Abstract
The National Human Genome Research Institute (NHGRI) Catalog of Published Genome-Wide Association Studies (GWAS) Catalog provides a publicly available manually curated collection of published GWAS assaying at least 100 000 single-nucleotide polymorphisms (SNPs) and all SNP-trait associations with P <1 × 10−5. The Catalog includes 1751 curated publications of 11 912 SNPs. In addition to the SNP-trait association data, the Catalog also publishes a quarterly diagram of all SNP-trait associations mapped to the SNPs’ chromosomal locations. The Catalog can be accessed via a tabular web interface, via a dynamic visualization on the human karyotype, as a downloadable tab-delimited file and as an OWL knowledge base. This article presents a number of recent improvements to the Catalog, including novel ways for users to interact with the Catalog and changes to the curation infrastructure.
INTRODUCTION
Genome-wide association studies (GWAS) assay at minimum hundreds of thousands of single-nucleotide polymorphisms (SNPs) to identify associations with complex clinical conditions and phenotypic traits. Unlike single gene disorders, such as Huntington’s disease, complex diseases are usually the result of a combination of genetic and environmental factors, each of which increases susceptibility to the condition. In the absence of a single causative variant, GWAS aim to identify SNPs in linkage disequilibrium with a gene or other regulatory element that might contain a causal variant contributing to a condition’s aetiology. GWAS are non-candidate gene-driven and use a whole-genome approach in large studies of up to hundreds of thousands of individuals investigating traits such as disease, response to drug and anthropometry (1).
The importance of GWAS in advancing scientific understanding of disease mechanisms and providing starting points for the development of medical treatments is well established. One of the first GWAS identified a polymorphism in complement factor H, part of complement-mediated inflammation, in a study on age-related macular degeneration. This association between the degenerative disease and inflammation pathways has since been verified in other studies and is now showing promising results as a therapeutic target (2). Recent discovery of shared loci in diseases previously thought not to have any common aetiology include gene CDKN2A/B in type II diabetes mellitus (3) and myocardial infarction (4) and CDKAL1 in Crohn’s disease (5) and type II diabetes mellitus (6).
GWAS data are typically reported in peer-reviewed publications as single studies or meta-analyses, and data deposited in resources such as the European Genome-phenome Archive (http://www.ebi.ac.uk/ega), or dbGaP (7), where users must apply for access to individual-level genotype and phenotype data and comply with a data access agreement. The National Human Genome Research Institute (NHGRI) Catalog of Published GWAS Catalog provides a publicly available manually curated collection of all published GWAS conforming to eligibility criteria including number of SNPs assayed and non-targeted study design. The eligibility criteria are public and available at http://www.genome.gov/27529028 (8). This article introduces a number of recent improvements including the automation of SNP extraction from papers, increasing the number of SNPs included per paper, review and re-annotation of trait designations using a dedicated GWAS ontology, delivery of a new automated GWAS visualization, delivery of automated PubMed searching to detect eligible papers and implementation of a curation tracking system. Together, these features support the curatorial staff in delivering more data for the Catalog, improve integration with other resources and provide improved visualization and queries for users.
CATALOG DATA
The GWAS Catalog data are extracted from the literature. Extracted information includes publication information, study cohort information such as cohort size, country of recruitment and subject ethnicity and SNP-disease association information including SNP identifier (i.e. Reference SNP cluster ID (RSID)), P-value, gene and risk allele. Each study is also assigned a trait that best represents the phenotype under investigation. When multiple traits are analysed in the same study, either multiple entries are created or individual SNPs are annotated with their specific traits. Traits are used both to query and visualize the data in the Catalog’s web form and diagram-based query interfaces.
Data extraction and curation for the GWAS Catalog is an expert activity; each step is performed by scientists supported by a web-based tracking and data entry system, which allows multiple curators to search, annotate, verify and publish the Catalog data. Papers that qualify for inclusion in the Catalog are identified through weekly PubMed searches, and then undergo two levels of curation. A detailed overview of the curation process can be found on the GWAS diagram website (www.ebi.ac.uk/fgpt/gwas/#curationtab).
Extraction and verification of data from the literature is time-consuming. GWAS include initial and replication data sets, and these are often not clearly separated in papers. Accurate description of the phenotype is also complex, as disease markers may be measured and implicit inferences are made by the authors from these. One of the most challenging areas of curation is the consistent extraction of ethnicity data. Many GWAS papers do not provide detailed descriptions of the ethnic background of their study cohorts, and even today there is no standardized way of reporting ethnicity. In some cases, inferences about the ethnicity of study subjects can be made based on the country of origin or recruitment using census information provided by the CIA world fact book (9). For example, subjects recruited in East Asian countries can usually be assumed to be of East Asian ancestry or subjects recruited in Scandinavian countries of European ancestry. However, such inferences cannot necessarily be made in more ethnically diverse countries such as the USA or the UK, which is where a disproportionately large number of GWAS are carried out.
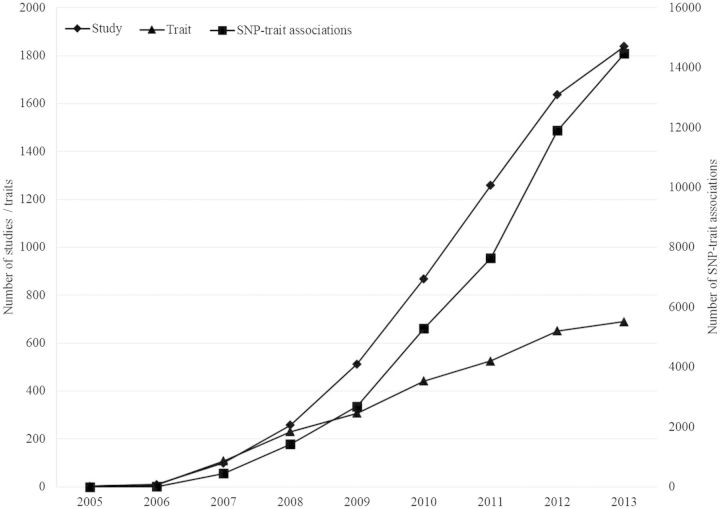
As the number of eligible published GWAS continues to rise (Figure 1), it proved essential to automate the curation process. Automation was achieved in 2013 by deploying an infrastructure to perform automatic searches of PubMed, tracking of papers through the curatorial pipeline and, for eligible papers, automatic extraction of citation information and batch extraction of SNP RSIDs, traits and P-value information from papers. Ease of extraction of large data sets was improved by the development of a spreadsheet-based batch SNP extractor to process all SNPs for one paper simultaneously. Historically, it was necessary for practical reasons to limit the number of extracted SNPs to 50 per paper. The SNP extractor allows curators to process all eligible SNP associations, typically those with a P <1 × 10−5, in a paper and has increased the number of SNPs curated and improved processing time for curators, as studies typically report larger numbers of SNPs and traits over time. Papers that were previously subject to the 50-SNP limit have been recently re-curated to extract the additional SNPs; in a small number of papers, >1000 significant SNPs were identified, thus greatly enriching the Catalog’s content.
Figure 1.
Studies, traits and SNP-trait associations from 2005–2013 reveal the growth in eligible studies.
A recent Catalog improvement has been to structure the trait annotations using an ontology and to use ontology and semantic web technologies to automate the process of providing the karyotype visualization. This involved developing an ontology to describe the traits, a knowledge base to contain the data, verifying all previous trait annotations and mapping ontology terms to them before delivering a new query interface for visualizing and querying the data.
ONTOLOGY
The utility of ontologies for providing formal data description frameworks in the biological domain is well established (10). This applies particularly to ontologies developed in an expressive knowledge representation language like OWL (11) whose rich semantic capabilities allow a much more natural modelling of complex concepts through axiomatization. This in turn allows for much richer querying than simple string searching. Mapping to ontologies also greatly facilitates data integration across heterogeneous data sources such as data extracted from the scientific literature. Until recently, the traits in the GWAS Catalog were available as an unstructured flat list, allowing only querying through direct string matching and therefore limiting the Catalog’s potential, both as a stand-alone resource and as an integral part of a wider network of genomic resources. Producing consistent high-quality phenotype mappings represents an essential yet challenging task. There is a high level of variation between studies in what constitutes a phenotype and how a phenotype is described. For example, a specific phenotype or disease may be studied explicitly or a phenotype considered to be a marker for disease, e.g. measurements of blood glucose concentration and body weight as risk factors for type II diabetes. To allow consistent groupings of studies and comparisons of results among studies, it is essential that all mappings are consistent but without making potentially incorrect assumptions. For example, a study exploring the effect of SNP level variation on blood glucose levels may use this as a risk indicator for type II diabetes, but unless this is explicitly stated a mapping to type II diabetes would be incorrect.
We evaluated several domain ontologies for coverage of the GWAS data, including Medical Subject Headings (MeSH), the Human Phenotype Ontology (12), the Disease Ontology (13) and the Experimental Factor Ontology (EFO) (14). No single ontology addresses all the representational needs of the GWAS Catalog. However, EFO has branches that can be extended to include new terms and allows modelling of tests, diseases, anatomy and anthropometry, imports terms from available domain-specific ontologies and uses axiomatization to maintain its structure. Many MeSH terms were approximately mapped and did not provide the precision required by the curators. For example, the annotation ‘C-reactive protein levels’ in the Catalog was mapped to MeSH ‘D002097 C-reactive protein’. But the implicit information in the Catalog annotation is that the protein levels are an indication of inflammatory response. When modelled in EFO, a new term, EFO: ‘EFO_0004458 C-reactive protein measurement’, with a parent class ‘inflammatory marker measurement’ allows queries for combinations of all inflammatory diseases and all inflammatory markers, and provides a more detailed query result for the user.
EFO is an application ontology that uses multiple reference ontologies such as chemical entities of biological interest (15) to produce a single ontology, which can be viewed under one hierarchy and can be applied to visualize a data set. Current GWAS traits range from simple disease descriptions, such as ‘breast cancer’, to measurements, such as ‘waist-hip ratio’ or ‘liver enzyme levels’, to compound traits like ‘body mass in chronic obstructive pulmonary disease’, which are hard to represent in an ontology, and trait definitions are often context-dependent. EFO not only contains disease categories but also phenotypic descriptions, compound treatments and so forth. Traits not present in EFO were added either by importing existing terms from reference ontologies including the Gene Ontology (GO) (16) and the Human Phenotype Ontology (12), or creating new concepts locally, or by requesting these from external ontologies. A new TermGenie template was provided by the GO editors to provide the required ‘response to drug’ terms for the Catalog. The process of building the ontology improved both the consistency of term assignment for existing studies, which were reviewed, and the query capability for the Catalog.
The availability of high-quality mappings between GWAS traits and ontology terms facilitates the integration of the GWAS Catalog with other resources. The Catalog is used by Ensembl (17) through direct import of the tab-delimited file of all the Catalog content that can be downloaded from the Catalog website. Ensembl also is in the process of mapping phenotypes to EFO, so adding the existing GWAS mappings to Ensembl would immediately add an entirely new layer of connectivity between concepts that are annotated with the same EFO terms. This is particularly useful at the cross-species mapping level, where mapping genetic and variation data from different species such as mouse and human to common phenotypes provides a new way of identifying related genes.
A schema ontology was designed to model the key concepts represented in the Catalog such as trait, SNP, study, gene and chromosome and relationships that link these concepts, e.g. ‘SNP part of gene’ or ‘study describes trait association’. This schema ontology forms the basis of an OWL knowledge base containing all the data in the GWAS Catalog in a format that can be processed by an ontology reasoner and queried. By representing the GWAS Catalog as an OWL knowledge base, and reasoning over the asserted axioms, it is possible to make inferences about the SNP-trait associations that are not possible in the relational database version of the Catalog. This enables more expressive queries at different levels of granularity and the ability to detect errors and inconsistencies in the data. Where previously, a comparison between gastric and oesophageal cancers would have required either all terms to be enumerated and searched separately, or a high-level search for all cancer-related traits, possible queries of the knowledge base range from ‘Find all SNPs that are associated with gastric cancer’ to ‘Find all SNPs that are associated with cancers located in the upper digestive tract’ to ‘Find all SNPs that are associated with cancer’. Synonyms are both imported from reference ontologies and added locally to support spelling variations and plurals. The knowledge base currently contains the data that are publicly available in the Catalog. To effectively represent the ethnicity information extracted by curators, an ethnicity ontology is currently under development. Furthermore, by extending the schema ontology to include additional concepts, it would be relatively easy to include extra information in a knowledge base, such as linkage disequilibrium information, or population-specific information, in the future. Equally, other ontologies could be imported without much difficulty to allow visualization of the data using alternative terminologies.
DATA ACCESS
GWAS Catalog data can be accessed in three ways. (i) Via the web interface hosted at the NHGRI, which provides access to data via a search form including traits and study publication information, as well as a tab-delimited file is available for download. (ii) Via a new dynamic query interface of traits visualized on the human karyotype, linked to literature, SNPs in Ensembl and ontology terms in the Experimental Factor Ontology was implemented (described later). (iii) GWAS Catalog data are available through commonly used data portals such as Ensembl (17), UCSC Genome Browser (18) and PheGenI (19) and many other community tools.
The GWAS diagram shown in Figure 2 is a visualization of all SNP-trait associations in the GWAS Catalog with the P < 5 × 10−8, mapped onto the human karyotype. Each dot represents an association between a SNP (or chromosomal region if multiple SNPs are in proximity) and a disease or trait. It has been produced quarterly by the GWAS Catalog team since 2006. Until mid-2012, this was done manually with the help of a medical illustrator and required a considerable number of person hours each month. The result was a static image distributed in PDF and PowerPoint formats. Each trait association on the diagram was represented by a circle of a different colour. While this generated an iconic diagram for the Catalog, the last published version of the hand-drawn diagram contained in excess of 200 colours. To maintain visual consistency in the redeveloped diagram, the new diagram retains the fundamental principles of the manually drawn version by reusing the same chromosomal ideograms and mapping SNP-trait associations to the level of cytogenetic bands. The crucial change from the old diagram is the design of the new colour scheme based on 17 colours representing larger, empirically determined and ontology-based trait categories. The high-level categories shown in the diagram were determined by analysis of the query logs to capture the most popular search terms, and assignment of colours by number of traits across the ontology hierarchy to ensure that colour distribution made the diagram comprehensible. Figure 2 shows that users can visualize all SNPs by a high-level term such as ‘metabolic disease’, and links are available to Ensembl, the Catalog interface and PubMed. Data can also be visualized by typing a query such as ‘C-reactive protein’ and using the NCBO autocomplete widget (20). An animation of SNPs and traits per Catalog release and downloadable images for common queries are provided for user download, as these are often used in slide presentations. It is possible to generate a customized diagram pertaining to a category of interest, e.g. cancer, and identify any zones of particularly high density for that trait. The nature of the categories gives an interesting insight into which GWAS traits produce an especially high number of strong SNP-trait associations, e.g. the category ‘liver enzyme measurement’ has a similar number of traits on the diagram as the much broader category ‘cardiovascular disease’. The visualization and ontology in combination allows users to access rich genomic information pertaining to a trait by a simple query for trait name, e.g. Alzheimer’s disease, using the hierarchy, e.g. all nervous system disease, and the diagram links to Ensembl for RSIDs, allowing access to genomic context, and population level information.
Figure 2.
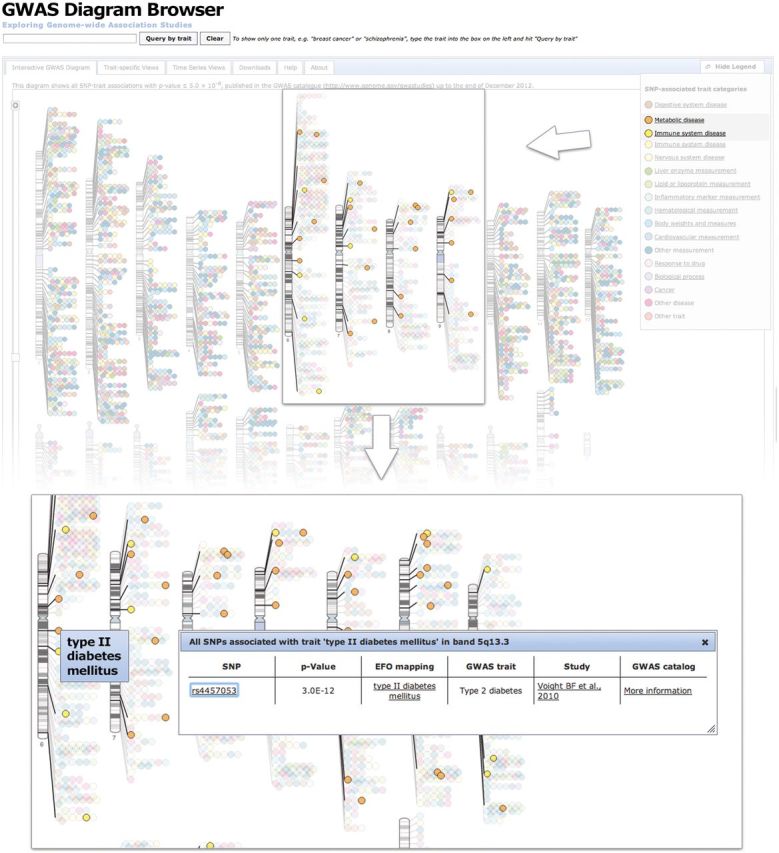
The interactive GWAS diagram is a visualization of all SNP-trait associations with P <5 × 10−8, mapped to the SNP's cytogenetic band. Visualizations of SNPs with metabolic or immune system disease are highlighted. Hovering over a SNP displays the trait name; clicking on it returns the individual SNP-trait associations, including P-value and publication, as well as links to the relevant entry in the GWAS Catalog, Europe PMC and Ensembl.
CONCLUSION
We present the GWAS Catalog, supporting infrastructure and new query, curatorial and visualization tools. The mapping of complex phenotypic traits to the Experimental Factor Ontology facilitates cross-study comparisons within the Catalog as well as the integration of Catalog data with other resources. It also supports the generation of a semantic web technology-driven interactive visualization of the GWAS Catalog data and offers community access. A number of improvements to data holdings, extraction processes and data access are planned based on recruitment of community feedback.
A user survey (performed at ASHG in 2012 and by emailing owners of data sets included in the Catalog) provided useful community feedback and allowed prioritization of future developments including: the provision and visualization of trait information at the SNP level, inclusion of queryable ethnicity information and enhancements to the usability of the web interfaces, such as deployment of the ontology in the form-based web portal. To ensure that the infrastructure and curation processes scale in the future, we are investigating inclusion of text mining to support curation, for example, to more easily identify ethnicity and population information and use of a triple stores for data storage to improve performance as data volumes grow.
A major challenge for the Catalog is the increasing complexity of studies, e.g. traits are more often compound, gene by environment studies are available and in the past year several gene by gene interaction studies were included in the Catalog. Therefore, the Catalog infrastructure must evolve to support these cases, improve data extraction times and address changing technologies for GWA studies, including the use of next-generation sequencing technology. A recent webinar organized by the Catalog explored these issues with a diverse range of researchers and provides background information on community use of and future directions for the Catalog (http://tinyurl.com/nbxxm2e).
Future technical developments include further enhancements to ethnicity extraction, including the development of an ontology to model ethnicity, improvements to the diagram to allow richer querying and filtering, e.g. by date ranges, publication and a combination of these. The diagram will also support better linking to external resources (Ensembl, dbSNP and PubMed) and more precise modelling of trait associations, to distinguish between the case where a single SNP is associated with multiple traits (myocardial infarction and high blood pressure), and the case where a SNP is shown to be associated with a trait in the context of a condition (e.g. myocardial infarction in a cohort of patients with high blood pressure). We plan to publish widgets that allow views of the GWAS diagram to be embedded in other resources. Finally, we would also like to improve the GWAS Catalog search interface to support ontology-driven querying and better integrate the visualization with the web form-based interface by developing a new user interface.
FUNDING
The National Institutes of Health [3U41-HG006104 to D.W. and J.M-.A.] and EMBL core funds supporting (to P.F., H.P. and T.B.); EMBL Core Funds and by the Health Thematic Area of the Cooperation Programme of the European Commission within the VII Framework Programme for Research and Technological Development supported (to J.M.); Grant number [200754] Gen2Phen. conducted this work as employees of the NHGRI, NIH (by H.A.J., P.H., K.A.H., T.A.M. and L.A.H.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Francis Rowland (EMBL-EBI) for advice on interface development and usability, Mary Todd Bergman and Spencer Phillips (EMBL-EBI Outreach) for assistance with figures, Fiona Cunningham (EMBL-EBI) and many users of the Catalog for their feedback. They also thank their colleagues at the NCBI, and Ensembl for distribution of data via their resources and feedback. They thank Darryl Leja for advice on development of the karyotype visualization and Mike Feolo and Ming Xu (NCBI) for informatics support.
REFERENCES
- 1.Manolio TA. Cohort studies and the genetics of complex disease. Nat. Genet. 2009;41:5–6. doi: 10.1038/ng0109-5. [DOI] [PubMed] [Google Scholar]
- 2.Troutbeck R, Al-Qureshi S, Guymer RH. Therapeutic targeting of the complement system in age-related macular degeneration: a review. Clin. Exp. Ophthalmol. 2012;40:18–26. doi: 10.1111/j.1442-9071.2011.02581.x. [DOI] [PubMed] [Google Scholar]
- 3.Voight B, Scott L, Steinthorsdottir V, Morris A, Dina C, Welch R, Zeggini E, Huth C, Aulchenko Y, Thorleifsson G, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schunkert H, König IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AFR, Barbalic M, Gieger C, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Circ. Cardiovasc. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franke A, McGovern DPB, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PIW, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 7.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The World Factbook 2013-14. Washington, DC: Central Intelligence Agency. 2013 [Google Scholar]
- 10.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith MK, Welty C, McGuinness DL. W3C Recommendation. 2009. Web ontology language. In: [Google Scholar]
- 12.Robinson PN, Mundlos S. The human phenotype ontology. Clin. Genet. 2010;77:525–534. doi: 10.1111/j.1399-0004.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- 13.Schriml LM, Arze C, Nadendla S, Chang Y-WW, Mazaitis M, Felix V, Feng G, Kibbe WA. Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012;40:D940–D946. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malone J, Rayner TF, Zheng Bradley X, Parkinson H. Proceedings of the Eleventh Annual Bioontologies Meeting. Toronto, Canada; 2008. Developing an application focused experimental factor ontology: embracing the OBO Community. [Google Scholar]
- 15.De Matos P, Dekker A, Ennis M, Hastings J, Haug K, Turner S, Steinbeck C. ChEBI: a chemistry ontology and database. J. Cheminform. 2010;2:P6. [Google Scholar]
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25: 25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karolchik D, Hinrichs AS, Kent WJ. The UCSC Genome Browser. Current Protocols in Bioinformatics. 2012;40:1.4.1–1.4.33. doi: 10.1002/0471250953.bi0104s40. [DOI] [PubMed] [Google Scholar]
- 19.Ramos EM, Hoffman D, Junkins HA, Maglott D, Phan L, Sherry ST, Feolo M, Hindorff LA. Phenotype-Genotype Integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur. J. Hum. Genet. EJHG. 2013;22:144–147. doi: 10.1038/ejhg.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, Musen MA. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011;39:W541–W545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]