Abstract
The Integrated Microbial Genomes (IMG) data warehouse integrates genomes from all three domains of life, as well as plasmids, viruses and genome fragments. IMG provides tools for analyzing and reviewing the structural and functional annotations of genomes in a comparative context. IMG’s data content and analytical capabilities have increased continuously since its first version released in 2005. Since the last report published in the 2012 NAR Database Issue, IMG’s annotation and data integration pipelines have evolved while new tools have been added for recording and analyzing single cell genomes, RNA Seq and biosynthetic cluster data. Different IMG datamarts provide support for the analysis of publicly available genomes (IMG/W: http://img.jgi.doe.gov/w), expert review of genome annotations (IMG/ER: http://img.jgi.doe.gov/er) and teaching and training in the area of microbial genome analysis (IMG/EDU: http://img.jgi.doe.gov/edu).
DATA SOURCES AND PROCESSING
The Integrated Microbial Genomes (IMG) system integrates genomes from all three domains of life, as well as viruses, plasmids and genome fragments (partial sequences of genomic regions of interest, such as biosynthetic clusters). Until 2012, IMG used NCBI’s RefSeq resource (1) as its main source of public genome sequence data and annotations consisting of predicted genes and protein products, with a RefSeq-specific pipeline used for retrieving new genomes from RefSeq’s ftp site. For non-public (i.e. ‘private’) datasets, the IMG ER Submission system allowed scientists to select their sequencing projects in GOLD (2) and then submit their genome sequence data for annotation and integration into the ‘Expert Review’ version of IMG, IMG/ER (http://img.jgi.doe.gov/er). Public and private genomes were processed using different annotation and data integration pipelines, and recorded in different databases.
In an effort to improve the efficiency of data processing and tracking, IMG’s genome submission, annotation and integration pipelines were consolidated in November 2012. The IMG ER Submission system (http://img.jgi.doe.gov/submit) and associated (submission, gene prediction, functional annotation and data integration) data processing pipelines were extended to handle both public and private genomes in a uniform manner. The pipelines use a common mechanism for tracking the processing status of genome datasets, GOLD provides the information needed for retrieving new public genomes from RefSeq or GenBank (3) and both public and private genomes are recorded in a common IMG data warehouse.
For every genome, the IMG data warehouse records primary genome sequence information including its organization into chromosomal replicons (for finished genomes) and scaffolds and/or contigs (for draft genomes), together with predicted protein-coding sequences, some RNA-coding genes and protein product names that are provided by the genome sequence centers or generated by IMG’s functional annotation pipeline.
Public and private genomes submitted for annotation and integration by IMG’s pipelines are first associated with sequencing projects in GOLD. Custom tools and metadata about the topology of contigs and scaffolds are used to identify the origin of replication of circular replicons and permute the corresponding scaffold or contig if necessary. To ensure accurate identification of partial genes bordering the gaps, gene models and other features are initially predicted on individual contigs and combined thereafter to generate scaffold-level structural annotation. CRISPR elements are detected using CRT (4) and PILERCR (5). Predictions from both methods are concatenated, and in case of overlapping elements, the shorter one is removed. Identification of tRNAs is performed using tRNAScan-SE-1.23 (6). Ribosomal RNA genes (5S, 16S and 23S) are predicted using hmmsearch against the custom models generated for each type of rRNA in bacteria and archaea (7,8). With the exception of tRNA and rRNA, all models from Rfam (9) are used to search the genome sequence. Sequences are first compared with a database containing all the non-coding RNA genes in the Rfam database using BLAST, then sequences that have hits to genes belonging to an Rfam model are searched using the program INFERNAL (10). Signal peptides are computed using SignalP (11), whereas transmembrane helices are computed using TMHMM (12). Protein-coding genes are predicted using Prodigal (13); models overlapping with CRISPRs and certain types of RNAs (e.g. rRNAs) are removed.
After a new genome is processed, protein-coding genes are compared with protein families and the proteome of selected publicly available ‘core’ genomes, with product names assigned based on the results of these comparisons. First, protein sequences are compared with COG (14) using RPS-BLAST, Pfam-A (15) using HMMER 3.0b2 executed inside Sanger’s pfam_scan.pl wrapper script and TIGRfam (16) databases using HMMER 3.0 (8), and associated with KEGG Orthology (KO) terms (17) using USEARCH (18). Genomes in IMG are associated with KEGG pathways using the assignment of KO terms to protein-coding genes, while their association with MetaCyc pathways (19) is based on correlating enzyme EC numbers in MetaCyc reactions with EC numbers associated with protein-coding genes via KO terms. Genes are further characterized using an IMG native collection of generic (protein cluster-independent) functional roles called IMG terms that are defined by their association with generic (organism-independent) functional hierarchies, called IMG pathways (20). IMG terms and pathways are specified by domain experts at DOE-JGI as part of the process of annotating specific genomes of interest, and are subsequently propagated to all the genomes in IMG using a rule-based methodology. Transporter genes are linked to the Transport Classification Database (21) based on their assignment to COG, Pfam or TIGRfam domains or IMG terms that correspond to transporter families.
The integration of new genomes into IMG involves computing protein sequence similarities between their genes and genes of all other (new or existing) genomes in the system, assigning IMG terms and protein product names to the genes of the new genomes, identifying fusions and computing conserved gene cassettes (putative operons). For each gene, IMG provides lists of related (e.g. homolog, paralog and ortholog) genes that are based on sequence similarities computed using USEARCH for protein-coding and RNA genes. A fused gene (fusion) is defined as a gene that is formed from the composition (fusion) of two or more previously separate genes (22). Fusions are identified based on computing USEARCH similarities between genes. Only genes from finished genomes are considered as putative components to avoid false predictions from fragmented genes in draft genomes. Furthermore, genes that frequently appear as fragmented in finished genomes, such as ‘transposases’ and ‘integrases’, as well as ‘pseudogenes’ are excluded from fusion calculations. Putative horizontally transferred genes are identified from the sequence similarity data. The phylogenetic distribution of best hits against a set of reference isolate genomes also provides additional information on possible horizontal gene transfers for isolates. A ‘chromosomal cassette’ is defined as a stretch of genes with intergenic distance ≤300 bp, whereby the genes can be on the same or different strands of the chromosome. Chromosomal cassettes with a minimum size of two genes common in at least two separate genomes are defined as ‘conserved chromosomal cassettes’. The identification of common genes across organisms is based on two gene clustering methods, namely, participation in COG and Pfam clusters (23).
Note that for public and private genomes that are already associated with genes and/or protein product names, the native gene and/or product names are preserved in IMG unless their replacement is explicitly requested at the time they are submitted for annotation and integration into IMG.
DATA CONTENT
Genomics data
The content of IMG has grown steadily since the first version released in March 2005, with the current version of IMG (as on 10 September 2013) containing 11 568 bacterial, archaeal and eukaryotic genomes, an increase of >300% since August 2011 (24). IMG also includes 2848 viral genomes, 1198 plasmids that did not come from a specific microbial genome sequencing project and 581 genome fragments, bringing its total content to 16 195 genome datasets with >42 million protein-coding genes.
The number of single cell genomes included into IMG has increased substantially: there are 1341 single cell genomes in the current version of IMG compared with only 21 in August 2011. Approximately 240 single cell genomes are part of the Microbial Dark Matter project that aims to expand the Genomic Encyclopedia of Bacteria and Archaea by targeting 100 single cell representatives of uncultured candidate phyla (25).
IMG has 13 342 genome datasets that are publicly available to all users without restrictions via the IMG/W datamart (http://img.jgi.doe.gov/w). Genomes that have not been yet published (also known as ‘private’) are password-protected and available only to the scientists who study (‘own’) them through the IMG/ER (‘Expert Review’) datamart (http://img.jgi.doe.gov/er). Private genomes are usually publicly released 6 months after the dataset becomes available in IMG.
IMG/ER allows individual scientists or groups of scientists to review and curate the functional annotation of microbial genomes in the context of IMG’s public genomes (26). Since August 2011, hundreds of private genomes have been reviewed and curated using IMG/ER, a relatively small fraction of the 9000 genomes that were processed by IMG’s data annotation and integration pipelines, as genome curation is a time-consuming process. Genome curation is usually carried out for identifying missing genes or for correcting functional annotations, e.g. as part of the process of curating IMG native terms and pathways.
Omics data
Proteomics datasets have been gradually included into IMG starting in 2009. Since August 2011, 64 new protein expression datasets (samples) that are part of two studies were included into IMG, bringing the total to 90 samples across five studies. The organization and analysis of proteomic data in IMG are discussed in (24).
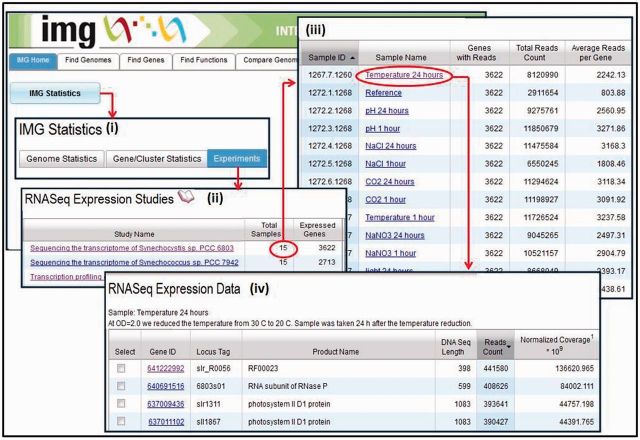
The first RNAseq (transcriptomic) datasets included into IMG in 2011 are part of the Synechococcus PCC study consisting of ∼40 samples (Billis,K., Billini,M., Kyrpides,N.C., and Mavromatis,K., submitted for publication). As of August 2013, IMG contains 99 samples across 10 RNASeq studies. A typical RNASeq study involves the sequencing of cDNA from a genome under different experimental conditions, with the effect of each experimental condition being captured by a sample. As part of RNASeq sequencing analysis, reads are mapped to the reference genome involved in the study, and the expressed genes in each sample are recorded with their observed read counts, mean, median and strand. RNA reads are mapped to reference genomes using Bowtie2 (27). The scope of mapping is determined by the type of cDNA sample (sscDNA/dscDNA) and the directionality of the libraries, whereby reads may map to a single strand or both strands of the reference sequence. Expression levels are normalized by computing RPKM (reads per kilobase per million), Quantile or Affine transformations and may need to be interpreted based on the type of cDNA in the sample. For genomes involved in RNASeq studies, the experiments/samples are recorded in IMG together with experimental conditions, and the read counts are organized per expressed gene, as illustrated in Figure 1.
Figure 1.
RNA-Seq data organization. (i) ‘Omics’ datasets generated can be accessed from ‘IMG Statistics’ on IMG’s front page, following the Experiments link available on the ‘IMG Statistics’ page. (ii) An RNA-Seq study is associated with samples and the number of genes expressed across all samples. (iii) Each sample is associated with the number of expressed genes, the total number of reads and the average number of reads per gene. (iv) An expressed gene is associated with a read count (total number of reads divided by the size of the gene) and normalized coverage (coverage for a gene in the experiment divided by the total number of reads in that experiment).
Biosynthetic clusters
IMG contains biosynthetic clusters of genes associated with pathways involved in the generation of secondary metabolites in isolate prokaryotic genomes. Experimentally validated biosynthetic clusters were identified by searching NCBI's nucleotide database for genome fragments (partially sequenced genomes) containing gene clusters associated with secondary metabolites/natural products (28). Additional biosynthetic clusters were predicted using ClusterFinder (Fischbach, submitted for publication). Biosynthetic clusters in IMG are associated with IMG, Metacyc and KEGG pathways as well as information available in GOLD on their natural products.
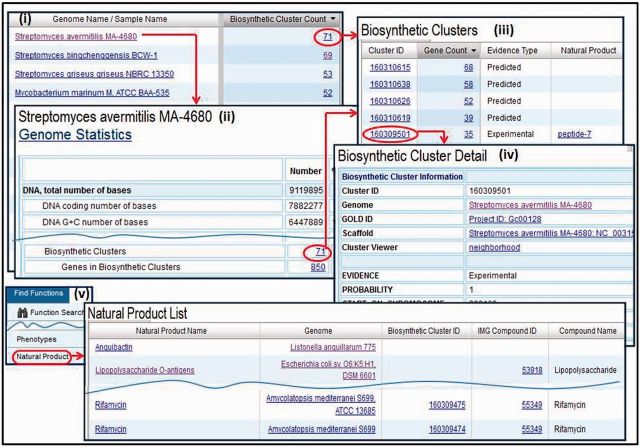
Genomes associated with biosynthetic clusters can be examined as illustrated in Figure 2, where these genomes are listed in descending order of the number of biosynthetic clusters present in them. Alternatively, IMG can be used to find genomes associated with natural products associated with genome fragments but not with biosynthetic clusters, as illustrated in Figure 2(v). Natural products are small metabolites found in nature, and although the biosynthetic clusters associated with the generation of natural products have been identified, there are still natural products whose production mechanisms in prokaryotes remain unknown.
Figure 2.
Biosynthetic clusters. (i) Genomes associated with biosynthetic clusters can be retrieved and examined using the ‘Genome Browser’. (ii) The number of biosynthetic clusters is provided in the ‘Genome Statistics’ section of the ‘Organism Detail’ page of a genome, together with a hyperlink to (iii) the list of biosynthetic clusters, whereby for each cluster the number of associated genes, the evidence type and the corresponding natural product are provided. (iv) A biosynthetic cluster can be examined using the ‘Biosynthetic Cluster Detail’ page, which includes information about the cluster. (v) ‘Natural Product List’ provides the list of the IMG genomes associated with natural products.
ANALYSIS TOOLS
Browsers and search tools allow finding and selecting genomes, genes and functions of interest, which can then be examined individually or analyzed in a comparative context. Gene content-based comparison of genomes is provided by the ‘Phylogenetic Profiler’ and the ‘Phylogenetic Profiler for Gene Cassettes’ tools that allow identifying genes in a query genome in terms of presence or absence of homologs in other genomes, or participation in conserved gene cassettes across other genomes (29,30). Function-based comparison of genomes is provided by the ‘Abundance Profile Overview’ and ‘Function Profile’ tools that allow comparing the relative abundance of protein families (COGs, Pfams, TIGRfams) and functional families (enzymes) across genomes. The composition of analysis operations is facilitated by genome, scaffold, gene and function ‘carts’ that handle lists of genomes, scaffolds, genes and functions, respectively. IMG analysis tools have been discussed in (24). Tools for identifying and correcting annotation anomalies, such as dubious protein product names, and for filling annotation gaps, such as genes that may have been missed by gene prediction tools or genes without predicted functions, are discussed in (26). IMG analysis tool extensions have addressed performance (30), data quality control, such as single cell data decontamination (31), and new data types, such as RNA-Seq and biosynthetic cluster data.
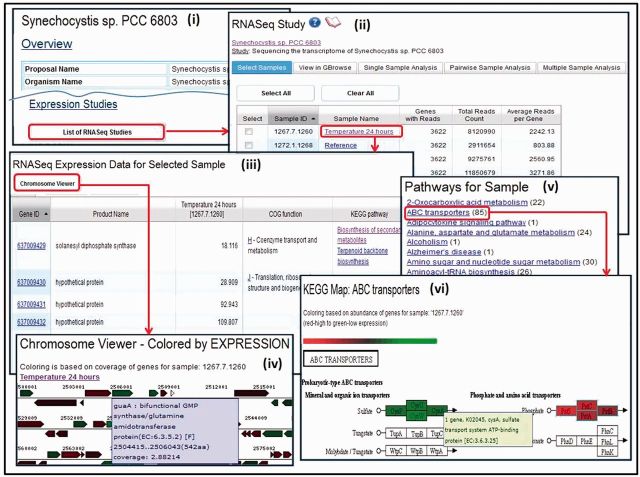
RNA-Seq studies can be accessed from the ‘Experiments Statistics’ section of the ‘IMG Statistics’ page or the ‘Organism Details’ pages of their associated genomes. For example, the ‘Expression Studies’ link in the ‘Organism Details’ page, such as that shown in Figure 3(i), leads to the list of associated RNA-Seq samples, as the list shown in Figure 3(ii).
Figure 3.
RNA-Seq data exploration. (i) The list of RNA-Seq studies associated with a genome can be accessed from its ‘Organism Details’, with each study associated with (ii) a list of RNA-Seq experiments (samples). Individual samples can be selected for further analysis, such as (iii) examining its expressed genes as a list or using the (iv) chromosome viewer. A sample can be also examined in the context of (v) pathways that have at least one enzyme associated with an expressed gene in the sample, whereby for each pathway (vi) enzymes are displayed with colors representing the level of expression for the associated genes; mousing over an enzyme shows the number of expressed genes associated with the enzyme.
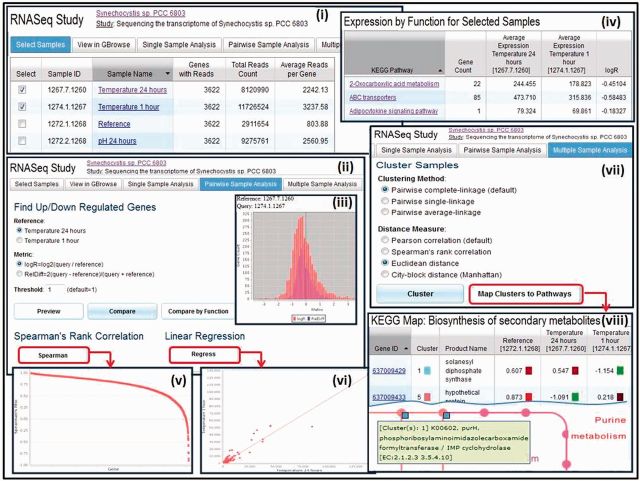
RNA-Seq studies associated with a genome can be compared using pairwise or multiple sample analysis tools as illustrated in Figure 4. After samples are selected for comparison (Figure 4(i)), pairs of samples can be compared in terms of up- or downregulation of genes, as illustrated in Figure 4(ii), with a threshold specified for the difference in gene expression. The difference in expression is computed using the logR = log2(query/reference) or the RelDiff = 2(query-reference)/(query + reference) metric. The comparison can be first previewed using a histogram, as illustrated in Figure 4(iii), which can help set the thresholds for the search of over-expressed or under-expressed genes between a pair of samples. The result of the comparison can be examined at the level of individual up- and downregulated genes, which can be selected for inclusion in the ‘Gene Cart’ for further analysis. Alternatively, the result of the comparison can be examined in terms of functions, as illustrated in Figure 4(iv), with genes associated with KEGG pathways or COG functions grouped together. Genes associated with a specific KEGG pathway can be examined in the context of the pathway, similar to the example shown in Figure 3(vi) earlier. The strength of the association between pairs of samples can be examined using ‘Spearman’s Rank Correlation’, as illustrated in Figure 4(v), whereas ‘Linear Regression’ analysis, illustrated in Figure 4(vi), helps determine whether two samples are technical replicates.
Figure 4.
RNA-Seq data comparison. (i) RNA-Seq sample comparison starts with the selection of samples of interest. (ii) ‘Pairwise Sample Analysis’ supports comparing samples in terms of up/downregulated genes, with (iii) a histogram preview helping setting the thresholds for comparison. (iv) The result of the comparison can be examined in terms of functions, whereby genes associated with KEGG pathways or COG functions are grouped together. (v) The strength of the association of gene expression between pairs of samples can be examined using ‘Spearman’s Rank Correlation’. (vi) ‘Linear Regression’ analysis helps estimate whether two samples are technical replicates. (vii) ‘Multiple Sample Analysis’ consists of clustering samples based on the abundance of expressed genes, using a variety of clustering methods. (viii) Clusters of samples can be examined in the context of pathways, whereby enzymes are displayed with colors representing the cluster.
Multiple RNA-Seq sample analysis usually involves clustering based on the abundance of expressed genes, where the proximity of grouping indicates the relative degree of similarity of samples to each other. There is a choice of clustering methods, such as pairwise complete linkage and pairwise single linkage, and distance measure, such as Pearson correlation, Spearman’s rank correlation and Euclidean distance, as illustrated in Figure 4(vii). The result of clustering is displayed as a hierarchical tree of samples and a normalized heat map of coverage values for each gene for each sample. Clusters of multiple samples can be also examined in the context of pathways, as illustrated in Figure 4(viii), whereby enzymes are displayed with colors representing the cluster.
FUTURE PLANS
IMG’s genome sequence data content is maintained through regular updates managed by the IMG submission system and involving new genomes sequenced at JGI, genomes sequenced at other organizations and submitted for inclusion into IMG by scientists worldwide and genomes from Genbank. For genomes with multiple submissions, only the latest version is kept in IMG. IMG genome data are distributed through genome data portals available at: http://genome.jgi.doe.gov/. IMG’s data annotation and integration pipelines have been automated, thus improving their ability to keep pace with the rapidly increasing number of sequenced genomes.
IMG’s integrated data framework allows assessing and improving the quality of genome annotations. Thus, the quality of gene models for genomes available in public resources is known to vary greatly depending on the quality of sequence and the software used for annotation. An analysis conducted at JGI of the protein-coding genes of microbial genes in Genbank indicates that ∼10% (>1 million) of predicted protein-coding are erroneous: they are false-positive genes, unidentified pseudogene fragments or genes with translational exceptions or have incorrectly predicted start sites. To improve the consistency of annotation and the quality of predicted genes, all public microbial genomes in IMG will be re-annotated using IMG’s annotation pipeline.
A rapidly increasing number of single cell genomes are included into IMG. Typically, the first version of a single cell genome is analyzed for identifying contigs that may come from contaminant (e.g. Pseudomonas, Ralstonia) organisms. The sequence of analysis steps needed to identify and remove contaminated contigs is described in (31).
The importance of functional genomics in validating gene function in an integrated comparative genomics context is also being underscored, pushing experimental data from methylomics and transposon mutagenesis experiments into IMG. Systematic paradigms for associating computationally predicted gene structural and functional information with experimental functional genomics are being constructed. Tools are being developed for mining and visualizing different types of Omics datasets in an integrated genomic context.
IMG’s users are faced with the increasing burden of analyzing a rapidly growing number of genomic datasets. This analytical challenge can be alleviated by synthesizing genomic data using the ‘pangenome’ conceptual abstraction (32). A pangenome consists of the core part of a species (i.e. the genes present in all of the sequenced strains or of all samples of a microbial community) and the variable part (the genes present in some but not all of the strains or samples). An experimental version of IMG has been extended with five pangenomes, as well as analysis tools and viewers that allow users to explore individual pangenomes and compare pangenomes and genomes. A public version of IMG containing pangenome data and analysis tools is expected to be released in the near future.
FUNDING
The Director, Office of Science, Office of Biological and Environmental Research, Life Sciences Division, U.S. Department of Energy under Contract No. [DE-AC02-05CH11231]; This research used resources of the National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. [DE-AC02-05CH11231]. Funding for open access charge: University of California.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pruitt KD, Tatusova T, Garth RB, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC. The Genomes on Line Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–D579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrahi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edgar RC. PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinformatics. 2007;8:18. doi: 10.1186/1471-2105-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 9.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2012;41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 12.Moller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 13.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Celments J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. TIGRFAMs and genome properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 19.Caspi R, Altman T, Dreher K, Fulcher AC, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanova NN, Anderson I, Lykidis A, Mavrommatis K, Mikhailova N, Chen IA, Szeto E, Palaniappan K, Markowitz VM, Kyrpides NC. 2007. Metabolic Reconstruction of Microbial Genomes and Microbial Community Metagenomes. Technical Report 62292, Lawrence Berkeley National Laboratory. http://img.jgi.doe.gov/w/doc/imgterms.html. [Google Scholar]
- 21.Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The transporter classification database: recent advances. Nucleic Acids Res. 2009;37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 23.Mavromatis K, Chu K, Ivanova N, Hooper SD, Markowitz VM, Kyrpides NC. Gene context analysis in the Integrated Microbial Genomes (IMG) data management system, accepted for publication. PLoS One. 2009;4:e7979. doi: 10.1371/journal.pone.0007979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markowitz VM, Chen IA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Biju J, Huang J, Williams P, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 26.Markowitz VM, Mavromatis K, Ivanova NN, Chen IA, Chu K, Kyrpides NC. IMG ER: a system for microbial annotation expert review and curation. Bioinformatics. 2009;25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 27.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh CT, Fischbach MA. Natural products version 2.0: connecting genes to molecules. J. Am. Chem. Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mavromatis K, Chu K, Ivanova N, Hooper SD, Markowitz VM, Kyrpides NC. Gene context analysis in the Integrated Microbial Genomes (IMG) data management system. PLoS One. 2009;4:e7979. doi: 10.1371/journal.pone.0007979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romosan A, Shoshani A, Wu K, Markowitz VM, Mavromatis K. 2013. Accelerating gene context analysis using bitmaps. Proceedings of the 25th International Conference on Scientific and Statistical Database Management (SSDBM 2013) [Google Scholar]
- 31.Clingenpeel S. 2012. JGI microbial single cell program: single cell data decontamination. http://img.jgi.doe.gov/w/doc/SingleCellDataDecontamination.pdf. [Google Scholar]
- 32.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl Acad. Sci. USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]