Abstract
SILVA (from Latin silva, forest, http://www.arb-silva.de) is a comprehensive resource for up-to-date quality-controlled databases of aligned ribosomal RNA (rRNA) gene sequences from the Bacteria, Archaea and Eukaryota domains and supplementary online services. SILVA provides a manually curated taxonomy for all three domains of life, based on representative phylogenetic trees for the small- and large-subunit rRNA genes. This article describes the improvements the SILVA taxonomy has undergone in the last 3 years. Specifically we are focusing on the curation process, the various resources used for curation and the comparison of the SILVA taxonomy with Greengenes and RDP-II taxonomies. Our comparisons not only revealed a reasonable overlap between the taxa names, but also points to significant differences in both names and numbers of taxa between the three resources.
IMPORTANCE OF A TAXONOMIC FRAMEWORK FOR MICROBIOLOGY
Most life on earth is microbial, belonging to the ‘Bacteria’ and ‘Archaea’ domains (1), and to numerous lineages of microbial ‘Eukaryota’ (e.g. protists) (2). Less than 1% of microbes are cultivable, and therefore diversity was vastly underestimated by traditional microbiological methods (3). The known extent of microbial diversity has grown and continues to grow rapidly as sequence-based methods are used to characterize microbes (4). One of the major breakthroughs in the study of the diversity of microbes was the use of the ribosomal rRNA (rRNA) gene sequences, particularly of the small subunit (SSU; also called 16S rRNA for Bacteria and Archaea and 18S rRNA for Eukaryota). For the first time, direct comparisons between divergent microbial lineages became possible and the evolutionary relationships among all microorganisms could be elucidated (1,5,6), leading to a unified three-domain taxonomy (7). In conjunction with this molecular framework for taxonomy, ecological surveys of rRNA gene diversity in the environment have made it possible to appreciate the extent of microbial diversity present on earth (8). Appropriate taxonomic classification in sequence databases is crucial for organizing and cataloging microbial diversity. The SILVA rRNA gene databases use a phylogenetic tree-guided manual curation approach for the taxonomy of Bacteria, Archaea and Eukaryota. The eukaryotic taxonomy has recently undergone extensive curation to reflect consensus views on evolutionary relationships among the major eukaryotic lineages, which are predominantly microbial.
With this article we would like to express our gratitude and honor to Prof. Dr Jean Euzéby for his tireless work in providing the ‘List of Prokaryotic Names with Standing in Nomenclature (LPSN)’. Since 1997, he has manually checked all issues of International Journal of Systematic and Evolutionary Microbiology (IJSEM) to extract the taxonomic information (such as new species, new combinations and emendations), classify it in an orderly manner and make it electronically available in LPSN (9). Furthermore, LPSN compiles information provided by: the Taxonomic Outline of Bacteria and Archaea (TOBA), the NCBI taxonomy (10), the Taxonomic Outlines of the Bergey's Manual of Systematic Bacteriology (11) and suggestions made by ‘The All-Species Living Tree Project (LTP)’ (12). Finally, we would also like to thank Dr Aidan C. Parte for taking over Prof. Dr Euzéby’s tasks, and continuing the LSPN resource (13).
NOMENCLATURE AND CLASSIFICATION
The current nomenclature of Bacteria and Archaea is officially regulated by the International Committee on Systematics of Prokaryotes (ICSP). The ICSP is the organization responsible for editing the Bacteriological Code (14), which is an official compilation of principles, rules and recommendations for naming new taxa and renaming existing taxa. Valid taxonomic ranks are subspecies, species, subgenus, genus, subtribe, tribe, subfamily, family, suborder, order, subclass and class. While subgenus, subtribe and tribe have fallen into disuse, the two categories phylum and domain are commonly used, although not officially covered by the Bacteriological Code. In addition, the ICSP enforced the publication of all new names and combinations of names in IJSEM to compile a valid standing in taxonomic records. Consequently, all new species or new combinations that have been published in other journals must finally appear in a Validation List published periodically by the IJSEM journal. Classification of taxa, however, occurs in parallel without official rules and is subject to personal opinion. Nevertheless, genealogical inferences of Bacteria and Archaea based on the 16S rRNA gene (15) are still regarded as the most accepted classification scheme (11).
BACTERIAL AND ARCHAEAL TAXONOMY IN SILVA
SILVA predominantly uses phylogenetic classification based on an SSU guide tree. Classification and clade names are informed by widely accepted sources, and discrepancies are resolved with the overall aim of making classification consistent with phylogeny. With release 100 in 2009, the SILVA full-length (>1200 bases for Bacteria/Eukaryota and >900 bases for Archaea) SSU gene guide tree went through a major manual curation effort to represent bacterial and archaeal taxa as groups in the tree. The core of this guide tree is based on the full-length sequence tree of the ARB 2004 release (curated and distributed by Wolfgang Ludwig), and is built by adding new sequences using the ARB parsimony tool in combination with filters to remove highly variable positions (16).
In the following releases, the curated classifications were extended to cover bacterial and archaeal full-length large subunit (LSU, 23S rRNA) and eukaryotic full-length SSU (18S rRNA) gene sequences. Finally, with the SILVA release 115 in August 2013, all quality-checked SSU and LSU rRNA gene sequences from all three domains of life were automatically classified based on the established SSU and LSU reference taxonomies. The bacterial and archaeal classification in SILVA is based on Bergey’s Taxonomic Outlines (17–20). Because both taxonomy and species are dynamic entities, changes are rapid and supplemental resources are required. In such cases, name changes and taxonomic outlines are adapted from LPSN (9). Moreover, the LPSN resource is also used to track down names without standing in nomenclature (i.e. not validly published taxa) and Candidatus taxa.
The curation of classification in SILVA uses a phylogenetic tree-based process, whereas LPSN and Bergey’s classifications are not explicitly phylogenetic; thus, topological differences between the SILVA Ref trees and other resources are expected. Most notably in Proteobacteria, where the Bergey’s taxonomic framework requires updates based on new phylogenetic findings, such discrepancies are observed. For example, the genus Ahrensia (type species accession: D88524) is classified under family Rhodobacteraceae of Alphaproteobacteria in LPSN; however, in the SSU Ref guide tree, this genus is grouped together with members of the family Phyllobacteriaceae. Normally, introducing polyphyletic groups accommodates such discrepancies, but in this case genus Ahrensia is kept under Phyllobacteriaceae because of high sequence identities (>94%) observed with other members of this family. Furthermore, in an effort to standardize the number of ranks to exactly six (domain, phylum, class, order, family and genus), subclasses and suborders have been omitted for some groups, as opposed to Bergey’s or LPSN’s recommendations.
In addition to this traditional taxonomic backbone, extensive effort is spent in every release to represent prominent clades known only from environmental sequences. The majority of these clades and groups are annotated in the guide tree based on literature surveys, and occasionally based on personal communications; therefore, not all of these clades are available in publications. Some examples are OCS116 clade (21), SAGMC and SAGME groups (22) and termite clusters (23). Supplementary Table S1 provides a full list of all such clades and groups that are part of the current SILVA taxonomy. We chose to name phylogenetically coherent groups above the family rank, consisting of only sequences from uncultured organisms, after the clone name of the earliest submitted sequence.
EUKARYOTIC TAXONOMY IN SILVA
Historically, eukaryotic classification has been more intensively studied than bacterial and archaeal taxonomy, and is governed by the zoological and botanical nomenclatural codes (ICZN and ICN). The taxonomic landscape for microbial Eukaryota is complicated because lineages are governed by one or both codes but fit neither (24,25), and classification has gone through major upheavals in recent times (26,27). These complications plus the fact that protists are infrequently included in microbial ecology studies (28) are most likely the reasons for SILVA being unique in its inclusion of sequences and taxonomies of all three domains of life, although a curated database dedicated exclusively to eukaryotic sequences has recently become available (29).
Eukaryotic taxonomy within the SILVA database has been significantly improved over the past years, after the inception of the Eukaryotic Taxonomy Working Group (ETWG; http://www.arb-silva.de/projects/eukaryotic-taxonomy). With SILVA release 111 in 2012, ETWG implemented a phylogenetic tree-guided curated taxonomy for Eukaryota based on the consensus views of the International Society of Protistologists (26,30), which focused on the higher taxonomic levels (e.g. Opisthokonta, Stramenopiles, Excavata). The taxonomy has been further improved with SILVA release 115 to adhere to the latest publication from the ISOP taxonomy committee (26). Moreover, higher-level ranks have been revised in SILVA release 115 for plants, fungi, and animals, although these groups are still to be considered ‘work in progress’ and their respective classifications should be viewed as provisional.
One aspect of taxonomy particularly important for integration with computational tools is the number of ranks used to classify an organism. Ideally, life would all be neatly classifiable into a fixed number of ranks (e.g. the seven Linnean ranks), implying that ranks are directly comparable. However, the different taxonomies (i.e. for plants and for Bacteria and Archaea) are constructed with different definitions of their units (e.g. species) and often, intermediate ranks are useful to better resolve the hierarchy of a particular group of organisms. Thus, the meaning of ranks and the number used to classify a taxon vary widely across the tree of life (30,31). Taxa such as vertebrate animals are represented by 15 levels of taxonomy, while some microbial lineages, such as some lineages in Amoebozoa (Fractovitelliida), are merely represented by three. Moreover, the degree of genetic divergence and evolutionary distance encompassed by a given rank also varies enormously, such that a genus can have no variation in the SSU rRNA gene at all (e.g. fungi) or more than 10% (in some protist lineages) (32).
To accommodate variations in the number of nested levels used to classify different lineages and to increase the stability of classification, ISOP has adopted rankless classification (24,25,30), i.e. the nested position of the taxon in the taxonomic hierarchy does not imply a Linnean rank. The SILVA eukaryotic classification reflects this fluidity by reporting the full taxonomic string for lineages regardless of the number of levels included. However, we recognized the difficulties this causes in computational analyses. To address this, a table of classification rank designations is provided with each full release of SILVA to help users address the practical challenges of using bioinformatic tools that are designed for, or require, explicit rank information (33). The highest-level groups of Eukaryota and their designated ranks are given in Table 1. Within the table, rank designations adhere to the overall goal of assigning the same rank to roughly equivalent evolutionary levels across the tree. Only taxonomic levels that are distinguishable in the SILVA guide tree (also included in releases) are included in the table file; therefore, several levels of animal, plant and fungal taxonomy are missing. These ranks serve as a guideline, and we welcome users to modify them according to their specific analysis needs. Like taxonomy as a whole, the curation of eukaryotic classification within SILVA is a work in progress and will continue to evolve. Suggestions for revisions to the taxonomy for eukaryotic clades are welcome, and users are encouraged to contact the SILVA team at contact@arb-silva.de.
Table 1.
Major eukaryotic lineages represented at the highest level of the taxonomic hierarchy in the current SILVA release, and a comparison to the highest level of the Protist Ribosomal Reference Database (PR2) database
| SILVA 115 | PR2 | Rank |
|---|---|---|
| Amoebozoa | Amoebozoa | Kingdom |
| Archaeplastida | Archaeplastida | Major clade |
| Apusozoa | ||
| Centrohelida | Hacrobia (Centroheliozoa) | Kingdom |
| Cryptophyceae | Hacrobia (Cryptophyta) | Kingdom |
| Excavata | Excavata | Major clade |
| Haptophyta | Hacrobia (Haptophyta) | Kingdom |
| Incertae Sedis | Kingdom | |
| Opisthokonta | Opisthokonta | Major clade |
| Picozoa | Hacrobia (Picobiliphyta) | Phylum |
| SAR (Stramenopiles, Alveolata, Rhizaria) | Alveolata | Major clade |
| Rhizaria | ||
| Stramenopiles |
The Rank column is provisional, as explained in the Eukaryotic Taxonomy section, and only refers to the lineages from SILVA 115 release.
LTP TAXONOMY
The LTP is an initiative for the development of highly curated 16S and 23S rRNA gene sequence databases, universal alignments and reference phylogenetic trees of all the type strains of Bacteria and Archaea (34). The LTP taxonomy represents the nomenclature and classification of all bacterial and archaeal taxa with validly published names as given in LPSN. Like SILVA, LTP is an authorized provider of the LPSN taxonomy. LPSN, as a partner of the LTP team, has facilitated the access to TOBA, the NCBI taxonomy and the Taxonomic Outlines of the Bergey's Manual of Systematic Bacteriology, which for the first time appear in a database-like format inside the LTP's ARB formatted and CSV exports. The several months of delay between IJSEM and LTP releases may cause slight variations between LTP's and LPSN's taxonomy, with LPSN having the latest updated taxonomy. The LTP taxonomy is distributed over four fields of information: (1) (fullname_ltp) corresponds to the species or subspecies name; (2) (high_tax_ltp) is the name of the next higher taxon above genus; (3) (type_ltp) contains information about type species that are the nomenclatural types for the higher taxa above species; and (4) (tax_ltp) contains the complete classification into higher ranks as it appears in LPSN. More information can be found at: http://www.arb-silva.de/projects/living-tree.
COMPARISON OF SILVA TAXONOMY WITH RDP-II AND GREENGENES
The SILVA, RDP-II and Greengenes databases have different approaches for obtaining a taxonomic hierarchy, and the subsequent classification of rRNA gene sequences (35,36). For example, Greengenes uses a mixture of different resources (own curation, NCBI, SILVA, RDP-II), while RDP-II is based on Bergey’s outline with minor additions from NCBI. Here, we provide a brief comparison of the three taxonomic hierarchies based on rank names at the phylum and genus level, based on the taxonomies taken from RDP-II and Greengenes as of May 2013.
SILVA release 115 consists of 46 taxa at phylum level, which includes both commonly agreed ‘named’ phyla and the widely accepted candidate divisions. All named phyla between the three databases are congruent, except for Greengenes, which is missing Korarchaeota (Table 2). Most disagreement occurred for candidate divisions. In SILVA, we currently designate 12 divisions, 6 of which are shared with RDP-II and 9 are shared with Greengenes. It is important to note that the same candidate divisions might be named differently in each database. In addition to named phyla and the candidate divisions, both SILVA and Greengenes have a number of phylum level taxa consisting only of environmental sequences. Supplementary Table S2 provides an overview of these. While some of these groups are quite small, comprising only few sequences that could be treeing artifacts, those that collect >100 sequences certainly require attention and sound coherence testing via different treeing approaches.
Table 2.
Comparison of the phyla and candidate divisions between SILVA, RDP-II and Greengenes
| SILVA | RDP-II | Greengenes |
|---|---|---|
| Acidobacteria | Acidobacteria | Acidobacteria |
| Actinobacteria | Actinobacteria | Actinobacteria |
| Aquificae | Aquificae | Aquificae |
| Armatimonadetes | Armatimonadetes | Armatimonadetes |
| Bacteroidetes | Bacteroidetes | Bacteroidetes |
| Caldiserica | Caldiserica | Caldiserica |
| Chlamydiae | Chlamydiae | Chlamydiae |
| Chlorobi | Chlorobi | Chlorobi |
| Chloroflexi | Chloroflexi | Chloroflexi |
| Chrysiogenetes | Chrysiogenetes | Chrysiogenetes |
| Crenarchaeota | Crenarchaeota | Crenarchaeota |
| Cyanobacteria | Cyanobacteria/ Chloroplast | Cyanobacteria |
| Deferribacteres | Deferribacteres | Deferribacteres |
| Deinococcus- Thermus | Deinococcus- Thermus | Thermi |
| Dictyoglomi | Dictyoglomi | Dictyoglomi |
| Elusimicrobia | Elusimicrobia | Elusimicrobia |
| Euryarchaeota | Euryarchaeota | Euryarchaeota |
| Fibrobacteres | Fibrobacteres | Fibrobacteres |
| Firmicutes | Firmicutes | Firmicutes |
| Fusobacteria | Fusobacteria | Fusobacteria |
| Gemmatimonadetes | Gemmatimonadetes | Gemmatimonadetes |
| Korarchaeota | Korarchaeota | |
| Lentisphaerae | Lentisphaerae | Lentisphaerae |
| Nanoarchaeota | Nanoarchaeota | Nanoarchaeota |
| Nitrospirae | Nitrospira | Nitrospirae |
| Planctomycetes | Planctomycetes | Planctomycetes |
| Proteobacteria | Proteobacteria | Proteobacteria |
| Spirochaetae | Spirochaetes | Spirochaetes |
| Synergistetes | Synergistetes | Synergistetes |
| Tenericutes | Tenericutes | Tenericutes |
| Thaumarchaeota | Thaumarchaeota | Thaumarchaeota |
| Thermodesulfobacteria | Thermodesulfobacteria | Thermodesulfobacteria |
| Thermotogae | Thermotogae | Thermotogae |
| Verrucomicrobia | Verrucomicrobia | Verrucomicrobia |
| Candidate division BRC1 | BRC1 | BRC1 |
| Candidate division JS1 | ||
| Candidate division KB1 | ||
| Candidate division OD1 | OD1 | ABY1_OD1 |
| Candidate division OP11 | OP11 | OP11 |
| Candidate division OP3 | OP3 | |
| Candidate division OP8 | OP8 | |
| Candidate division OP9 | OP9 | |
| Candidate division SR1 | SR1 | SR1 |
| Candidate division TM7 | TM7 | TM7 |
| Candidate division WS3 | WS3 | WS3 |
| Candidate division WS6 | WS6 |
Differences are marked in bold.
In the next step, we investigated shared named and Candidatus taxa at the genus level. Here, the results were quite different from the phylum-level comparison, as both the amount of taxa and their names differed between the three databases (Figure 1).
Figure 1.
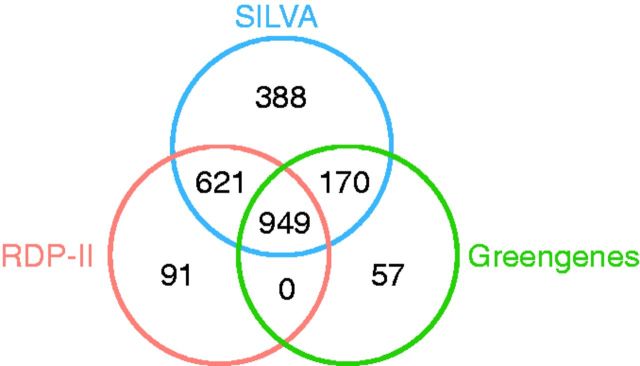
Venn diagram showing the number of shared taxa at genus level between SILVA, RDP-II and Greengenes. Only taxa containing sequences from cultivated organisms are included in this comparison. The overlapping part in the middle shows the number of all taxa jointly shared by all three databases; the other overlaps show taxa shared between two databases, but not the third. RDP-II and Greengenes share no other taxa in addition to the 949 shared jointly by all three databases.
The RDP-II and SILVA projects share the highest amount of taxa at the genus level, while SILVA and Greengenes share the least. Furthermore, SILVA has the highest number of unique taxa included in the classification, followed by Greengenes and RDP-II. The higher number of unique taxa in SILVA can be attributed to the inclusion of Candidatus taxa and taxa without standing in nomenclature taken from LPSN. This comparison also illustrates the differences (and similarities) in the curation procedure and the resources used for curation; SILVA and RDP-II appear to use the same resources and follow the same guidelines. Finally, it is important to point out that despite the differences between curation methods used, the three databases still share a sizeable number of taxa with each other.
OUTLOOK
SILVA and LTP taxonomies will be maintained with high diligence into the future. A primary focus of the taxonomic efforts in SILVA will be continual improvements of the specific eukaryotic groups, i.e. fungi, plants and animals.
A further objective is the reconciliation of the classifications used by all rRNA gene databases. The aim of such a project would be, at the very least, to provide users with the exact same name for a given taxon, regardless of the data set and classification method used. Talks are underway to implement reconciliation for Bacteria and Archaea. As there is good agreement on named taxa, such reconciliation should be relatively straightforward and mainly involving better sharing of data. Reconciliation of clades and unnamed groups, however, will require more efforts. As the ETWG working group also consists of Greengenes and RDP-II colleagues, such reconciliation will not be necessary for Eukaryota.
With the newly developed SILVA-NGS pipeline, we will grant easy access to the SILVA taxonomy for the classification of rRNA gene amplicon data. SILVA-NGS accepts any kind of short- and long-read sequence rRNA gene data in FASTA format and performs quality control, alignment and classification of rRNA genes based on the curated SILVA taxonomy. All steps (upload, progress monitoring, visualization of results and download of data) can be geared via the SILVA-NGS web-interface. The system is available at www.arb-silva.de/ngs.
Finally, we would like to emphasize that the SILVA taxonomy curation is an open and transparent process, and input from users and experts is highly appreciated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by EU’s Seventh Framework Program funds BioVeL (www.biovel.eu) [283359], SYMBIOMICS (www.symbiomics.de) [264774] and the German Research Foundation [GL-553/4-1]. Funding for open access charge: German Research Foundation [GL-553/4-1].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the contributions of the ETWG and LTP working group members on eukaryotic taxonomy and database maintenance. They would also like to thank several colleagues for providing bacterial, archaeal and eukaryotic clades: Max Planck Institute for Marine Microbiology: colleagues Bernhard Fuchs, Katrin Knittel, Emil Ruff, Hanno Teeling and Tim Richter-Heitmann; Max Planck Institute for Terrestrial Microbiology: colleagues Andreas Brune and Tim Köhler; Iñaki Ruiz-Trillo and Gemma Henderson. Finally, they would like to thank members of the RDP-II and Greengenes teams for fruitful discussions.
REFERENCES
- 1.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 2.Patterson DJ. The diversity of eukaryotes. Am. Nat. 1999;154:S96–S124. doi: 10.1086/303287. [DOI] [PubMed] [Google Scholar]
- 3.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert JA, Dupont CL. Microbial metagenomics: beyond the genome. Ann. Rev. Mar. Sci. 2011;3:347–371. doi: 10.1146/annurev-marine-120709-142811. [DOI] [PubMed] [Google Scholar]
- 5.Caron DA. New accomplishments and approaches for assessing protistan diversity and ecology in natural ecosystems. Bioscience. 2009;59:287–299. [Google Scholar]
- 6.Moreira D, Lopez-Garcia P. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 2002;10:31–38. doi: 10.1016/s0966-842x(01)02257-0. [DOI] [PubMed] [Google Scholar]
- 7.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in-situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Euzéby JP. List of bacterial names with standing in nomenclature: a folder available on the internet. Int. J. Syst. Bacteriol. 1997;47:590–592. doi: 10.1099/00207713-47-2-590. [DOI] [PubMed] [Google Scholar]
- 10.Federhen S. The NCBI taxonomy database. Nucleic Acids Res. 2012;40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodfellow M, Kampfer P, Busse HJ, Trujillo ME, Suzuki KI, Ludwig W, Whitman WB. Bergey's Manual of Systematic Bacteriology. New York: Springer; 2012. [Google Scholar]
- 12.Munoz R, Yarza P, Ludwig W, Euzeby J, Amann R, Schleifer KH, Glockner FO, Rossello-Mora R. Release LTPs104 of the all-species living tree. Syst. Appl. Microbiol. 2011;34:169–170. doi: 10.1016/j.syapm.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Parte AC. LPSN—list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2013;42:D613–D616. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapage SP, Sneath PHA, Lessel E, Skerman V, Seeliger H, Clark W. International Code of Nomenclature of Bacteria: Bacteriological Code, 1990 Revision. Washington: ASM Press; 1992. [PubMed] [Google Scholar]
- 15.Garrity GM, Jonson KL, Bell J, Searles DB. Taxonomic Outline of the Prokaryotes. 2nd edn. New York: Springer; 2002. [Google Scholar]
- 16.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boone DR, Castenholz RW, Garrity GM, Stanley JT. The Archaea and the Deeply Branching and Phototrophic Bacteria. New York: Springer; 2001. [Google Scholar]
- 18.Brenner DJ, Krieg NR, Garrity GM, Staley JT. The Proteobacteria. New York: Springer; 2005. [Google Scholar]
- 19.Vos PD, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB. The Firmicutes. New York: Springer; 2009. [Google Scholar]
- 20.Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB. The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes. New York: Springer; 2010. [Google Scholar]
- 21.Morris RM, Vergin KL, Cho JC, Rappe MS, Carlson CA, Giovannoni SJ. Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the bermuda atlantic time-series study site. Limnol. Oceanogr. 2005;50:1687–1696. [Google Scholar]
- 22.Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK. Archaeal diversity in waters from deep South African gold mines. Appl. Environ. Microbiol. 2001;67:5750–5760. doi: 10.1128/AEM.67.21.5750-5760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Köhler T, Stingl U, Meuser K, Brune A. Novel lineages of Planctomycetes densely colonize the alkaline gut of soil-feeding termites (Cubitermes spp.) Environ. Microbiol. 2008;10:1260–1270. doi: 10.1111/j.1462-2920.2007.01540.x. [DOI] [PubMed] [Google Scholar]
- 24.Adl SM, Leander BS, Simpson AG, Archibald JM, Anderson OR, Bass D, Bowser SS, Brugerolle G, Farmer MA, Karpov S, et al. Diversity, nomenclature, and taxonomy of protists. Syst. Biol. 2007;56:684–689. doi: 10.1080/10635150701494127. [DOI] [PubMed] [Google Scholar]
- 25.Lahr DJ, Lara E, Mitchell E. Time to regulate microbial eukaryote nomenclature. Biol. J. Linn. Soc. 2012;107:469–476. [Google Scholar]
- 26.Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parfrey LW, Barbero E, Lasser E, Dunthorn M, Bhattacharya D, Patterson DJ, Katz LA. Evaluating support for the current classification of eukaryotic diversity. PLoS Genet. 2006;2:e220. doi: 10.1371/journal.pgen.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caron DA, Worden AZ, Countway PD, Demir E, Heidelberg KB. Protists are microbes too: a perspective. ISME J. 2009;3:4–12. doi: 10.1038/ismej.2008.101. [DOI] [PubMed] [Google Scholar]
- 29.Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L, Boutte C, Burgaud G, de Vargas C, Decelle J, et al. The protist ribosomal reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013;41:D597–D604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 31.Avise JC, Liu J. On the temporal inconsistencies of Linnean taxonomic ranks. Biol. J. Linn. Soc. 2011;102:707–714. [Google Scholar]
- 32.Pawlowski J, Audic S, Adl S, Bass D, Belbahri L, Berney C, Bowser SS, Cepicka I, Decelle J, Dunthorn M, et al. CBOL protist working group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol. 2012;10:e1001419. doi: 10.1371/journal.pbio.1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH, Ludwig W, Glöckner FO, Rossello-Mora R. The all-species living tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 2008;31:241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 35.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2011;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]


