Abstract
Recent studies have found that uncontrolled diabetes and consequential hyperglycemic conditions can lead to increased incidence of osteoporosis. Osteoblasts, adipocytes, and mesenchymal stem cells (MSCs) are all components of the bone marrow microenvironment and thus may have an effect on diabetes-related osteoporosis. However, few studies have investigated the influence of these three cell types on each other, especially in the context of hyperglycemia. Thus, we developed a hydrogel-based 3D culture platform engineered to allow live-cell retrieval in order to investigate the interactions between MSCs, osteoblasts, and adipocytes in mono-, co-, and tri-culture configurations under hyperglycemic conditions for 7 days of culture. Gene expression, histochemical analysis of differentiation markers, and cell viability were measured for all cell types, and MSC-laden hydrogels were degraded to retrieve cells to assess colony-forming capacity. Multivariate models of gene expression data indicated that primary discrimination was dependent on neighboring cell type, validating the need for co-culture configurations to study conditions modeling this disease state. MSC viability and clonogenicity were reduced when mono- and co-cultured with osteoblasts in high glucose levels. In contrast, MSCs had no reduction of viability or clonogenicity when cultured with adipocytes in high glucose conditions and adipogenic gene expression indicated that cross-talk between MSCs and adipocytes may occur. Thus, our unique culture platform combined with post-culture multivariate analysis provided novel insight into cellular interactions within the MSC microenvironment and highlights the necessity of multi-cellular culture systems for further investigation of complex pathologies such as diabetes and osteoporosis.
Introduction
Diabetes is associated with insulin deficiency (Type I) or resistance (Type II) and consequential dysregulation in adipose tissue and energy metabolism.1 Notably, both type I and II diabetes are associated with increased risk of osteoporosis, a skeletal disorder characterized by low bone mass and microarchitectural deterioration of bone.2 Among other cell types, adipocytes and osteoblasts are dysregulated during the progression of diabetes and resulting secondary osteoporosis.3 As both cell types are differentiated from mesenchymal stem cells (MSCs) and are components of the bone marrow microenvironment,1,3,4 it is possible that the progression of these diseases involves altered MSC behavior.3
The stem cell microenvironment, where stem cells derive signals from the extracellular matrix (ECM), cellular contacts, and both short and long range soluble factors,5,6 has been seen to change in disease states and has recently gained interest as a potential new target for disease therapies.5,6 Within the bone marrow compartment, MSCs are directed to differentiate to osteoblasts or adipocytes, a process that is tightly regulated, partially by cellular communication between MSCs and the osteoblasts and adipocytes in the immediate microenvironment.3 Irregular MSC behavior has been observed in abnormal environments, such as the tumor microenvironment, where MSCs home and potentially participate in tumor pathogenesis.7 Similarly, in an in vitro model of Gaucher disease, MSCs were seen to have reduced proliferative capacity and may contribute to increased bone resorption.8 As it has been hypothesized that alterations in the MSC microenvironment both contribute to and result from interactions with bone and adipose tissues,3 understanding how environmental changes inherent to diabetes impact these interactions may provide insight into the role MSCs play in the progression of diabetes and concomitant osteoporosis.
Clinically, diabetes is often associated with hyperglycemic conditions due to the body’s inability to properly regulate the amounts of glucose in the blood.4 Studies have shown that elevated glucose levels have negative effects on MSCs, adipocytes and osteoblasts, all of which are cell types that influence the MSC microenvironment. Data suggest that at high glucose levels, MSCs undergo increased apoptosis and senescence as well as lose colony forming capacity and osteogenic potential.9–12 Adipocytes have demonstrated decreased insulin sensitivity, unregulated triglyceride storage, increased production of reactive oxygen species and pro-inflammatory cytokines, and decreased adiponectin secretion when cultured in high glucose conditions.13–15 Finally, osteoblasts cultured in high glucose have shown reduced proliferative capacity, mineralization capabilities, collagen I synthesis, and expression of differentiation markers.16–19 However, how these individual consequences impact cellular cross-talk between all three cell types remains to be fully understood, though previous work has shown that intercellular communication is affected in the context of diabetes. For instance, murine osteoblasts in co-culture with bone marrow cells from diabetic mice undergo increased cell death as compared to those co-cultured with bone marrow cells from normal mice.20 This indicates that MSCs derived from diabetic tissues may have an altered secretome, but how these changes influence interactions between MSCs and neighboring cell types in the bone marrow niche remains largely unexplored. Understanding how hyperglycemic conditions influence MSCs both directly and indirectly (through soluble signaling from neighboring osteoblasts and adipocytes) may provide insight into how the altered stem cell microenvironment contributes to tissue dysregulation, particularly in the development of diabetes-related osteoporosis.
To gain such biological insight, it is necessary to use an in vitro culture system that permits the co-culture of multiple cell types but still allows specific cell population analyses. As opposed to in vivo experiments, in vitro systems can be advantageous by eliminating the confounding factors present in animal models and by permitting use of human cells. In past in vitro studies, both 2D techniques, such as transwell-based systems and cell patterning,21,22 and 3D techniques, involving various biomaterial-based scaffolds,23,24 have provided useful information about cellular interactions. However, in many of these systems, both separation of unique cell populations for analysis and retrieval of live cells for further experimentation are difficult. Therefore, this study demonstrates the development and implementation of a 3D tri-culture platform that permits individual cell type analysis 25 and on-demand retrieval and further analysis of live MSCs post-culture. To achieve live cell retrieval, a matrix metalloprotease (MMP)-degradable poly(ethylene-glycol) (PEG)-based material was included in the culture system. This material has been used previously to encapsulate MSCs with no decrease in cell viability.26–28 With this culture system, we were able to co-culture MSCs, osteoblasts, and adipocytes in normal and high glucose conditions, and then analyze each cell type separately for gene expression, histological markers, and cell viability.
In many in vitro studies of intercellular cross-talk, only single outcome measures are reported, which often does not elucidate how genes, proteins, and other measure of cell response interact and contribute to an observed cell behavior.29 In recent years, some of these shortcomings have been addressed by developing heat-maps of gene and protein arrays, modeling proposed signaling networks by integrating results from many different experiments, and constructing multivariate statistical models.25,30,31 In particular, multivariate statistical analysis techniques can provide insight into the greatest sources of variance in complex biological systems and can elucidate how outcome measures correlate with different environmental and culture conditions.32 We have previously used multivariate statistical techniques to evaluate gene expression data from simultaneous co-culture of three cell types, showing in a proof-of-concept experiment that these models can provide valuable biological insight in a multicellular in vitro system.25 In this study, these techniques were further utilized to gain insights into how neighboring cell type and environment interact to influence MSC behavior and gene expression in an in vitro model of hyperglycemia. Specifically, we studied cellular response to elevated glucose levels in three different culture configurations: monoculture (one cell type), co-culture (two different cell types) and tri-culture (three different cell types) (Fig. 1). Cellular response was analyzed by measuring gene expression levels, viability, histological markers, and MSC colony-forming potential over a seven day time period using both univariate and multivariate analysis approaches.
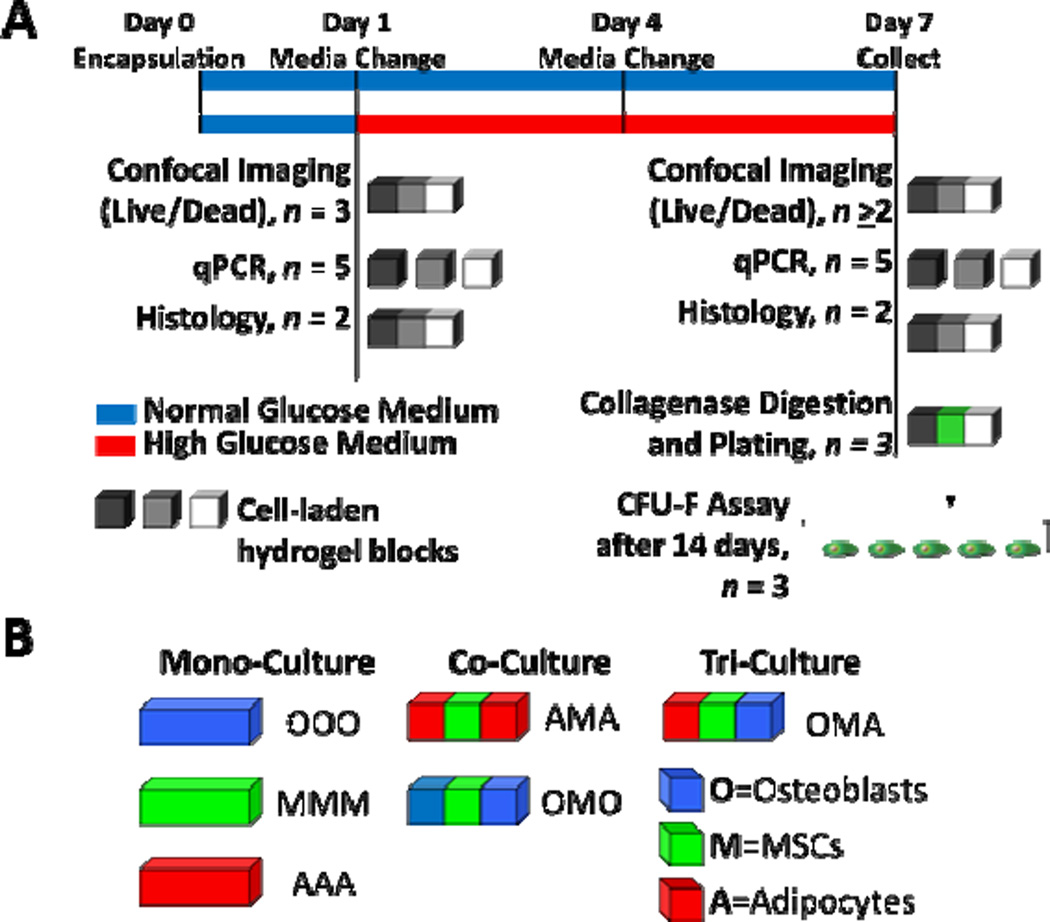
Fig. 1.
Six culture configurations were evaluated in this study. (A) Cells were cultured in mono-, co-, or tri-culture hydrogel constructs for up to 7 days and then analyzed as depicted above. After one day in culture, media was changed and half of the constructs were switched to high glucose. Shades of gray and white blocks represent hydrogel blocks of different types of cells in either mono-, co-, or tri-culture. (B) Tri-culture configurations consisted of adipocytes, MSCs, and osteoblasts (designated OMA); co-culture configurations consisted of osteoblasts and MSCs or adipocytes and MSCs (designated OMO and AMA, respectively); and mono-culture configurations consisted of osteoblasts, MSCs or adipocytes (designated OOO, MMM, and AAA, respectively).
Results
Adipocyte Response to Culture Conditions and Glucose Levels
Adipocytes were cultured under normal and high glucose conditions in three different culture configurations: mono-culture with only adipocytes (AAA), co-culture with MSCs (AMA), or tri-culture with MSCs and osteoblasts (OMA), where the underlined letter represents the cell type being discussed from a particular culture configuration (Fig. 1). Lipid deposition was observed using Nile Red staining at all time-points and experimental conditions (Fig. 2A). Lipid deposition was constant throughout the duration of culture and no changes in lipid deposits were observed regardless of time, glucose level, or culture configuration (data not shown).
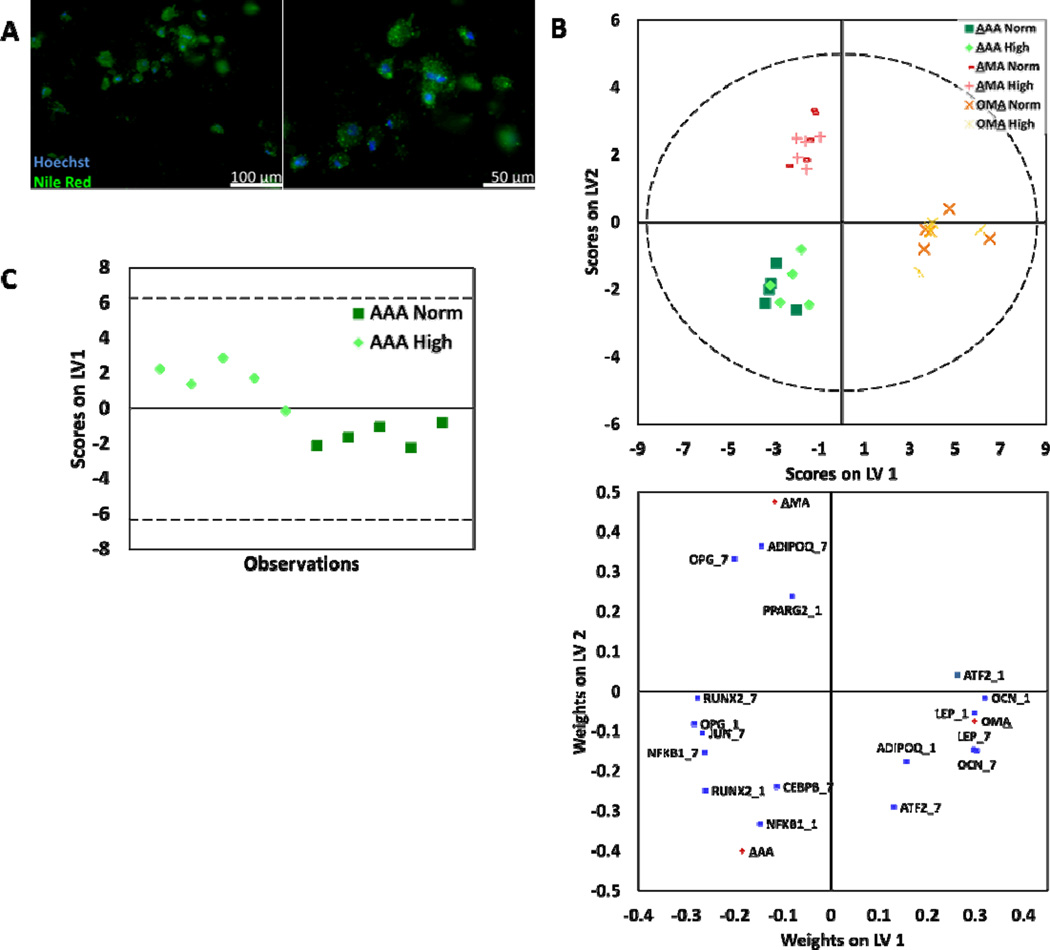
Fig. 2.
Adipocytes were assessed for response to culture conditions. (A) Representative images of adipocytes stained with Nile Red, specific for lipids (green) and counterstained with Hoechst, specific for nuclei (blue). (B) PLS-DA was used to build models for adipocytes assigned to classes by culture configuration. Models yielded two latent variables that discriminated adipocytes by culture configuration (R2Y=0.93 and Q2= 0.92). (C) For AAA culture, classes based on normal and high glucose levels were discriminated using PLS-DA (AAA: R2Y= 0.79 and Q2= 0.70). The corresponding weight plot can be found in the supplementary information (Fig. S1C). In all models, dashed lines represent the 95% confidence limit of the distribution of weights.
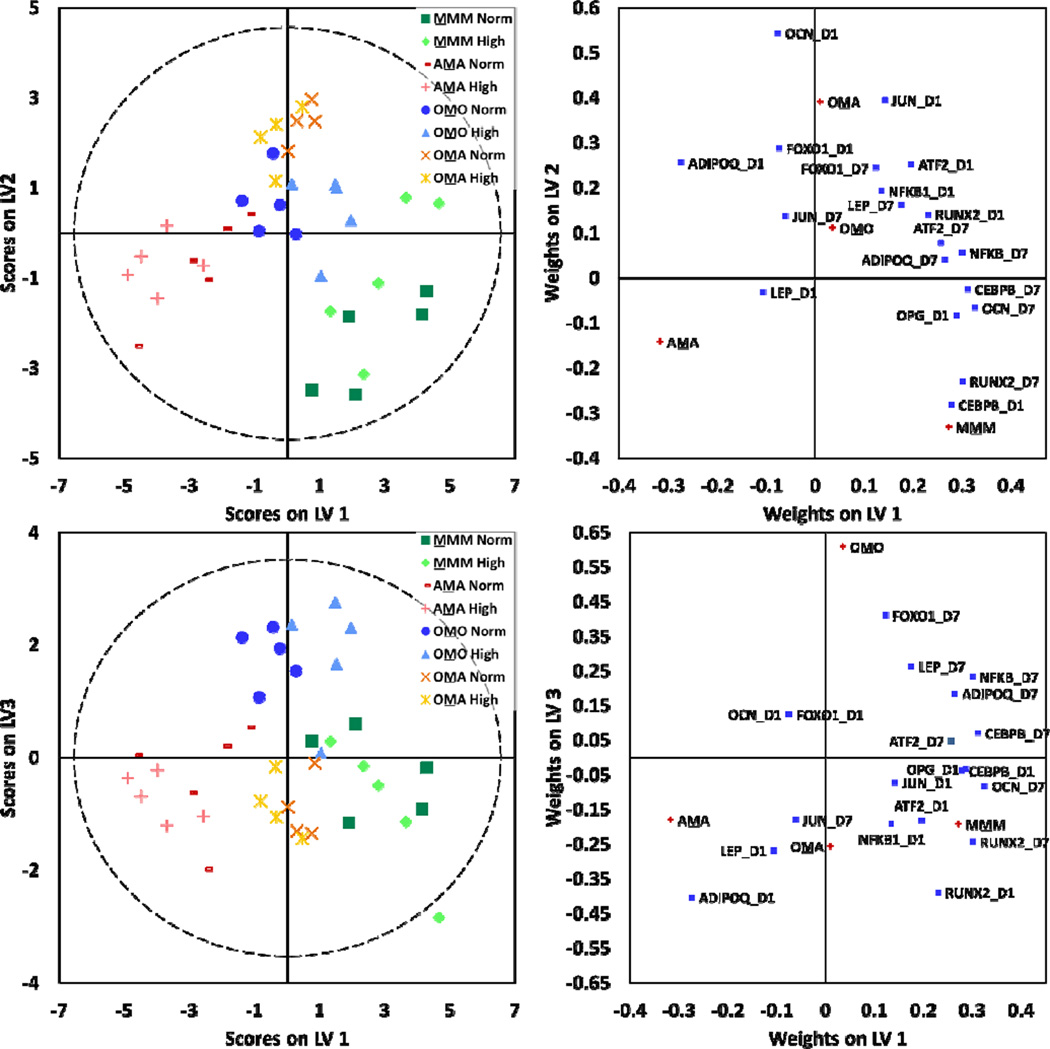
Expression levels for a variety of genes that act as markers of adipogenesis and energy metabolism (Table S1, ESI) were measured for each condition using qPCR. Using this data, Principal Component Analysis (PCA) was conducted to model adipocyte gene expression data that included all experimental conditions (Fig. S1A, ESI). Based on observed clustering patterns in PCA, Partial Least Squares Discriminate Analysis (PLS-DA) was used to build a model for adipocytes assigned to classes by culture configuration (Fig. 2B). The PLS-DA model yielded two latent variables (LV) that discriminate adipocytes by culture configuration (R2Y=0.93 and Q2= 0.92). The first LV discriminates OMA (or osteoblast-containing cultures) from AAA and AMA (or non-osteoblast-containing cultures). The second LV discriminates AMA from AAA. The weight plot showed significant correlation of osteocalcin (OCN), leptin (LEP), adiponectin (ADIPOQ), and ATF2 with OMA cultures; peroxisome proliferator-activated receptor γ (PPARγ2), ADIPOQ, and osteoprotegrin (OPG) with AMA cultures; and JUN, NFKB1, CEBPB, and RUNX2 with AAA cultures. Then, the dataset was further divided and sub-models of gene expression data from AAA, AMA, and OMA cultures were used in PCA (Fig. S1B, ESI). In the case of the AAA culture configuration, observations clustered by glucose level and classes assigned by normal and high glucose levels were further discriminated using PLS-DA (Fig. 2C; S2C, ESI).
Confocal microscopy was used to image hydrogel blocks of adipocytes stained with LIVE/DEAD reagents in each culture condition at each time point (representative images in Fig. S2A, ESI). Numbers of live and dead cells were quantified and the fraction of viable cells is reported normalized to day 1, normal glucose (Fig. 3). Adipocytes maintained viability across the entire experiment, and no statistically significant differences were observed within each culture configuration.
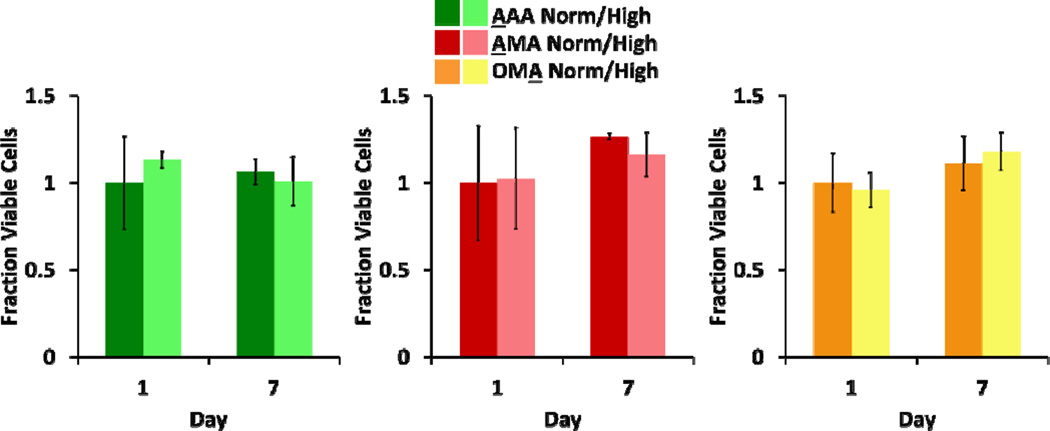
Fig. 3.
Viability of adipocytes was assessed in each culture configuration at normal and high glucose levels. Fraction viable cells (via LIVE/DEAD staining) for each culture condition is presented (n = 3 for AAA, AMA; n ≥2 for OMA; no statistically significant differences were seen, p < 0.05).
Osteoblast Response to Culture Conditions and Glucose Levels
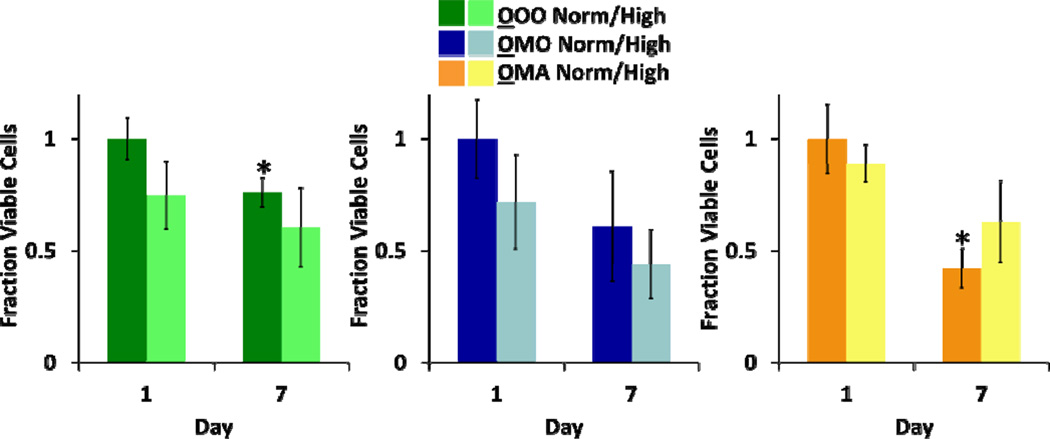
Osteoblasts were cultured under normal and high glucose conditions in three different culture configurations: mono-culture (OOO), co-culture with MSCs (OMO), or tri-culture with MSCs and adipocytes (OMA) (Fig.1). Alkaline phosphatase (ALP) activity was evaluated using ALP substrate stain in all experimental conditions across all time points. Similar levels of production were seen for normal and high glucose conditions (data not shown) and representative images from each culture configuration at both time points are presented (Fig. 4A). Low production was seen throughout the duration of the culture and a transiently higher amount of ALP production was seen in osteoblasts from OMA cultures at day 1 (Fig. 4A).
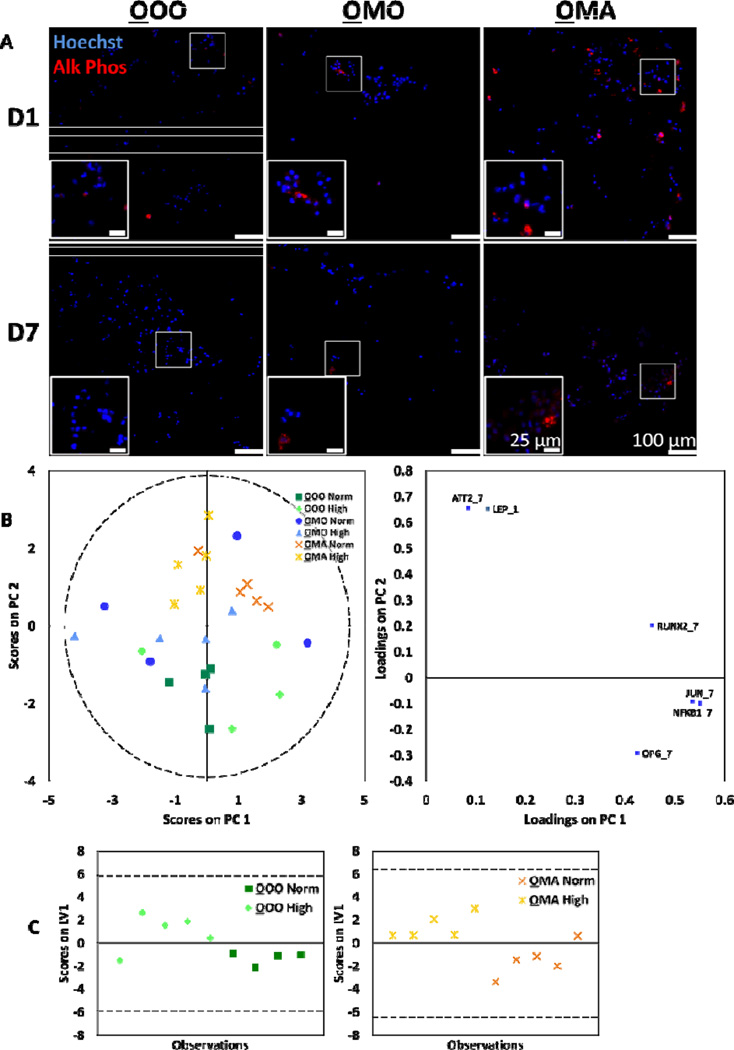
Fig. 4.
Osteoblasts were assessed for response to culture conditions. (A) Representative images of osteoblasts stained for ALP, a marker of osteogenic differentiation (red), and counterstained with Hoechst for nuclei (blue). Noticeably higher amount of ALP production was seen in osteoblasts from OMA cultures at day 1. (B) PCA was applied to the gene expression data of the global osteoblast population, yielding two principal components (R2X= 0.81 and Q2= 0.56). (C) For OOO and OMA cultures, classes based on normal and high glucose levels were discriminated using PLS-DA (OOO: R2Y= 0.5 and Q2= 0.43; OMA: R2Y=0.62 and Q2= 0.55). The corresponding weight plots can be found in the supplementary information (Fig. S3B). In all models, dashed lines represent the 95% confidence limit of the distribution of scores.
Expression levels for a variety of genes that act as markers of osteogenesis and energy metabolism (Table S1, ESI) were measured for each condition using qPCR. Using this data, PCA was conducted to model osteoblast gene expression data that included all experimental conditions (Fig. 4B). The PCA yielded two principal components (R2X= 0.81 and Q2= 0.56). The first component indicates presence of random variance that can be attributed neither to culture configuration nor to glucose condition, possibly due to the genes measured in the experiment. However, the second component separates OMA cultures from OOO and OMO cultures. Further, three separate PCA models were built within each osteoblast culture configuration (Fig. S3A, ESI). In cases that the majority of observations clustered by glucose level in PCA (OOO and OMA cultures), classes based on normal and high glucose levels were further discriminated using PLS-DA (Fig. 4C; S3B, ESI). Notably, a different set of genes served to separate glucose classes in OOO and OMA cultures configurations. PCA for osteoblasts in the OMO culture configuration indicated that primary variance in gene expression was not due to glucose level and did not warrant further discrimination using PLS-DA (Fig. S3A, ESI).
Numbers of live and dead osteoblasts were quantified from confocal microscope images, as described above (Fig. S2B, ESI). Viability decreased for both OOO and OMA cultures in normal glucose at Day 7, while viability was maintained in the OMO cultures (Fig. 5).
Fig. 5.
Osteoblast viability was measured in response to culture conditions. Fraction viable cells (via LIVE/DEAD staining) for each culture condition is presented (n = 3 for OOO, OMO; n ≥2 for OMA;* = Significantly different from same culture configuration and glucose condition on Day 1; p < 0.05).
MSC Response to Culture Conditions and Glucose Levels
MSCs were cultured under normal and high glucose conditions in four different culture configurations: mono-culture (MMM), co-culture with osteoblasts (OMO), co-culture with adipocytes (AMA), or tri-culture with osteoblasts and adipocytes (OMA) (Fig. 1). Expression levels for a variety of genes that act as markers of osteogenesis, adipogenesis, and energy metabolism (Table S1, ESI) were measured for each condition using qPCR. Using this data, PCA was conducted to build a model of MSC gene expression data that included all experimental conditions (Fig. S4, ESI). Based on observed clustering patterns, a PLS-DA model was built for MSCs assigned to classes by culture configuration (Fig. 6). Models yielded three LV that discriminated MSCs by culture configuration (R2Y=0.73 and Q2= 0.63). The first LV discriminates MMM and AMA, the second discriminates MMM and OMA, and the third discriminates OMA and OMO. Then, four separate PCA models were built for each MSC culture configuration (Fig. S5, ESI). In the cases of OMO and AMA, observations clustered by glucose level in PCA and classes based on normal and high glucose levels were further discriminated using PLS-DA (Fig. S6, ESI). Similarly to osteoblast cultures that could be discriminated by glucose level, a different set of genes acted to separate MSCs by glucose level in OMO culture configurations as compared to AMA culture configurations.
Fig. 6.
MSCs were assessed for gene expression changes response to culture conditions. PLS-DA models were constructed for MSCs assigned to classes by culture configuration. Models yielded three latent variables that discriminated MSCs by culture configuration (R2Y=0.73 and Q2= 0.63). In all models, dashed lines represent the 95% confidence limit of the distribution of weights.
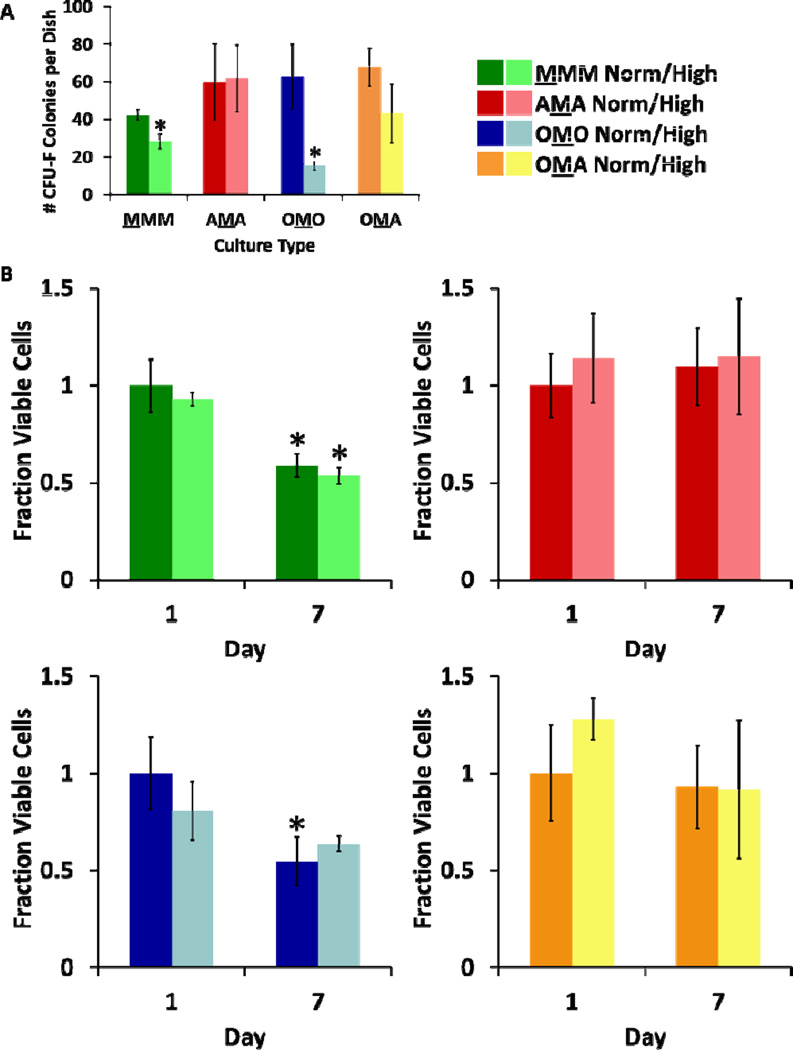
After 7 days of culture in normal and high glucose conditions, the degradable MSC block in each culture configuration was exposed to collagenase and degraded. Live cells were recovered and colony forming ability was assessed after 14 days of growth (Fig. 7A). Significantly fewer colonies were seen in MMM and OMO cultures in high glucose conditions as compared to normal glucose, but number of colonies were similar regardless of glucose level in in AMA and OMA cultures.
Fig. 7.
MSC clonogenicity was measured post-culture using colony forming assays (A) Number of colony-forming units greater than 2 mm in diameter per dish (n = 3) (* = Significantly different from same culture configuration; p < 0.05). (B) MSC viability was measured in response to culture conditions. Fraction viable cells (via LIVE/DEAD staining) for each culture condition is presented (n = 3 for MMM, OMO, AMA; n ≥2 for OMA; * = Significantly different from same culture configuration and glucose condition on Day 1; p < 0.05).
Numbers of live and dead MSCs were quantified from confocal microscope images, as described above (Fig. S2C, ESI). Decreases in viability were seen for MSCs in both high and normal glucose MMM cultures at Day 7 and also in normal glucose OMO cultures at Day 7 (Fig. 7B).
Discussion
As stem cell microenvironments have been observed to undergo changes that affect resident stem cells in disease states,7 the correlation between diabetes and osteoporosis may be better explained by understanding the MSC response to the environmental changes inherent to diabetes. Thus, we were interested to characterize how MSCs are affected by soluble factors secreted by osteoblasts and adipocytes in hyperglycemic conditions. As experimental models available to date have not been satisfactory in addressing this technical need, we expanded upon a model system of the MSC microenvironment previously developed in our laboratory to include a degradable polymer for live-cell retrieval.25,26 In this model system, we focused on decoupling the effects of paracrine signals from other types of signaling that may occur in the bone marrow niche. Within the bone marrow microenvironment, information is thought to be transmitted over relatively short length scales (on the order of hundreds of microns) given the well-defined, compact architecture,33,34 so we believe the geometry of our tri-culture system sufficiently mimics this aspect of the structure of the bone marrow while allowing for separation of cell types post-culture. From this system, we analyzed gene expression, histological markers, cell viability, and MSC colony forming ability after 7 days of culture in normal or high glucose conditions.
Encapsulated Cells Maintain Viability and Phenotype
Previous experiments provided evidence that our culture platform was suitable for at least 18 days of culture.25 In the experiments presented here, cell viability was generally maintained for the duration of the experiment in all cell types (Fig. 3, 5, and 7B). However, osteoblasts in OOO and OMA culture configurations under normal glucose conditions exhibited significantly lower viability by day 7. The decreased viability observed in the mono- and tri-culture conditions remains unclear, but is possibly due to interactions with neighboring cell types. MSCs may have promoted osteoblast viability in the co-culture, but these effects could have been interrupted by the presence of adipoctyes in the tri-culture. Clinically, osteoblast turnover is thought to occur on the order of days,35 suggesting that osteoblasts need constant replenishment from the stem cell compartment. While soluble signals from stem cells are present in this system, direct replenishment via differentiation is impossible. Thus, the maintenance of viability observed in OMO cultures might be a result of communication with MSCs that encouraged osteoblast survival. Further refinement of our culture platform could improve osteoblast viability by including more soluble signals known to contribute to normal osteoblast function in vivo.
Osteoblast production of ALP, a marker for osteogenic phenotype, persisted at generally low levels throughout the duration of culture time (Fig. 4A). Similarly, adipocyte lipid deposition was evident in adipocytes throughout the duration of culture in all experimental conditions (Fig. 2A). Taken together, the maintenance of viability and cell type-specific markers indicate that the culture system is a valid platform for analysis of interactions between these three cell types, even under hyperglycemic conditions.
Multivariate Models Allow Discrimination by Neighboring Cell Type and Glucose Level
Multivariate statistical modeling was used to better understand maximum sources of variance within the gene expression of each cell type as well as to inform future co-culture experimental design. In global multivariate models for all cell types, the maximum source of variance originated from differences in neighboring cell types, referred to as culture configuration (i.e. mono-, co-, or tri-culture), as determined by clustering patterns and discrimination across principal components and latent variables (Fig. 2B, 4B, and 6). In adipocyte global models, the first LV discriminated osteoblast containing cultures from non-osteoblast containing cultures. From this, we know that the presence of osteoblasts influences adipocyte behavior, an important consideration in designing future co-culture studies that aim to understand adipocyte behavior within the bone marrow microenvironment. Osteoblast models showed less distinct clustering patterns, possibly indicating a large amount of random variance in the system. Alternatively, these results may be indicative of osteoblast stability regardless of neighboring cell type. Overall, future co-culture experiments may need to analyze different genes or other output measures to gain a better understanding osteoblast behavior. Finally, MSC global models discriminated each culture type from the other except for MMM and OMO culture configurations. This suggests that interactions with adipocytes significantly influence MSCs, as was evident in other analyses and will be discussed further. As osteoblast presence did not seem to impact MSCs as strongly, at least based on gene expression data, future co-culture studies could aim to gain a deeper understanding of MSC and adipocyte communication and the mechanisms in which this communication influences MSC behavior.
Clustering by glucose level was not often observed in global models, indicating that neighboring cell types had a greater influence on gene expression than glucose conditions. However, this may have been partially due to the genes analyzed in this study, and by including more genes, discrimination by glucose in global models may have been possible. When sub-groups of the dataset were modeled to understand primary variance within each culture configuration, it was found that in several cases discrimination by glucose level could be achieved (Fig. 2C and 4C; Fig. S6, ESI). In other cases, it was more difficult to definitively determine if culture configurations could be separated by glucose level, and whether this is due to the choice of genes analyzed or the influence of the culture configuration on cellular response to glucose remains unclear. Overall, it was evident that the response of individual cell type to glucose level was unique to each culture configuration and thus interactions with neighboring cell types. This provides further incentive to probe cellular response to pathological conditions within a co-culture setting, as it better represents the simultaneous cell response to neighbors and environment that occurs in vivo.
The fact that correlations in the multivariate models were seen primarily based on type of neighboring cells strongly supports the idea that traditional monoculture platforms are not able to mimic/reproduce important features of systems-level cell-cell communication that occurs in disease states. Other indirect (no cell-cell contact) co-culture studies with bone marrow cells also report that cellular behavior changes when in communication with other orthopaedic-derived cells. Increased osteoblast death has been observed in response to diabetic bone marrow cells20 and reduced MSC hypertrophy and mineralization was achieved when in co-culture with chondrocytes.36 These studies and the work presented here provide strong motivation to include multiple cell types in in vitro studies of the bone marrow microenvironment in order to gain more relevant biological insights for diseases affecting the entire tissue. Thus, our culture platform provides a novel technology that is well-suited for future studies in which multiple co-cultured cell types responding both to each other and to environmental changes may be required to adequately recapitulate a complex in vivo disease state.
Adipocyte Co- and Tri-culture Conditions Correlate with Genes Related to Energy Metabolism and Adipogenesis
Multivariate models were also used to understand the correlations between gene expression and culture configuration. In the adipocyte-only model, markers of adipogenesis and energy metabolism (OCN, LEP, ADIPOQ, PPARγ2, OPG) showed statistically significant correlation with AMA and OMA cultures while markers of inflammation and oxidative stress (JUN, NFKB1, CEBPB) showed correlation with AAA cultures (Fig. 2B). This suggests that presence of MSCs may promote adipocytes to up-regulate genes related to adipogenesis and energy metabolism. Interestingly, these differences in gene expression did not manifest in adipocyte viability data or levels of lipid deposition, as both were constant in all conditions (Fig. 2A and 3). It is possible that adipocytes were robust to environmental changes, or that gene expression data provided an early view of adipocyte response to neighboring cells that may manifest in viability differences or changes in lipid at a later time. These results are consistent with experiments that have been done previously, in which adipocytes have been observed to secrete markers of inflammation and to undergo hypertrophy rather than apoptosis under hyperglycemic conditions.37 Overall, these results indicate the presence of cross-talk between adipocytes and MSCs, which was further confirmed in additional studies discussed below.
MSCs Modulate Osteoblast Response to Glucose
Cell viability and MSC clonogenicity studies suggested that MSCs exert modulatory effects on osteoblasts at the expense of their own viability and clonogenicity, trends that were not seen when MSCs were co-cultured with adipocytes. MSCs in OMO and MMM cultures demonstrated decreased viability and clonogenicity after 7 days of culture, while both viability and clonogenicity were maintained in AMA and OMA cultures (Fig. 7). Similar to MSCs in OMO and MMM cultures, previous work has also shown reduction of MSC clonogenicity and viability under hyperglycemic conditions.12 Osteoblast viability, on the other hand, was unlike that of MSCs, as only osteoblasts in the OMO cultures showed constant cell viability for the duration of culture (Fig. 5), whereas MSCs from OMO cultures showed decreased viability over time in high glucose conditions (Fig. 7B). These opposing trends were also seen in gene expression data, where MSCs from OMO cultures were seen to cluster by glucose level in the global multivariate model and culture configuration sub-model (Fig. 6; S6, ESI) while osteoblasts from the OMO culture configuration could not be discriminated by glucose level (Fig. S3A, ESI).This complementing relationship may indicate that MSCs somehow act to modulate the response of osteoblasts to glucose in a way that changed their own behavior. As MSCs are known to secrete therapeutic soluble factors,38 this work further suggests that the secretion of soluble factors in hyperglycemic conditions may have had positive effects on osteoblasts but was detrimental to some MSC functions. However, this does not appear to be a universal consequence of MSC co-cultured with differentiated cells, as these detrimental effects were not seen in MSCs cultured with adipocytes in any configuration, discussed below.
Adipocytes Promote MSC Viability and Clonogenicity and Early Osteoblast ALP Activity
Unlike MSCs in monoculture or co-culture with osteoblasts, MSCs in co-culture with adipocytes showed maintenance of viability and clonogenicity in high glucose conditions. Changes in adipocyte gene expression in the presence of MSCs also provides evidence that interactions between MSCs and adipocytes occurred, possibly due to an increase in expression of LEP and ADPIQ by adipocytes, which is known to act upon other cell types.37 Furthermore, it appears that adipocytes also modulated osteoblast behavior, especially in regards to ALP production (Fig. 4A). As ALP is a marker for osteogenesis, higher levels in osteoblasts co-cultured with adipocytes after one day may indicate that interactions with adipocytes promoted their activity, although high ALP staining was not observed by day 7 for any sample types. Thus, it appears that adipocytes may “buffer” MSCs from the reduction of clonogenicity and viability induced by hyperglycemic conditions. Analyzing these results alongside gene expression data, the ability of adipocytes to influence other cell types may correlate with upregulation of genes for adipogenesis and energy metabolism in adipocytes.
The idea that adipocytes may act to modulate MSC response to glucose has not, to our knowledge, been reported in other experiments. It is well known that adipocytes and osteoblasts co-exist in the bone marrow microenvironment, and that the ratio of adipocytes to osteoblasts increases with age.1,39 It also has been observed that an increase of adipocytes in the marrow coincides with the onset of disease like osteoporosis and anorexia nervosa, but the direct impact of higher numbers of adipocytes on disease progression has yet to be determined.1,40 It has been hypothesized that increased marrow fat may be a compensatory mechanism in times of disease,40 or be a direct cause of reduced MSC osteoblastogenesis.40,41 Our results suggest it is possible that, under hyperglycemic conditions, an increased number of adipocytes in the bone marrow may exert compensatory mechanisms to maintain MSC function and possibly delay the onset of osteoporosis. Thus, our system provides important insight into a complex biological system that has not been achieved through other mono- and co-culture experiments and highlights the interconnected nature of these three cell types in pathological conditions.
Implications for Future Co-Culture Studies
As a whole, the study presented illustrates a platform that facilitated elucidation of non-intuitive cellular behavior in a model of hyperglycemia. This tri-culture platform permitted the study of reciprocal cellular interactions, often impossible in other in vitro culture systems. Specifically, the tri-culture allows simultaneous analysis of cellular response to both pathological conditions and neighboring cell types. Also unique to our culture platform is the ability for on-demand retrieval of live cells, allowing for further analysis of individual cell response to their environment. In our model of the hyperglycemic bone marrow microenvironment, it was observed that neighboring cell type was the primary source of variance in our system and that disease-relevant environmental alterations can best be understood within the context of multi-cell systems. It was also found that while MSC clonogenicity and viability decreased when in culture with osteoblasts, both were maintained when in culture with adipocytes, regardless of the presence of osteoblasts. One possible explanation for this can be derived from adipocyte gene expression data, which indicates that cross-talk between adipocytes and MSCs resulted in an altered reaction to high glucose levels, at least at early stages of hyperglycemia.
These results provide specific avenues of study that can follow. Further studies to understand mechanistic reasons for why MSC viability and colony forming ability is decreased in the presence of osteoblasts but not adipocytes could inform future therapies aimed to target diseases like osteoporosis. Also, as increased marrow adipocyte levels coincide with osteoporosis,1 it would be interesting to further investigate if the relative quantities of osteoblasts and adipocytes change MSC response to culture conditions. The culture system described here could be used to investigate these questions and to generate further hypotheses that could be studied in vivo. Overall, the technological innovations in our in vitro tri-culture platform permitted a deeper understating of cellular response to hyperglycemic conditions that may be used to direct future research into cell-based therapies for diabetes and its secondary pathologies.
Materials and Methods
Preliminary Cell Culture
All cell culture reagents were obtained from Mediatech unless otherwise specified. Primary human MSCs were obtained from Texas A&M Health Sciences Center and expanded in Minimal Essential Medium-Alpha (αMEM) with 16.5% fetal bovine serum (FBS; Hyclone), 1 g/L glucose, 2 mM L-glutamine, 1% amphotericin B, and 0.1% gentamicin and cultured at 37°C and 5% CO2 in a humidified incubator. Primary human osteoblasts (Lonza) were expanded to 4 population doublings in OGM Osteoblast Growth Medium (Lonza) containing 10% FBS, ascorbic acid, 50 μg/mL gentamicin, and 37 ng/mL amphotericin B. Primary human subcutaneous pre-adipocytes (Lonza) were expanded to 1–2 population doublings according to the manufacturer’s protocol in PGM-2 Basal Medium (Lonza) containing 10% FBS, 2 mM L-glutamine, 50 μg/mL gentamicin, and 37 ng/mL amphotericin B. Cultures at 80% confluence were differentiated into adipocytes for 9 days in Dulbecco’s Modified Eagle Medium (DMEM) with 10% FBS, 1 g/L glucose, 60 µM indomethacin, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 0.5 µM dexamethasone, and 45 pM insulin.
Fabrication of Culture System
Cells were encapsulated in PEG-based materials (Sigma-Aldrich, unless otherwise noted). PEG-diacrylate (PEG-DA) (Mn = 8kDa) was synthesized by combining acryloyl chloride and PEG according to previous methods.42
To promote cell adhesion and viability, PEG-based materials containing fibronectin-derived GRGDS (Bachem) and laminin-derived YIGSR (Anaspec) were synthesized from Acryl-PEG-succinimidyl valerate (SVA) (Mn=3.4kDa; Laysan Bio) according to previous methods25,42 to obtain Acryl-PEG-RGDS and Acryl-PEG-YIGSR. To allow on-demand degradation of particular blocks in the culture system, a collagenase-degradable peptide sequence, GGGLGPAGGK (abbreviated LGPA), was bi-functionalized with Acryl-PEG-SVA (Mn=3.4kDa) according to previous methods28,43 to obtain PEG-LGPA-DA polymer chains.
To fabricate cultures, cells were suspended in precursor hydrogel solutions at 15×106 cells/mL. Solutions were formulated with 10% w/w PEG-DA (osteoblasts and adipocytes) or PEG-LGPA-DA (MSCs) and 0.05% w/w D2959 photoinitiator (Ciba), with 1 mM Acryl-PEG-RGDS and Acryl-PEG-YIGSR for MSCs and adipocytes, respectively. Precursor hydrogel solutions were photopatterned into 1.5 × 1.5 × 1 mm laminated gel structures using a PDMS mold and UV light according to previous methods (Fig. 1).25 An opaque photomask was used in subsequent steps to prevent any further UV light exposure and crosslinking. Each subsequent layer is laminated to the prior and constructs do not separate over the course of the experiment or experience changes in tensile properties, as demonstrated in prior work.25,26,44 Laminated constructs were extracted from the device and sectioned with a scalpel perpendicular to the long axis of the laminate to yield up to eighteen 1.5 mm-wide mono-, co-, and tri-culture constructs (Fig. 1).
Culture of Encapsulated Cells
Gel structures were cultured in individual wells of a 12 well plate in DMEM with 10% FBS, 5.5 mM glucose (100 mg/dL, similar concentration to normal fasting serum glucose level in humans), 2 mM L-glutamine, 70 µM L-ascorbate, 45 pM insulin (6.4 μU/mL, normal fasting serum insulin level), 1% amphotericin B, and 0.1% gentamicin. After 1 day, a subset of constructs was switched to 22.3 mM glucose (400 mg/dL, similar to hyperglycemic glucose level in poorly-controlled diabetic patients). All constructs were then cultured for a total of 7 days with an additional media change at Day 4 (Fig. 1).
Cell Viability and Image Analysis
Hydrogel constructs (n = 3 for all mono-culture, co-culture, and day 1 (D1) tri-cultures; n = 2 for day 7 (D7) tri-cultures) were analyzed on Days 1 and 7 culture using a LIVE/DEAD assay (Invitrogen). Constructs were rinsed in sterile phosphate buffer saline (PBS) at 37 °C for 30 minutes and then incubated in LIVE/DEAD dyes (1 µM calcein AM, 1 µM ethidium homodimer-1) for 45 minutes at 37 °C. Constructs were then rinsed with PBS for 15 minutes and imaged with confocal microscopy (10× objective, LSM 700; Zeiss). For each construct, 1 image stack was collected for each cell type present (dimensions: 693 × 693 μm; stack depth = 0 – 800 µm with 10-µm intervals). Image stacks were analyzed using ImageJ software (version 1.46a; NIH). Each stack was split into green (calcein) and red (EthD-1) channels. Every fifth image was analyzed for total number of live and dead cells. Particles of diameter greater than 94 μm2 and 12 μm2 were counted for the live and dead cell analysis, respectively, using the Particle Analysis macro. Subsequently, fraction viable cells (# live cells/# total cells counted) was calculated for each stack and normalized to Day 1 normal glucose conditions.
Colony Forming Unit-Fibroblasts (CFU-F) Analysis
To evaluate clonogenicity, at day 7, MSC-containing PEG-LGPA-DA hydrogel constructs (n = 3) were incubated in a 1,100 U/mL collagenase (Gibco) solution in MSC expansion medium for 1 hour. Fractions of the media containing the recovered cells (100 μL for MMM, 200 μL for OMO and AMA, and 300 μL for OMA; volumes determined in order to seed approximately the same number of cells for each construct type) were plated into 15-cm tissue culture dishes (Corning) containing 20 mL of MSC expansion medium. Media was changed 1 day after seeding and cells were then fed every 3 days for 14 days of culture. Clonogenicity was evaluated by counting colonies ≥ 2 mm (stained with 3% crystal violet (Sigma-Aldrich), 100% methanol (BDH)) after 14 days.
Histological Analysis
For histological analysis, all culture configurations under both normal and high glucose conditions were infiltrated by graded concentrations of sucrose in PBS followed by graded concentrations of optimal cutting temperature compound (OCT; Sakura Finetek), frozen in liquid nitrogen, and stored at −80°C until sectioning. Embedded gel constructs were serially cryosectioned at a 20 µm thickness (Microm HM 560 Cryostat; Thermo Scientific) and mounted on Superfrost Plus slides (Fisher). Sections were stained either for lipids (10 µg Nile Red/mL in 1% acetone) or alkaline phosphatase (ALP) activity according to the manufacturer’s protocol (Vector Red Alkaline Phosphatase Substrate Kit; Vector Labs). In both cases, sections were counterstained with Hoechst 33258 (0.25 mg/mL in PBS for 5 min; Molecular Probes). Sections were visualized with epifluorescence microscopy under FITC filters for Nile Red, Texas Red Filters for ALP, and DAPI filters for Hoechst.
mRNA Isolation and qPCR
Hydrogel constructs (n = 5) were rinsed in PBS and blocks containing individual cell populations were separated from each other using a scalpel for gene expression analysis by qPCR after 1 and 7 days in mono-, co-, or tri-culture. Gel blocks containing the same cell type were pooled from 2 co-culture constructs or 3 tri-culture constructs of the same culture configuration and glucose condition to provide sufficient and equivalent amounts of mRNA for quantification. Pooled blocks were homogenized in microcentrifuge tubes with pellet grinders and mRNA was extracted using a QIAshredder tissue homogenizer and RNeasy kit with DNase I digestion (Qiagen). cDNA was generated using SuperScript III Reverse Transcriptase (Invitrogen) with Oligo(dT)15 primers and dNTPs (Promega). Gene expression was analyzed with quantitative PCR amplification performed on a StepOnePlus™ Real-Time PCR System (Applied Biosystems) in the presence of SYBR Green/ROX master mix (Applied Biosystems). In addition to endogenous controls (40S ribosomal protein S18 (RPS18) and beta-actin), gene expression was measured for genes indicating osteogenesis (runt-related transcription factor 2 (RUNX2), Osteocalcin (OCN), and Osteoprotegerin (OPG)); adipogenesis (peroxisome proliferator-activated receptor γ (PPARγ2), CCAAT/enhancer-binding protein beta (CEBPB), Leptin (LEP), and adiponectin (ADIPOQ)); and glucose-responsive transcription factors (activating transcription factor 2 (ATF2), forkhead box protein O1 (FOX01), c-jun (JUN), and nuclear factor kappa B (NFKB1)). Sequences for custom-designed primers (Invitrogen) can be found in the supplementary information.
To analyze PCR amplification data, the raw fluorescence data was processed using LinRegPCR (v12.11; http://www.hartfaalcentrum.nl). Starting amplicon number (No) was calculated based on mean efficiencies (E) and cycle threshold (Ct) using the formula No = Nt/ECt, where Nt is the amplicon number at the cycle threshold. No for each target gene were normalized to a geometric mean of the starting amplicon numbers of the endogenous controls to obtain relative expression values.
Univariate Statistical Analysis of Cell Viability and Colony Formation
Viability and CFU-F results are depicted as mean ± standard deviation. Prior to statistical analysis, all data were transformed with a Box-Cox transformation. Where significant factors and interactions were identified by ANOVA, Tukey’s post hoc test (significance level p < 0.05) was used to generate pairwise comparisons between means of individual sample groups and determine statistically significant differences.
Multivariate Models of Gene Expression
Statistical modeling was performed using SIMCA-P+ software (version 12.0.1.0, Umetrics) on gene expression results. All Box-Cox-transformed data were mean-centered and scaled to unit variance prior to analysis as a means of normalization to allow all variables to be considered equally scaled in the principal components or latent variables.32 For PCA, N × K X-matrices were generated of N culture condition observations and K time-variant gene expression responses. PCA was conducted to determine the source of maximum variation in the dataset, resulting in clusters of similar observations separated across one or more principal components. Based on clustering, partial least squares discriminate analysis (PLS-DA) was performed to find LV that served as discriminating factors to best separate observations.25,45 For PLS-DA, N × M Y-matrices were generated of N culture condition observations and M responses, which were based on assigned classes (culture configuration or glucose level). To optimize model quality, pruning processes were performed to remove observations that lied outside of the 95% confidence interval and variables that were not influential in model generation. The overall quality of each model is summarized by two parameters: R2X (PCA) or R2Y (PLS-DA) provide a measure of the extent that the model explains the variation in data matrices and indicates a goodness of fit; and Q2 provides a measure of the extent to which the variation of a future experimental data set may be predicted by the model and indicates a goodness of prediction.45 For further model quality assessment, model validation based on permutation testing was performed in SIMCA-P. In brief, this technique randomly shuffles the positions of variables in the Y-block and then fits a new PLS model to the permuted Y-block. Cross-validation methods are used to generate R2Y and Q2 values for new models. If the R2Y and Q2 values of the new models are lower than R2Y and Q2, the model is valid.46
Supplementary Material
Insight, Innovation, Integration.
In this work, the utility of biomaterial-based platforms with on-demand degradation in combination with multivariate statistical analysis to model perturbations in the stem cell microenvironment is demonstrated. Within a 3D hydrogel platform, we co-cultured MSCs, adipocytes, and osteoblasts, three cell types implicated in diabetes-related osteoporosis, under normal and hyperglycemic conditions, to understand how cell-derived soluble factors and environmental perturbations influence the stem cell microenvironment. The implementation of an enzymatically-degradable component within the platform allowed retrieval of viable MSCs for further culture and clonogenicity analysis. This system provides unique capabilities that will better enable future studies to elucidate how complex interactions between various cellular components at the tissue and organ levels result in pathology progression or tissue healing.
Acknowledgements
The authors wish to acknowledge funding from NIH R21 EB009153 as well as the NSF Stem Cell Biomanufacturing IGERT (DGE 0965945) to TER. Primary human MSCs were provided by the Texas A&M Health Science Center College of Medicine, Institute for Regenerative Medicine at Scott and White Healthcare through a grant from NCRR of the NIH (P40RR017447).
Footnotes
Electronic Supplementary Information (ESI) Available
References
- 1.de Paula FJA, Horowitz MC, Rosen CJ. Novel insights into the relationship between diabetes and osteoporosis. Diabetes Metab. Res. Rev. 2010;26:622–630. doi: 10.1002/dmrr.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazziotti G, Bilezikian J, Canalis E, Cocchi D, Giustina A. New understanding and treatments for osteoporosis. Endocrine. 2012;41:58–69. doi: 10.1007/s12020-011-9570-2. [DOI] [PubMed] [Google Scholar]
- 3.Rosen CJ, Bouxsein ML. Mechanisms of disease: is osteoporosis the obesity of bone? Nat. Clin.Pract. Rheum. 2006;2:35–43. doi: 10.1038/ncprheum0070. [DOI] [PubMed] [Google Scholar]
- 4.Giaccari A, Sorice G, Muscogiuri G. Glucose toxicity: the leading actor in the pathogenesis and clinical history of type 2 diabetes - mechanisms and potentials for treatment. Nutrition, Metabolism, and Cardiovascular Diseases. 2009;19:365–377. doi: 10.1016/j.numecd.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Jones DL, Wagers AJ. No place like home: anatomy and function of the stem cell niche. Nat. Rev. Mol. Cell Biol. 2008;9:11–21. doi: 10.1038/nrm2319. [DOI] [PubMed] [Google Scholar]
- 6.Minguell JJ, Erices A, Conget P, Onget PAC. Mesenchymal Stem Cells. Exp. Biol. Med. 2001;226:507–520. doi: 10.1177/153537020122600603. [DOI] [PubMed] [Google Scholar]
- 7.Kuhn NZ, Tuan RS. Regulation of stemness and stem cell niche of mesenchymal stem cells: implications in tumorigenesis and metastasis. J. Cell. Physiol. 2010;222:268–277. doi: 10.1002/jcp.21940. [DOI] [PubMed] [Google Scholar]
- 8.Lecourt S, Vanneaux V, Cras A, Freida D, Heraoui D, Herbi L, Caillaud C, Chomienne C, Marolleau JP, Belmatoug N, Larghero J. Bone marrow microenvironment in an in vitro model of Gaucher disease: consequences of glucocerebrosidase deficiency. Stem Cells Dev. 2012;21:239–248. doi: 10.1089/scd.2011.0365. [DOI] [PubMed] [Google Scholar]
- 9.Cramer C, Freisinger E, Jones RK, Slakey DP, Dupin CL, Newsome ER, Alt EU, Izadpanah R. Persistent high glucose concentrations alter the regenerative potential of mesenchymal stem cells. Stem Cells Dev. 2010;19:1875–1884. doi: 10.1089/scd.2010.0009. [DOI] [PubMed] [Google Scholar]
- 10.Gopalakrishnan V, Vignesh RC, Arunakaran J, Aruldhas MM, Srinivasan N. Effects of glucose and its modulation by insulin and estradiol on BMSC differentiation into osteoblastic lineages. Biochem. Cell Biol. 2006;84:93–101. doi: 10.1139/o05-163. [DOI] [PubMed] [Google Scholar]
- 11.Li, Y-M, Schilling T, Benisch P, Zeck S, Meissner-Weigl J, Schneider D, Limbert C, Seufert J, Kassem M, Schütze N, Jakob F, Ebert R. Effects of high glucose on mesenchymal stem cell proliferation and differentiation. Biochem. Biophys. Res. Commun. 2007;363:209–215. doi: 10.1016/j.bbrc.2007.08.161. [DOI] [PubMed] [Google Scholar]
- 12.Stolzing A, Coleman N, Scutt A. Glucose-induced replicative senescence in mesenchymal stem cells. Rejuv. Res. 2006;9:31–35. doi: 10.1089/rej.2006.9.31. [DOI] [PubMed] [Google Scholar]
- 13.Gagnon A, Sorisky A. The effect of glucose concentration on insulin-induced 3T3-L1 adipose cell differentiation. Obes. Res. 1998;6:157–163. doi: 10.1002/j.1550-8528.1998.tb00330.x. [DOI] [PubMed] [Google Scholar]
- 14.Lin Y, Berg AH, Iyengar P, Lam TKT, Giacca A, Combs TP, Rajala MW, Du X, Rollman B, Li W, Hawkins M, Barzilai N, Rhodes CJ, Fantus IG, Brownlee M, Scherer PE. The hyperglycemia-induced inflammatory response in adipocytes: the role of reactive oxygen species. J. Biol. Chem. 2005;280:4617–4626. doi: 10.1074/jbc.M411863200. [DOI] [PubMed] [Google Scholar]
- 15.Sun J, Xu Y, Dai Z, Sun Y. Intermittent high glucose stimulate MCP-l, IL-18, and PAI-1, but inhibit adiponectin expression and secretion in adipocytes dependent of ROS. Cell Biochem. Biophys. 2009;55:173–180. doi: 10.1007/s12013-009-9066-3. [DOI] [PubMed] [Google Scholar]
- 16.Botolin S, McCabe LR. Chronic hyperglycemia modulates osteoblast gene expression through osmotic and non-osmotic pathways. J. Cell. Biochem. 2006;99:411–424. doi: 10.1002/jcb.20842. [DOI] [PubMed] [Google Scholar]
- 17.García-Hernández A, Arzate H, Gil-Chavarría I, Rojo R, Moreno-Fierros L. High glucose concentrations alter the biomineralization process in human osteoblastic cells. Bone. 2012;50:276–288. doi: 10.1016/j.bone.2011.10.032. [DOI] [PubMed] [Google Scholar]
- 18.Inaba M, Nishizawa Y, Shioi a, Morii H. Importance of sustained high glucose condition in the development of diabetic osteopenia: Possible involvement of the polyol pathway. Osteoporosis Int. 1997;7:209–212. doi: 10.1007/BF03194374. [DOI] [PubMed] [Google Scholar]
- 19.Zayzafoon M, Stell C, Irwin R, McCabe LR. Extracellular Glucose Influences Osteoblast Differentiation and c–Jun Expression. J. Cell. Biochem. 2000;79:301–310. doi: 10.1002/1097-4644(20001101)79:2<301::aid-jcb130>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 20.Coe LM, Irwin R, Lippner D, McCabe LR. The bone marrow microenvironment contributes to type I diabetes induced osteoblast death. J Cell Physiol. 2011;226:477–483. doi: 10.1002/jcp.22357. [DOI] [PubMed] [Google Scholar]
- 21.Nakaoka R, Hsiong SX, Mooney DJ. Regulation of Chondrocyte Differentiation Level via Co-culture with Osteoblasts. Tissue Eng. 2006;12:2425–2433. doi: 10.1089/ten.2006.12.2425. [DOI] [PubMed] [Google Scholar]
- 22.Tsuda Y, Kikuchi A, Yamato M, Chen G, Okano T. Heterotypic cell interactions on a dually patterned surface. Biochem Biophys Res Commun. 2006;348:937–944. doi: 10.1016/j.bbrc.2006.07.138. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Cuddihy MJ, Kotov NA. Three-dimensional cell culture matrices: state of the art. Tissue Eng. Pt. B.-Rev. 2008;14:61–86. doi: 10.1089/teb.2007.0150. [DOI] [PubMed] [Google Scholar]
- 24.Leng L, McAllister A, Zhang B, Radisic M, Günther A. Mosaic hydrogels: one-step formation of multiscale soft materials. Adv. Mater. 2012;24:3650–3658. doi: 10.1002/adma.201201442. [DOI] [PubMed] [Google Scholar]
- 25.Hammoudi TM, Rivet CA, Kemp ML, Lu H, Temenoff JS. Three-dimensional in vitro tri-culture platform to investigate effects of crosstalk between mesenchymal stem cells, osteoblasts, and adipocytes. Tissue Eng. Pt. A. 2012;18:1686–1697. doi: 10.1089/ten.tea.2011.0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton SK, Bloodworth NC, Massad CS, Hammoudi TM, Suri S, Yang PJ, Lu H, Temenoff JS. Development of 3D hydrogel culture systems with on-demand cell separation. Biotechnol. J. 2013;8:485–495. doi: 10.1002/biot.201200200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterson J, Hubbell JA. Enhanced proteolytic degradation of molecularly engineered PEG hydrogels in response to MMP-1 and MMP-2. Biomaterials. 2010;31:7836–7845. doi: 10.1016/j.biomaterials.2010.06.061. [DOI] [PubMed] [Google Scholar]
- 28.Yang PJ, Levenston ME, Temenoff JS. Modulation of mesenchymal stem cell shape in enzyme-sensitive hydrogels is decoupled from upregulation of fibroblast markers under cyclic tension. Tissue Eng. Pt. A. 2012;18:2365–2375. doi: 10.1089/ten.tea.2011.0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosgrove BD, Griffith LG, Lauffenburger DA. Fusing Tissue Engineering and Systems Biology Toward Fulfilling Their Promise. Cellular and Molecular Bioengineering. 2008;1:33–41. [Google Scholar]
- 30.Kirouac DC, Ito C, Csaszar E, Roch A, Yu M, Sykes EA, Bader GD, Zandstra PW. Dynamic interaction networks in a hierarchically organized tissue. Mol Syst Biol. 2010;6:417. doi: 10.1038/msb.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivet Ca, Hill AS, Lu H, Kemp ML. Predicting cytotoxic T-cell age from multivariate analysis of static and dynamic biomarkers. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.003921. M110.003921-M003110.003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaudet S, Janes KA, Albeck JG, Pace EA, Lauffenburger DA, Sorger PK. A compendium of signals and responses triggered by prodeath and prosurvival cytokines. Mol. Cell. Proteomics. 2005;4:1569–1590. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Wasnik S, Tiwari A, Kirkland MA, Pande G. ch. 3. In: Jeon KW, editor. International review of cell and molecular biology. 1st edn. vol. 298. Academic Press; 2012. pp. 95–133. [DOI] [PubMed] [Google Scholar]
- 34.Travlos GS. Normal structure, function, and histology of the bone marrow. Toxicol Pathol. 2006;34:548–565. doi: 10.1080/01926230600939856. [DOI] [PubMed] [Google Scholar]
- 35.Manolagas SC. Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr Rev. 2000;21:115–137. doi: 10.1210/edrv.21.2.0395. [DOI] [PubMed] [Google Scholar]
- 36.Sheehy EJ, Vinardell T, Buckley CT, Kelly DJ. Engineering osteochondral constructs through spatial regulation of endochondral ossification. Acta Biomater. 2013;9:5484–5492. doi: 10.1016/j.actbio.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 37.Cusi K. The role of adipose tissue and lipotoxicity in the pathogenesis of type 2 diabetes. Curr. Diabetes Rep. 2010;10:306–315. doi: 10.1007/s11892-010-0122-6. [DOI] [PubMed] [Google Scholar]
- 38.Caplan AI. Adult Mesenchymal Stem Cells for Tissue Engineering Versus Regenerative Medicine. J. Cell. Physiol. 2007;213:341–347. doi: 10.1002/jcp.21200. [DOI] [PubMed] [Google Scholar]
- 39.Fazeli PK, Horowitz MC, MacDougald OA, Scheller EL, Rodeheffer MS, Rosen CJ, Klibanski A. Marrow fat and bone--new perspectives. J Clin Endocrinol Metab. 2013;98:935–945. doi: 10.1210/jc.2012-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bredella MA, Fazeli PK, Miller KK, Misra M, Torriani M, Thomas BJ, Ghomi RH, Rosen CJ, Klibanski A. Increased bone marrow fat in anorexia nervosa. J Clin Endocrinol Metab. 2009;94:2129–2136. doi: 10.1210/jc.2008-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffith JF, Yeung DK, Ma HT, Leung JC, Kwok TC, Leung PC. Bone marrow fat content in the elderly: a reversal of sex difference seen in younger subjects. J Magn Reson Imaging. 2012;36:225–230. doi: 10.1002/jmri.23619. [DOI] [PubMed] [Google Scholar]
- 42.Hahn MS, Taite LJ, Moon JJ, Rowland MC, Ruffino KA, West JL. Photolithographic patterning of polyethylene glycol hydrogels. Biomaterials. 2006;27:2519–2524. doi: 10.1016/j.biomaterials.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 43.Moon JJ, Saik JE, Poche RA, Leslie-barbick JE, Smith AA, Dickinson ME, West JL, Poché Ra, Lee S-H. Biomimetic hydrogels with pro-angiogenic properties. Biomaterials. 2010;31:3840–3847. doi: 10.1016/j.biomaterials.2010.01.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Temenoff JS, Athanasiou KA, LeBaron RG, Mikos AG. Effect of poly(ethylene glycol) molecular weight on tensile and swelling properties of oligo(poly(ethylene glycol) fumarate) hydrogels for cartilage tissue engineering. J Biomed Mater Res. 2002;59:429–437. doi: 10.1002/jbm.1259. [DOI] [PubMed] [Google Scholar]
- 45.Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikstrom C, Wold S. Multi- and Megavariate Data Analysis Part I: Basic Principles and Applications. 2nd edn. Umea, Sweden: Umetrics, AB; 2006. [Google Scholar]
- 46.Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikstrom C, Wold S. Multi- and Megavariate Data Analysis Part II: Advanced Applications and Method Extensions. 2nd edn. Umea, Sweden: Umetrics; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.