Abstract
Recent evidence has implicated APOBEC3B as a source of mutations in cervical, bladder, lung, head and neck, and breast cancers. APOBEC enzymes normally function in innate immune responses, including those that target retroviruses, suggesting links between mutagenesis, immunity and viral infection in the process of cancer development.
Human cancer genomes are exceptionally complex, as they contain tens of thousands of mutations. Despite this complexity, the observed nucleotide substitutions may provide fingerprints to indict specific mechanisms in carcinogenesis. In this issue of Nature Genetics, two papers, one from Reuben Harris and colleagues1 and a second from Dmitry Gordenin, Gad Getz and colleagues2, demonstrate that a gene encoding one member of the APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) family of cytidine deaminases, APOBEC3B, leaves a tell-tale mutation signature that is enriched in the genomes of many human cancers. APOBEC family members normally function as part of the innate immune system that protects against retrovirus and retrotransposon propagation, such as restricting HIV-1 viral reverse transcription3. However, these enzymes can also deaminate cytosines in the host genome and generate C→T mutations4–6.
In a previous study, Burns et al.4 provided evidence that APOBEC3B is overexpressed in breast tumors and cell lines and that the APOBEC3B mutation signature is statistically more prevalent in the breast tumor database of The Cancer Genome Atlas (TCGA) than is expected4. In this issue, Burns et al.1 extend their study to include 19 different human tumors. They systematically analyzed mutation frequencies, spectra and sequence contexts using whole-exome mutation data from TCGA. Their RNA sequencing (RNA-seq) studies indicated that APOBEC3B is overexpressed in several human cancer types, and this overexpression correlates with the presence of the APOBEC3B mutation signature. These analyses, together with evidence from previous studies4,6,7, suggest that APOBEC3B may contribute to the cytosine mutation clusters observed in many cancers5,6.
The paper by Roberts et al.2 provides an in-depth analysis of both whole-genome and whole-exome sequencing data sets from various sources, including TCGA. Using whole-genome data sets, the authors found that many mutations are clustered and localized at breakpoints of chromosomal rearrangements, which is consistent with the known specificity of APOBEC family members3,6 for single-stranded DNA. Interestingly, the APOBEC3B mutation signature was detectable in colorectal and prostate cancers only when whole-genome, but not whole-exome, data were used, suggesting a tissue-specific bias against enrichment of mutations by APOBEC3B in coding regions. In addition, their analysis showed that the HER2-enriched subtype of breast cancer harbors an exceptionally high frequency of APOBEC3B-associated mutations as compared with other breast cancer subtypes. Both studies from Burns et al.1 and Roberts et al.2 independently reached the same conclusion that the APOBEC3B mutation signature is specifically enriched in six types of cancers, including those of the cervix, bladder, lung (adeno and squamous cell), head and neck, and breast.
APOBEC3B mutation signature
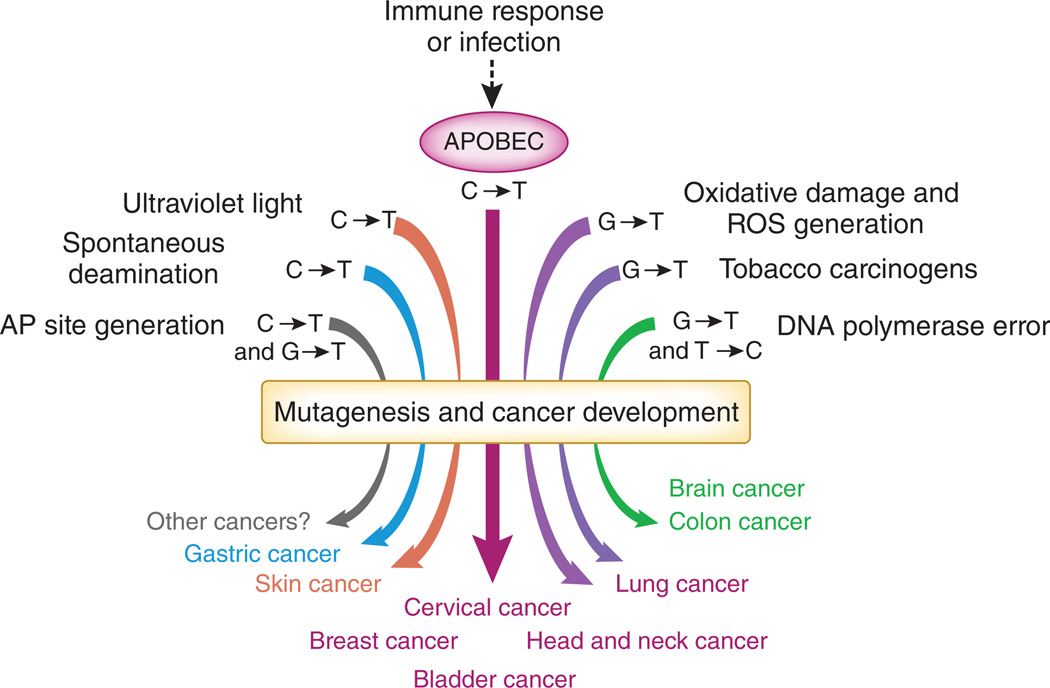
Mutation signatures have aided in the identification of environmental mutagens and carcinogens (Fig. 1)7,8. C→T transitions in cervix, bladder, lung, head and neck, and breast cancers have been suggested to be caused by APOBEC3B on the basis of the positive association of the distinct mutation signature and overexpression of APOBEC3B1,2. Cytosine deamination by APOBEC3B results in a uracil residue. Excision of the uracil residue by uracil DNA glycosylase generates an apurinic (AP) site, and insertion of adenine opposite the AP site results in C→T transitions9. Alternatively, DNA polymerases can replicate across the uracil residue directly and insert an adenine, which also results in C→T transitions9. AP sites and C→T mutations can also arise by other processes (Fig. 1), including spontaneous or chemical-induced cytosine deamination, error-prone bypass after ultraviolet light damage or DNA polymerase errors7,9. The contribution of other mutagenic processes in the cancers that are associated with APOBEC3B mutagenesis is still unclear. One can partially discriminate between the various events leading to cytosine substitutions using the sequence of the adjacent nucleotides. APOBEC3B preferentially deaminates cytosine residues when it is adjacent to a 5′ thymine6,10,11 and a 3′ thymine or adenine4. In the current studies, only cytosine substitutions that occur within the trinucleotide TCA or TCT sequence context are attributed to APOBEC3B mutagenesis1,2. However, the contribution of other mutagens to cytosine substitutions in these sequence contexts cannot be ruled out. Further studies are also warranted to identify the translesion polymerase that contributes to the C→G and C→A mutations observed at these sequence motifs1,2.
Figure 1.
Mutation signatures and cancer. There are many sources of mutagenic activity that contribute to tumorigenesis. APOBEC3B, which has a known role in innate immune responses, is now implicated as a source of mutation in cervical, bladder, lung, head and neck, and breast cancers. ROS, reactive oxygen species.
Immunity, mutagenesis and cancer
Other APOBEC enzymes could also act as mutators. Overexpression of two other cytidine deaminases has been shown to cause cancer in transgenic mice: overexpression of Apobec1 causes hepatocellular carcinoma12, whereas overexpression of activation-induced deaminase (Aid) causes T cell lymphomas13. Overexpression of Apobec3b in animal models may confirm its tumor-type specificity.
Elucidation of the signals that lead to overexpression of APOBEC family members in cancer cells may reveal how viral infection and immune responses are associated with carcinogenesis. Expression of AID and mutagenesis have been shown to be induced by Helicobacter pylori infections in normal gastric epithelia14. Intriguingly, both the current studies1,2 and recent work from Lawrence et al.7 separately showed that the APOBEC3B mutation signature is enriched in cervical and head and neck cancers, and a major risk factor for the development of these cancers is infection by human papilloma virus. It will be interesting to determine whether viral infections can trigger APOBEC3B mutagenesis and whether variation in infection and immune status can explain why APOBEC3B is associated with cancers in some tissues but not others.
On the basis of these studies, it is reasonable to hypothesize that inhibitors of APOBEC3B may prevent mutation accumulation in specific human cancers. Alternatively, it is postulated that APOBEC-mediated mutagenesis of viral DNA may result in an increase in viral mutation load to a level that exceeds the threshold for viral viability3,10. Accordingly, induction of APOBEC family members in certain human tumors with an existing high mutation load may similarly increase mutation numbers to a point that exceeds tumor viability15.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Burns MB, Temiz NA, Harris RS. Nat. Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts SA, Getz G, Gordenin DA. Nat. Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris RS, Liddament MT. Nat. Rev. Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 4.Burns MB, et al. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lada AG, et al. Biol. Direct. 2012;7:47. doi: 10.1186/1745-6150-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor BJ, et al. eLife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrence MS, et al. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nik-Zainal S, et al. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeb LA, Preston BD. Annu. Rev. Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 10.Bishop KN, et al. Curr. Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 11.Beale RC, et al. J. Mol. Biol. 2004;337:585–596. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 12.Yamanaka S, et al. Proc. Natl. Acad. Sci. USA. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbiani DF, Nussenzweig MC. Annu. Rev. Pathol. 2013;8:79–103. doi: 10.1146/annurev-pathol-020712-164004. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto Y, et al. Nat. Med. 2007;13:470–476. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- 15.Prindle MJ, Fox EJ, Loeb LA. Curr. Drug Targets. 2010;10:1296–1303. doi: 10.2174/1389450111007011296. [DOI] [PMC free article] [PubMed] [Google Scholar]