Nature. 1958;182:421–422. doi: 10.1038/182421a0. [DOI] [PubMed] [Google Scholar]
- 16.Yap YG, Camm AJ. Potential cardiac toxicity of H1-antihistamines. Clin. Allergy Immunol. 2002;17:389–419. [PubMed] [Google Scholar]
- 17.Dorman G, Prestwich GD. Using photolabile ligands in drug discovery and development. Trends Biotechnol. 2000;18:64–77. doi: 10.1016/s0167-7799(99)01402-x. [DOI] [PubMed] [Google Scholar]
- 18.Simplicio AL, Clancy JM, Gilmer JF. Beta-aminoketones as prodrugs with pH-controlled activation. Int. J. Pharm. 2007;336:208–214. doi: 10.1016/j.ijpharm.2006.11.055. [DOI] [PubMed] [Google Scholar]
- 19.Huttunen KM, Mahonen N, Raunio H, Rautio J. Cytochrome P450-activated prodrugs: targeted drug delivery. Curr. Med. Chem. 2008;15:2346–2365. doi: 10.2174/092986708785909120. [DOI] [PubMed] [Google Scholar]
- 20.Topf N, Worgall S, Hackett NR, Crystal RG. Regional ‘pro-drug' gene therapy: intravenous administration of an adenoviral vector expressing the E. coli cytosine deaminase gene and systemic administration of 5-fluorocytosine suppresses growth of hepatic metastasis of colon carcinoma. Gene Ther. 1998;5:507–513. doi: 10.1038/sj.gt.3300611. [DOI] [PubMed] [Google Scholar]
- 21.Ortiz de Montellano PR. Cytochrome P450-activated prodrugs. Future Med. Chem. 2013;5:213–228. doi: 10.4155/fmc.12.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bressler R. Grapefruit juice and drug interactions. Exploring mechanisms of this interaction and potential toxicity for certain drugs. Geriatrics. 2006;61:12–18. [PubMed] [Google Scholar]
- 23.Uno T, Yasui-Furukori N. Effect of grapefruit juice in relation to human pharmacokinetic study. Curr. Clin. Pharmacol. 2006;1:157–161. doi: 10.2174/157488406776872550. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sim SC, Ingelman-Sundberg M. The Human Cytochrome P450 (CYP) Allele Nomenclature website: a peer-reviewed database of CYP variants and their associated effects. Hum. Genomics. 2010;4:278–281. doi: 10.1186/1479-7364-4-4-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preissner S, Kroll K, Dunkel M, Senger C, Goldsobel G, Kuzman D, Guenther S, Winnenburg R, Schroeder M, Preissner R. SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions. Nucleic Acids Res. 2010;38:D237–D243. doi: 10.1093/nar/gkp970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Eichborn J, Bourne PE, Preissner R. Cobweb: a Java applet for network exploration and visualisation. Bioinformatics. 2011;27:1725–1726. doi: 10.1093/bioinformatics/btr195. [DOI] [PubMed] [Google Scholar]
- 29.Madadi P, Hildebrandt D, Gong IY, Schwarz UI, Ciszkowski C, Ross CJ, Sistonen J, Carleton BC, Hayden MR, Lauwers AE, et al. Fatal hydrocodone overdose in a child: pharmacogenetics and drug interactions. Pediatrics. 2010;126:e986–e989. doi: 10.1542/peds.2009-1907. [DOI] [PubMed] [Google Scholar]


