Summary
Here we report a comprehensive characterization of our newly developed inhibitor MM-401 that targets the MLL1 H3 lysine (K) 4 methyltransferase activity. MM-401 is able to specifically inhibit MLL1 activity by blocking MLL1-WDR5 interaction and thus the complex assembly. This novel targeting strategy does not affect other MLL family HMTs, revealing a unique regulatory feature for the MLL1 complex. Using MM-401 and its enantiomer control MM-NC-401, we show that inhibiting MLL1 methyltransferase activity specifically blocks proliferation of MLL leukemia cells by inducing cell cycle arrest, apoptosis and myeloid differentiation without general toxicity to normal bone marrow cells or non-MLL leukemia cells. More importantly, transcriptome analyses show that MM-401 induces similar changes in gene expression as MLL1 deletion, supporting a predominant role of MLL1 activity in regulating MLL1-dependent leukemia transcription program. We envision broad applications for MM-401 in basic and translational research.
Introduction
MLL1 (also called MLL, KMT2A, HRX, HTRX and ALL1) is one of the six MLL family histone methyltransferases (HMT) in mammals (Dou et al., 2005; Milne et al., 2002; Nakamura et al., 2002). It catalyzes mono-, di- and tri-methylation of histone H3 on K4 through the evolutionarily conserved SET domain. Both MLL1 and H3 K4 methylation (H3K4me) localize across gene promoters, transcription start sites (TSS) and 5’ transcribed regions of target genes and facilitate transcription initiation (Guenther et al., 2005; Lauberth et al., 2013). Deregulation of MLL1 accounts for 5–10% of acute myeloid leukemia (AML) in adults and almost 70% of acute lymphoblastic leukemia (ALL) in infants (Ayton and Cleary, 2001). The most common MLL1 rearrangements are balanced MLL1 translocations, in which one MLL1 allele is truncated and fused in frame with over 70 partners to produce oncogenic MLL1 fusion proteins (e.g. MLL-AF9, MLL-ENL) (Ayton and Cleary, 2001; Bernt and Armstrong, 2011; Dou and Hess, 2008) Mechanistic studies showed that at least some MLL1 fusion proteins, especially those forming the EAP complex (Mueller et al., 2007), are recruited to Hoxa9 and directly stimulate transcription elongation by recruiting cofactor complexes such as PAFc (Milne et al., 2010; Muntean et al., 2010), DOT1L (Jo et al., 2011; Krivtsov et al., 2007; Okada et al., 2005) as well as pTEFb/BRD4 (Lin et al., 2010; Yokoyama et al., 2010). In addition to MLL1 rearrangement, MLL1 tandem duplication and amplification are also reported in subpopulations of MLL leukemia. With rare exception (Ohyashiki et al., 1986), one common feature of the MLL1 abnormality in leukemia is the preservation of at least one wild type MLL1 allele with the intact SET domain.
Mouse genetic studies have shown that MLL1 is essential for fetal and adult hematopoiesis by regulating expression of Hox genes (e.g. Hoxa9) and other transcription cofactors promoting hematopoietic stem cell expansion (Artinger et al., 2013a; Jude et al., 2007; McMahon et al., 2007; Yu et al., 1995). In addition to regulating normal hematopoiesis, wild type MLL1 is also required for MLL1 fusion protein mediated leukemogenesis in vivo (Thiel et al., 2010). It has been shown that knocking out wild type MLL1 allele leads to loss of leukemic transformation capability of MLL-AF9 cells even in the presence of the onco-driving MLL1 fusion protein (Thiel et al., 2010). These genetic studies, however, have not specifically examined the role of MLL1 mediated H3K4me. Indeed, deleting MLL1 SET domain in mice does not lead to gross defects in hematopoietic development (Terranova et al., 2006), raising the questions on whether H3K4me by MLL1 plays an important role in normal hematopoiesis. The importance of H3K4me by MLL1 has not been directly tested in MLL leukemia.
Regulation of MLL1 activity is unique since the MLL1 SET domain has extremely low HMT activity, which is dramatically enhanced upon assembly into a core complex with three other proteins WDR5, ASH2L and RbBP5 (WAR) (Dou et al., 2006). All three proteins contribute to the optimal activity of the MLL1 complex, albeit through different mechanisms (Cao et al., 2010). Depleting any of these three proteins leads to drastic reduction of overall activity of the MLL1 complex. Importantly, it is shown that the interaction between MLL1 and WDR5 is critical for the integrity of the MLL1 complex and therefore, its methyltransferase activity (Dou et al., 2006; Patel et al., 2008; Song and Kingston, 2008). We, and others, further exploit this feature to develop inhibitors of the MLL1 methyltransferase activity. Blocking MLL1-WDR5 interaction by small molecule inhibitors leads to potent inhibition of the MLL1 methyltransferase activity in vitro (Karatas et al., 2013; Senisterra et al., 2013). However, these inhibitors are not characterized for their inhibition of other MLL family HMTs and their effects on H3K4me and MLL1-dependent transcriptome in cells.
Here we report a comprehensive characterization of our newly developed inhibitor, MM-401, for MLL1 methyltransferase activity. MM-401 is able to inhibit MLL1 activity by blocking MLL1-WDR5 interaction. Importantly, our targeting strategy does not affect other MLL family HMTs, revealing a unique regulatory feature of the MLL1 complex. Furthermore, MM-401, but not its enantiomer control MM-NC-401, specifically inhibits growth of MLL leukemia cells by inducing cell cycle arrest, apoptosis and myeloid differentiation without general toxicity to normal bone marrow cells or inhibition of non-MLL leukemia cells. At the molecular level, RNA-sequencing analyses show that MM-401 induces similar changes in gene expression as MLL1 deletion, supporting a major role of H3K4me by MLL1 in MLL leukemia.
Results
Development of MM-401 for inhibiting MLL1 methyltransferase activity
Here we report the development of a cyclic compound MM401 that targets MLL1-WDR5 interaction interface required for the integrity of the MLL1 core complex (Figure 1A). The design of MM-401 is based on the linear peptidomimetic MM-101 we previously reported (Karatas et al., 2013). Compare to MM-101, MM-401 maintained high binding affinity to WDR5 (Ki< 1 nM) as detected by the label-free BioLayer Interferometry (BLI/OctetRED) assay (Supplemental Figure 1A). Binding equilibrium fitting of MM-401 as well as binding kinetics for MM-401 were summarized in Supplemental Figure 1B–1D. The high binding affinity of MM-401 was further confirmed by the competitive fluorescence polarization (FP) experiment, which showed that MM-401 had a half-maximum inhibitory concentration (IC50) of 0.9 nM in disrupting WDR5-MLL1 interaction (Supplemental Figure 1E and 1F). This represents over 3 and 700 folds more potency relative to MM-101 (IC50 = 2.9 nM) and the WDR5-interacting motif in MLL1 (WIN, IC50 = 750 nM) (Supplemental Figure 1F). We also synthesized MM-NC-401, the enantiomer of MM-401, which had identical physiochemical properties and chemical structure with the exception of stereochemistry for four chiral centers (Figure 1A). MM-NC-401 had no detectable binding to WDR5 (IC50 > 100 µM, Ki> 10 µM) (Supplemental Figure 1C, 1D and 1F) and was used as the negative control for assays described herein. The synthesis of both compounds is described elsewhere.
Figure 1. Structure based design of the MLL1-derived peptidomimetics MM-401 and MMNC-401.
A. Chemical structures of MM-401 and MM-NC-401. MM-401 is significantly improved from MM-101. IC50 and Ki values of each compound for binding to WDR5 and MLL1 inhibition were summarized in Supplemental Figure 1F. B. Crystal structure of WDR5/MM-401 complex. The electron density (2Fo-Fc) map, contoured to 1δ is shown for MM-401. WDR5 is in green and MM-401 is in yellow. C. Comparison of MM-401 (yellow) and MLL1 peptide (pink) in WDR5 complexes (green). MM-401 interacts with WDR5 through inserting the Arg side chain into WDR5’s central channel, similar to that of the MLL1 peptide D. View of hydrogen bonding (dotted lines) and conserved water molecules (brown) in WDR5 binding pocket (green). MM-401 is in blue. E. View of MM-401 (yellow) specific interaction network in the WDR5 (green) binding pocket. See also Figure S1.
We first determined the co-crystal structure of MM-401 in complex with WDR5 at a resolution of 2.1 Å to confirm our specific targeting strategy (Figure 1B–1D). Data collection and refinement statics were provided in Table 1. Analysis of the WDR5/MM-401 complex structure confirmed that the interaction network between WDR5 and MM-401 was well conserved compared to that of WDR5-MLL1 interaction (Figure 1C). The interactions were mediated by a series of hydrogen bond and van der Waals interactions (Figure 1D). The Arg guanidinium moiety of MM-401 was sandwiched between two aromatic rings from F133 and F263 by cation-π stacking interactions (Figure 1E). Further arginine-specific recognition is achieved by extensive inter-molecular hydrogen bonding, either directly or indirectly to the nitrogen atoms in Arg guanidinium group (Figure 1D). The indirect hydrogen bonds were mediated by two conserved water molecules, which were also observed in the WDR5-MLL1 structure. Besides the guanidinium group, all the backbone amide and carbonyl of MM-401 formed either intra- or inter-molecular hydrogen bonds (Figure 1D), which assures specific recognition of MM-401 by WDR5. Compared to the MLL1 peptide, MM-401 made more extensive hydrophobic contacts with WDR5. One unique interaction was the phenyl group of MM-401 stacking with the phenyl group of WDR5 Y260 (Figure 1E). Similarly, the aliphatic four-carbon linker and 3rd residue side chain (an ethyl group) also made additional contacts with Y131, F149 and S49 of WDR5, which were absent from the MLL1-WDR5 interaction (Figure 1E). These additional hydrophobic interactions and restricted conformation underlie the enhanced binding of MM-401 to WDR5.
Table 1.
Data collection and refinement statistics
| WDR5-MM-401 | |
|---|---|
| Data collection | |
| Space group | P1 |
| Cell dimensions | |
| a, b, c (Å) | 46.904, 47.242, 68.896 |
| α, β, γ (°) | 88.50, 89.50, 74.53 |
| Resolution (Å) | 2.1 |
| Rsym (Å) (high res. Shell) | 0.060(0.167) |
| I/σI (high res. Shell) | 14.2 (3.9) |
| Completeness (%)(high res. Shell) | 97.8 (94.9) |
| Redundancy (high res. Shell) | 1.9 (1.7) |
| Refinement | |
| Resolution (Å) | 37.8–2.1 |
| No. reflections | 32442 |
| R work/R free (%) | 15.9/20.8 |
| No. atoms | |
| WDR5 | 4799 |
| MM-401 | 84 |
| Water | 523 |
| B-factor (Å2) | |
| WDR5 | 17.719 |
| MM-401 | 14.53 |
| Water | 25.401 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.919 |
MM-401 specifically inhibits MLL1 activity but does not affect other MLL family HMTs
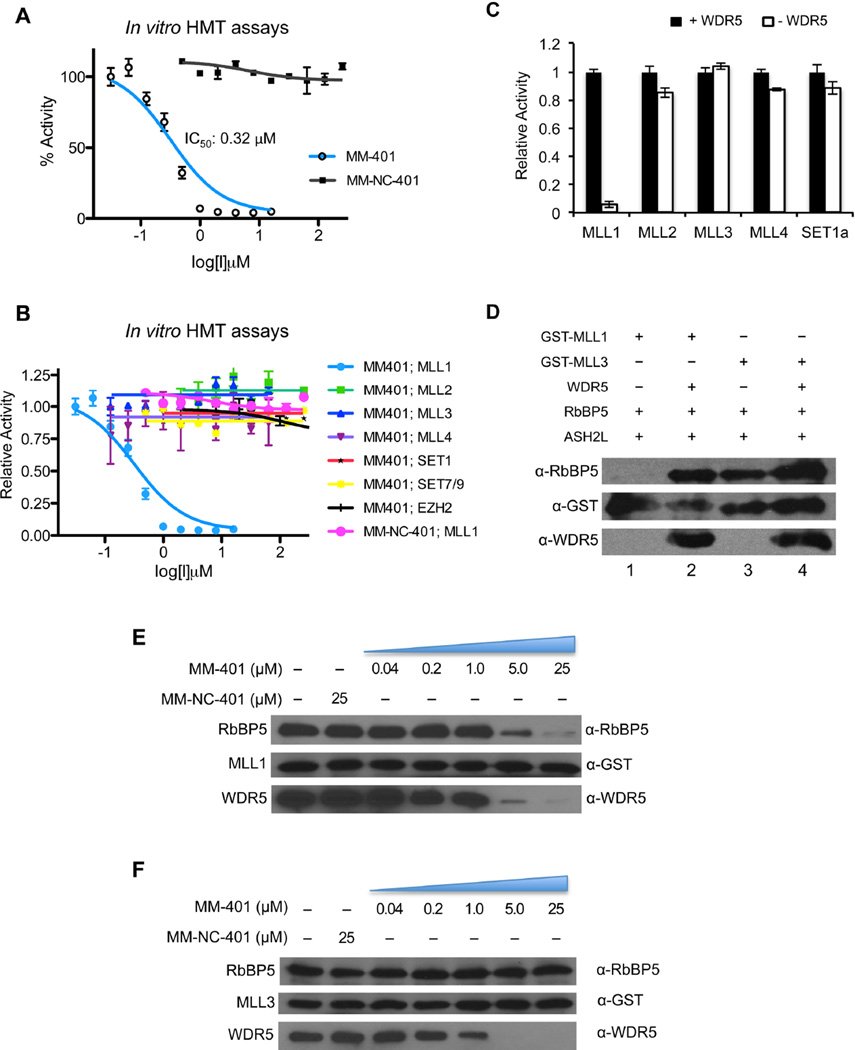
After establishing MM-401 as a potent inhibitor for the MLL1-WDR5 interaction, we next tested MM-401 for its inhibition of MLL1 activity in the in vitro histone methyltransferase (HMT) assay. As shown in Figure 2A, MM-401 was able to inhibit MLL1 (0.5 µM) activity with IC50 of 0.32µM. In contrast, no inhibition was detected with control MM-NC-401 at concentrations of up to 250µM. Of note, IC50 of MM-401 in the in vitro HMT assay is higher than that of protein binding described above. This is mostly due to higher MLL1 complex concentration in the assay that is required for robust detection of H3K4me. In fact, since 0.5 µM of the MLL1 complex was used in the HMT assay, IC50 of 0.32µM by MM-401 was close to the theoretical limit (0.25µM), supporting that MM-401 is a highly effective inhibitor for MLL1 in vitro. Mechanistic studies for MM-401 inhibition showed that it is a non-competitive inhibitor (Supplemental Figure 2A and 2B), consistent with its targeting of MLL1-WDR5 interaction interface instead of the catalytic SET domain.
Figure 2. MM-401 specifically inhibits activity of the MLL1 complex through disrupting complex assembly.
A. In vitro HMT assay to evaluate potency of MM-401 and MM-NC-401. IC50 for MM-401 are presented as mean ± s.d. B. In vitro HMT assay to evaluate selectivity of MM-401 among histone methyltransferases as indicated on bottom. IC50 for HMTs except MLL1 was not determined due to lack of inhibition. C. In vitro HMT assay for MLL family HMT complexes with (black) or without WDR5 (white). Y-axis, relative activity with WDR5 containing complex for each MLL family HMT arbitrarily set as 1. Data represent mean values for triplicates ± s.d. D. In vitro GST pull down for the MLL1 or MLL3 complexes with or without WDR5 as indicated on top. MLL1/MLL3, RbBP5 and WDR5 in the bound fraction were detected in immunoblots using antibodies as indicated on left. E–F. MM-401 specifically disrupts MLL1 complex assembly. GST-MLL1 (E) or GST-MLL3 (F) and HeLa nuclear extracts were mixed together with increasing concentration of MM-401 or 25 µM MM-NC-401 as indicated on top. MLL1 or MLL3, WDR5 and RbBP5 in bound fraction were detected by respective antibody as indicated. See also Figure S2 and S3.
We next tested MM-401 specificity against other histone lysine methyltransferases (HKMTs). As shown in Figure 2B, MM-401 did not inhibit SET7/9, a non-MLL family H3K4 mono-methyltransferase, or the H3 K27 methyltransferase EZH2 complex at concentration of up to 500 µM. It also had no inhibition for an expanded panel of HKMTs including H3K9 HMTs G9a and Suv39h1, H3K36 HMT MMSET as well as H3K79 HMT DOT1L (Supplemental Figure 2C). To our surprise, MM-401 had no inhibition for other MLL family HMTs as well (Figure 2B and Supplemental Figure 2D), despite the conservation of the core components (Dou et al., 2006) as well as the WIN motif in different MLLs (Cosgrove and Patel, 2010).
Since WIN motifs from different MLL1 interact with WDR5 with similar affinities (Supplemental Figure 3A), we decided to examine whether WDR5 was required for activities of all MLL family HMTs. To this end, we reconstituted all MLL complexes with or without WDR5 and tested their methyltransferase activities. We found that only MLL1 need WDR5 for its full HMT activity (Figure 2C). In contrast, WDR5 was dispensable for MLL2-4 and hSET1 complexes (Figure 2C). Furthermore, WDR5 did not affect the methylation state specificity of other MLLs (Supplemental Figure 2E and data not shown). The essential role of WDR5 in regulating MLL1 complex activity was probably due to the unique requirement of WDR5 in maintaining integrity of the MLL1 complex. As shown in Figure 2D, the MLL1 SET domain was not able to interact with RbBP5 in the absence of WDR5 (lane 1 vs. 2). In contrast, SET domains of both MLL3 and MLL4 (WBP7, the closest homologue of MLL1) were able to directly interact with RbBP5 in the absence of WDR5 (Figure 2D and Supplemental Figure 3B). WDR5 was also dispensable for assembly of other MLL family HMTs (data not shown). These results suggest that despite conservation of core components, the MLL1 complex has distinct biochemical features that allow for specific targeting. To further confirm the mechanism of action by MM-401, we incubated 10 µM GST-MLL1, GST-MLL3 or His-MLL4 (WBP7) protein with HeLa nuclear extract in the presence of increasing concentration of MM-401 and examined pull down efficiency of RbBP5 and WDR5 after GST or Ni-NTA purification. As shown in Figure 2E, increasing concentration of MM-401 (0.04 to 25 µM) was able to gradually disrupt MLL1 bindings to both WDR5 and RbBP5. In contrast, although MM-401 effectively dissociated WDR5, it did not affect MLL3 or MLL4 binding to RbBP5 even at highest concentration (Figure 2F and Supplemental Figure 3C). In all cases, 25µM MM-NC-401 had no effect on the integrity of the complexes (Figure 2E, 2F and Supplemental Figure 3C). Since RbBP5 and ASH2L are essential for all MLL family HMTs (Supplemental Figure 3D and data not shown), specific dissociation of RbBP5 from the MLL1 complex after MM-401 treatment underlies its inhibition specificity for MLL1.
MM-401 specifically inhibits MLL1-dependent H3K4 methylation in cells
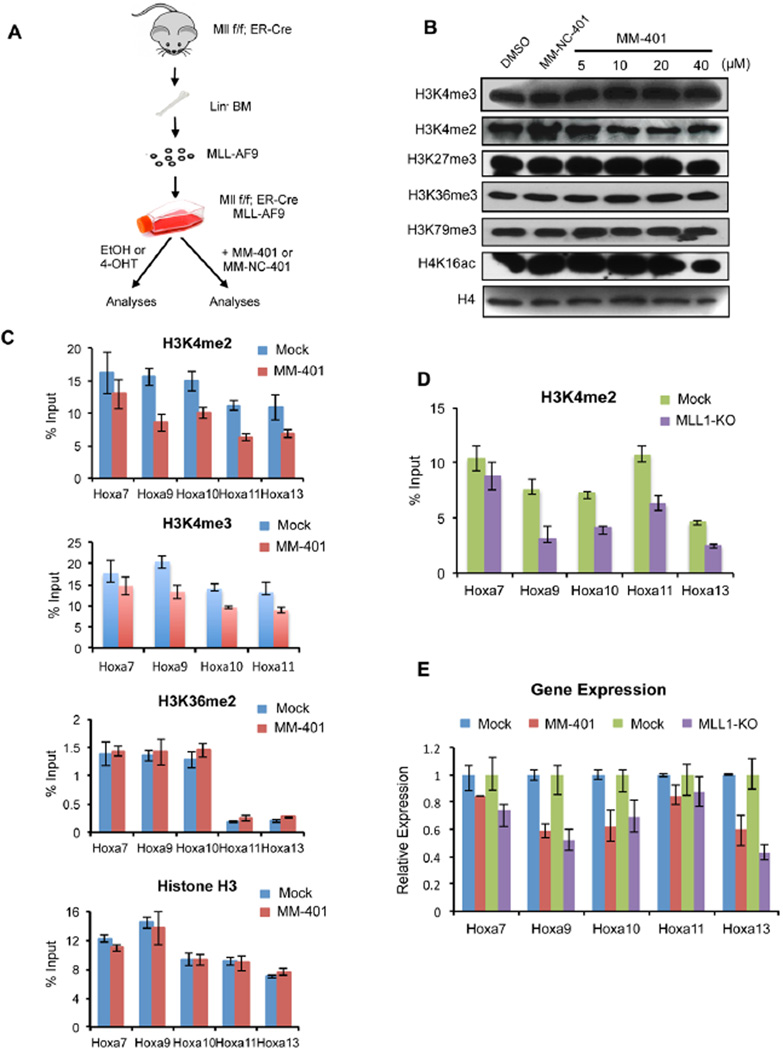
After characterization of MM-401 in vitro, we examined its cellular activities in transduced murine MLL-AF9 cells. In order to compare the effects of MM-401 to MLL1 loss of function mutation, we also established the MLL-AF9; Mll1f/f; ERT-Cre cell line by transducing MLL-AF9 into bone marrow progenitor cells (BMPCs) isolated from Mll1f/f; ERT-Cre mice (Figure 3A) (McMahon et al., 2007). In this cell line, wild type MLL1 gene can be effectively deleted after 4-OHT treatments (Supplemental Figure 3E). Consistent with the observed specificity of MM-401 in vitro, there was no change in global H3K4me in murine MLL-AF9 leukemia cells treated with a range of concentrations of MM-401 for four days (Figure 3B). Furthermore, MM-401 had no effects on other histone modifications such as H3K27me3, H3K36me3 or H3K79me3 (Figure 3A). MM-401 also had no effects on H4K16ac, catalyzed in part by a WDR5-containing MOF complex (Dou et al., 2005). In contrast, MM-401 (20µM) was able to reduce MLL1-dependent H3K4me2 and H3K4me3 across 5’ Hox A loci (i.e. Hoxa9 to Hoxa13 loci) in MLL-AF9 cells after 48hrs of MM-401 treatment (Figure 3C). As controls for ChIP assays, no differences in H3K36me2 and total H3 were detected at these Hox A loci (Figure 3C, bottom two panels). Similar reduction (~30–50%) of H3K4me2 was also observed in the MLL-AF9; Mllf/f; ERT-Cre cells after MLL1 deletion, supporting its biological relevance (Figure 3D). Reduction of H3K4me in these cells was not due to changes in MLL1, RbBP5 or WDR5 protein levels (Supplemental Figure 3F), or the binding of WDR5 and RbBP5 at Hox A loci (Supplemental Figure 3G) or the nuclear distribution of MLL1 (Supplemental Figure 3H). Thus, it is likely that MM-401 only perturbed the interface that was causally linked to MLL1 methyltransferase activity. Corresponding to changes in H3K4me, expression of 5’ Hox A genes, especially Hoxa9 and Hoxa10, was significantly decreased after cells were treated with MM-401 (Figure 3E). The levels of reduction were similar to those after MLL1 gene deletion, supporting the functional relevance. As control, the enantiomer MM-NC-401 had no effects on Hox gene expression. Of note, reduction of Hox gene expression was detected as early as 2 days after MM-401 treatment. This is in contrast to the DOT1L inhibitor, which requires 7-10 day treatments before detecting change in Hoxa9 expression (Daigle et al., 2011b).
Figure 3. MM-401 specifically inhibits H3K4me at 5’ Hox A loci.
A. Schematic for using MLL-AF9; Mllf/f, ERT-Cre cells for ChIP and gene expression analyses. B. Immunoblots for global histone modifications (indicated on left). Histone H4 was used as the loading control. C. ChIP analyses at Hoxa7 to Hoxa13 promoters in murine MLL-AF9 cells with or without MM-401 treatment. Antibodies are indicated on top. D. CHIP assays using MLL-AF9 transduced Mll1f/f; ERT-Cre+ BM cells with or without 4OHT treatment. For C-D, Y-axis represents relative value to 5% input. E. RT-PCR for Hoxa7-Hoxa13 in MLL-AF9 transduced Mll1f/f; ERT-Cre+ BM cells treated with DMSO, MM-401 (20µM), Mock or 4-OHT (MLL-KO) as indicated on top. Gene expression was normalized against Gapdh and presented as fold change against DMSO treated cells, which was arbitrarily set at 1. For C–E, data are presented as mean ± s.d. from three experiments. See also Figure S3 and S4.
To establish specificity of MM-401 for MLL1-dependent H3K4me in cells, we decided to examine H3K4me at MLL1 independent gene loci in MLL-AF9 cells after MM-401 treatment. Since specific distribution of MLL family HMTs in the genome was not defined in MLL-AF9 cells, we selected 10 MLL1-independent genes based on three criteria: 1) highly expressed in MLL-AF9 cells; 2) no MLL1 binding was detected (Supplemental Figure 4A); and 3) no expression change after MLL1 deletion (Supplemental Figure 4B). As shown in Supplemental Figure 4C, neither H3K4me2 nor H3K4me3 were affected at these gene promoters. H3K4me2 at 3’ Hox A loci (Hoxa1-a6), which were not MLL1 direct targets (Supplemental Figure 4A), was also not changed upon MM-401 treatment (Supplemental Figure 4E). Relative expression of Hox A genes were shown in Supplemental Figure 4D. These results support that MM-401 specifically targets H3K4me at MLL1 regulated genes in cells.
MM-401 induces similar changes in MLL-AF9 transcriptome as the MLL1 deletion
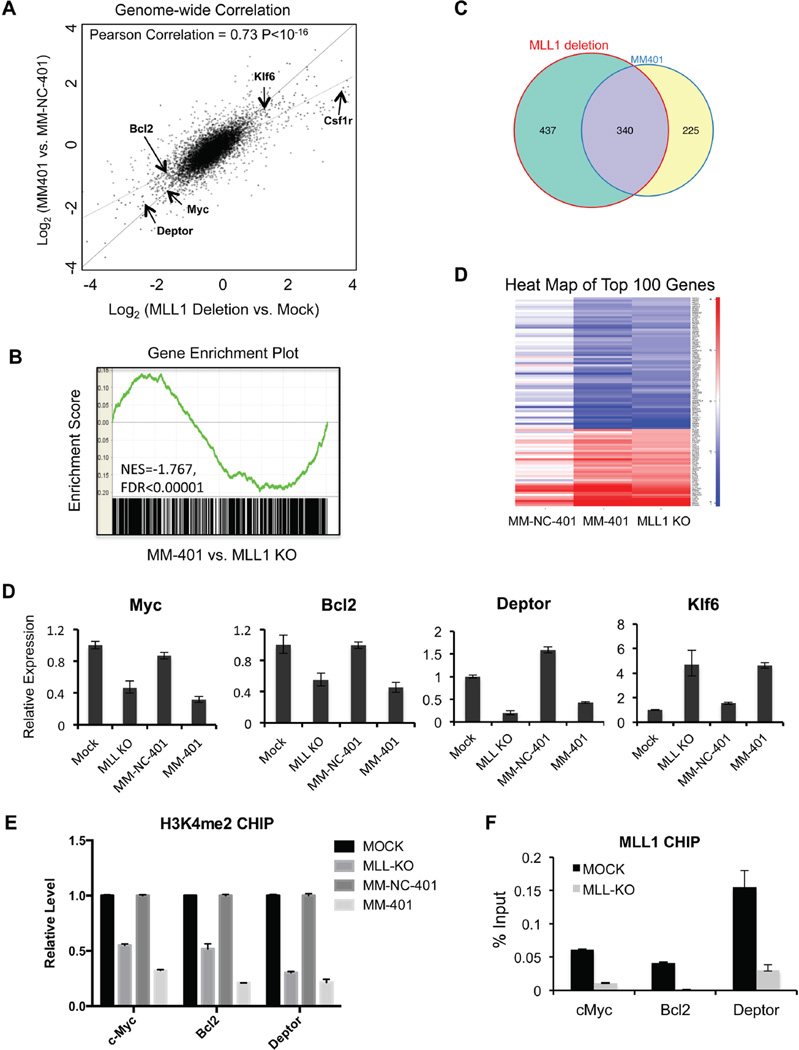
To examine the regulation of MM-401 on MLL1 dependent transcriptome, we compared global gene expression profiles in MLL-AF9 cells after MM-401 treatment or MLL1 deletion. Specifically, the MLL-AF9; Mll1f/f; ERT-Cre cells were treated with either 4-OHT for MLL1 deletion or with MM-401 for inhibiting H3K4me. Vehicle (ethanol for 4-OHT) or MM-NC-401 treated cells were used as respective controls. RNA isolated from these cells was subject to Illumina sequencing (RNA-seq). Sequencing reads from RNA-seq were mapped to ~8000 genes in each comparison group (MLL1 deletion vs. mock and MM-401 vs MM-NC-401). Remarkably, genome-wide transcription profiles of these two comparison groups were highly correlated (Figure 4A, Pearson correlation coefficient = 0.73, p< 10−16). Gene set enrichment analysis (GSEA) also documented significant overlap between these two transcriptome (Figure 4B). Among 8000 genes sequenced in each comparison group, only a small subset of genes showed differential expression in either MLL1 deletion or MM-401 treated cells relative to their respective controls. 777 genes (p< 0.05, Log2 (fold change)>1) were differentially expressed in MLL-AF9 cells after MLL1 deletion (Supplemental Table 1) and 564 genes (p< 0.05, Log2 (fold change)>1) altered expression upon MM-401 treatment (Supplemental Table 2). In both cases, ~60% genes were down regulated, consistent with the role of MLL1 activity in transcriptional activation. Among the differentially expressed genes above, 340 genes were commonly found in both MLL1 deletion and MM-401 treatment as illustrated in the Venn diagram (Figure 4C, Supplemental Table 3 and 4), which is highly significant (p< 10−10). Furthermore, heat map of top 100 genes that changed expression upon MM-401 treatment showed comparable changes in MLL1 knockout cells, whereas little correlation was found with enantiomer MM-NC-401 treated samples (Figure 4D). The corresponding changes in MLL-AF9 transcriptome upon MLL1 deletion or MM-401 treatment strongly argue that MM-401 is able to modulate MLL1-dependent gene transcription and that the methyltransferase activity plays a predominant role in MLL1 function in MLL-AF9 leukemia.
Figure 4. MM-401 induces similar changes in gene expression as MLL1 deletion in MLL-AF9 cells.
A. Correlation of log2 fold change between MLL1 knockout vs. wild type and MM-401 vs. MM-NC-401 treated MLL-AF9 cells across all genes. Lines represent the identity line (solid) and the line of best fit (dotted) respectively. Pearson Correlation was obtained using R program with p<10−16 (two-tailed t-test). B. GSEA analysis for transcriptome of MM-401 treated and MLL1 deleted MLL-AF9 cells. The gene set from MM-401 treatment was ranked and enrichment of gene set from MLL1-depletion was indicated by FDR. C. Venn diagram for genes that have > 2 fold changes in expression in MLL1 deletion or MM-401 treated samples. C. Heat map of top 100 genes that showed expression changes following treatment of MM-401 in different cells as indicated on bottom. D. Q-PCR confirmation for MLL1 targets as indicated on top. Gene expression was normalized against GAPDH and presented as fold change against mock treated cells, which is arbitrarily set at 1. Data are presented as mean ± s.d. from three experiments. E–F. CHIP assays for H3K4me2 (E) and MLL1 (F) at genes indicated on bottom. Y-axis represents relative value to % input in MOCK or MM-NC-401 treatment. Data are presented as mean ± errors from three experiments. See also Figure S5 and Table S1–5.
Wild type MLL1 is required for expression of Myc and Bcl2 in MLL leukemia
Recently, it is demonstrated that MLL1 is required to regulate gene expression beyond Hox clusters in normal hematopoiesis (Artinger et al., 2013b). We decided to further analyze genes that changed expression upon MLL1 deletion or MM-401 treatment in MLL-AF9 leukemia cells. GO term analyses showed that gene pathways that were commonly down regulated included embryo- and multi-organ developments as well as multi-lineage cell differentiation while gene pathways that were commonly up regulated included development and activation of mature myeloid cells such as macrophages and neutrophils (Supplemental Figure 5 and Supplemental Table 5). These transcriptome changes suggest a shift towards more differentiated phenotypes (see below). Two genes, Bcl2 and Myc, were identified by RNA-seq among the commonly down-regulated genes. Both genes were proposed to be key targets of the BRD3/4 inhibitor in MLL leukemia (Dawson et al., 2011a; Delmore et al., 2011; Zuber et al., 2011), which disrupts the downstream gene pathway of pTEFb that interacts with MLL1 fusion proteins. We also identified other novel MLL1 targets such as Deptor (Figure 4D), an antagonist for mTOR, which was recently shown to be critical in hematopoiesis and leukemogenesis evoked by loss of Pten (Kentsis and Look, 2012). Down regulation of these genes were confirmed by real-time PCR in both MLL1 knockout cells and MM-401 treated cells (Figure 4D). As shown in Figure 4F, Myc, Bcl2 and Deptor were MLL1 direct targets. Knockout MLL1 led to lower levels of H3K4me2 at these gene promoters, which were phenocopied by MM-401 treatment (Figure 4E). We also found that up-regulation of a tumor suppressor Klf6 (Narla et al., 2001) (Figure 4D), which was likely due to indirect effects of MLL1 deletion/inhibition (data not shown). The lists of genes that changed expression (> 2 fold) upon MLL1 deletion and MM-401 treatment were included in Supplemental Table 3 and 4. It will be interesting to examine the functional implications of novel MLL1 targets in normal and malignant hematopoiesis in future.
Targeting MLL1 methyltransferase activity by MM-401 selectively inhibits MLL leukemia
Earlier mouse genetic studies have shown that wild type MLL1 is essential for development of MLL-AF9 leukemia in vivo (Thiel et al., 2010). However, whether MLL1 methyltransferase activity is required for MLL leukemia remains unclear. The fact that MM-401 blocks Hoxa9, Myc and Bcl2 expression implies that MLL1 activity probably plays an important role in MLL leukemia. Consistent with this, knock down WDR5 by shRNA in MLL-AF9 cells led to significantly reduced transformation capability of the MLL-AF9 cells similar to MLL1 gene deletion (Supplemental Figure 6B and 6C).
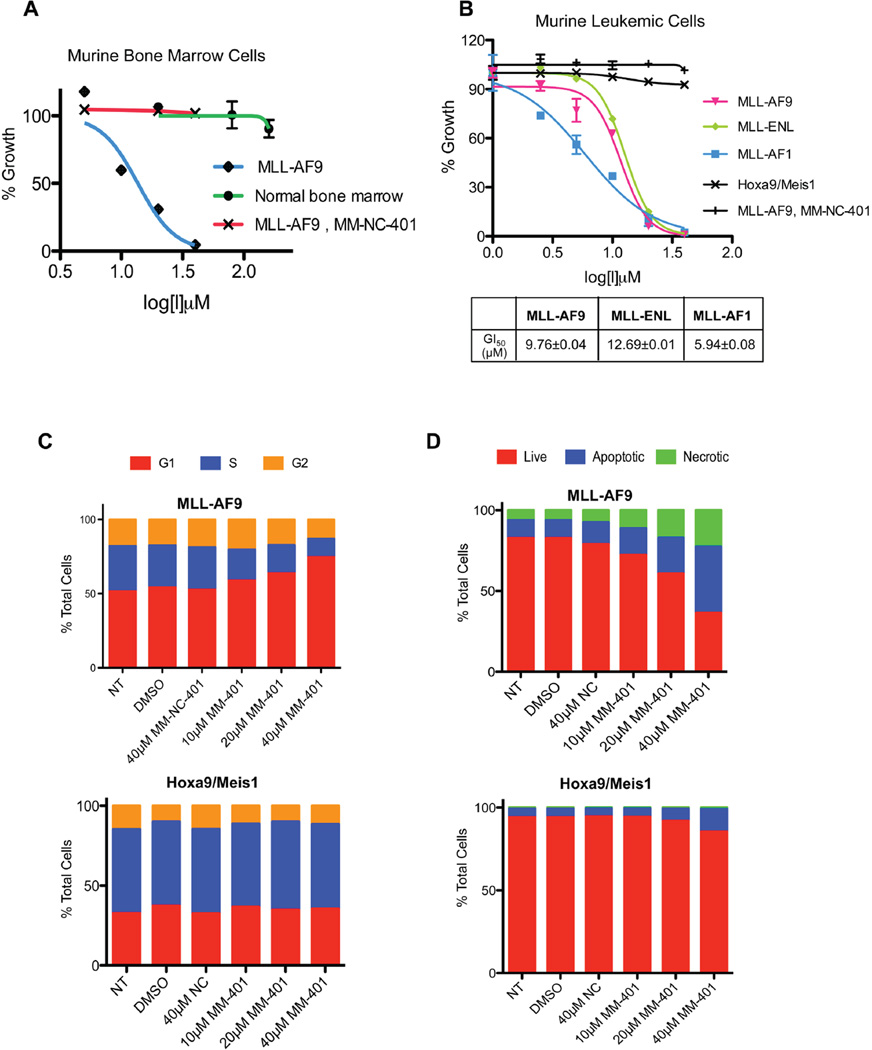
To directly test whether targeting MLL1 activity by MM-401 inhibits proliferation of MLL-AF9 leukemia cells, we treated primary BMPCs with or without MLL-AF9 with MM-401. MM-401 had profound effects on proliferation of MLL-AF9 cells in liquid culture, with half maximum growth inhibition (GI50) value of ~10µM (Figure 5A). In contrast, negative control MM-NC-401 had minimal effects on MLL-AF9 cells. Consistent with MM-401 functioning through MLL1, it did not cause further growth inhibition to MLL-AF9; Mllf/f; ERT-Cre cells after MLL1 deletion (data not shown). Interestingly, despite the importance of MLL1 in normal hematopoiesis (Hess et al., 1997; Jude et al., 2007; McMahon et al., 2007; Yu et al., 1995), MM-401 did not affect proliferation of primary BMPCs in liquid culture at concentrations of up to 160µM (Figure 5A, see discussion). Colony formation of normal BMPCs also remained unchanged after MM-401 treatment (Supplemental Figure 5). These results suggested that MM-401 specifically inhibited MLL-AF9 leukemia without affecting normal BMPCs. To further assess the selectivity of MM-401 for MLL leukemia, we tested several murine cell lines transformed with different oncogenic drivers. As shown in Figure 5B, MM-401 effectively inhibited growth of MLL-ENL and MLL-AF1 leukemia cells while had no effects on leukemia cells overexpressing exogenous Hoxa9 and Meis1.
Figure 5. MM-401 specifically inhibits murine MLL leukemia cells.
A. Growth inhibition of normal or MLL-AF9 transduced murine bone marrow cells by MM-401 or MM-NC-401. Cell growth was determined by the CellTiterGlo assay and presented as % of mock treated cells. B. Growth inhibition of murine MLL-AF9, MLL-ENL, MLL-AF1 or Hoxa9/Meis1 leukemia cells by MM-401 or MM-NC-401. GI50 was summarized below and with data presented as mean ± s.d. from three experiments. C. Dose-dependent effects of MM-401 on cell cycle progression analyzed by FACS. D. Dose-dependent effects of MM-401 on apoptosis analyzed by Annexin V/PI staining. For C–D, compound treatments of murine MLL-AF9 (top) or Hoxa9/Meis1 (bottom) cells were indicated. Y-axis is % cells in each category as indicated on top. See also Figure S6.
MM-401 induces cell cycle arrest, apoptosis and differentiation of MLL leukemic blasts
To investigate the action of MM-401 in cells, we analyzed cell cycle progression, apoptosis and differentiation of murine MLL-AF9 and Hoxa9/Meis1 cells with or without MM-401 treatment. We found that MM-401 was able to induce prominent G1/S arrest in MLL-AF9 cells in a concentration dependent manner (Figure 5C, top panel), consistent with the cell cycle defects incurred by acute loss of MLL1 (Liu et al., 2008). In contrast, cell cycle index of Hoxa9/Meis1 cells remained largely unchanged by MM-401 treatment (Figure 5C, bottom panel). MM-401 also specifically induced apoptosis of MLL-AF9 cells (top) as early as 48-hour post treatment whereas it had minimal effects on Hoxa9/Meis1 cells (bottom) (Figure 5D).
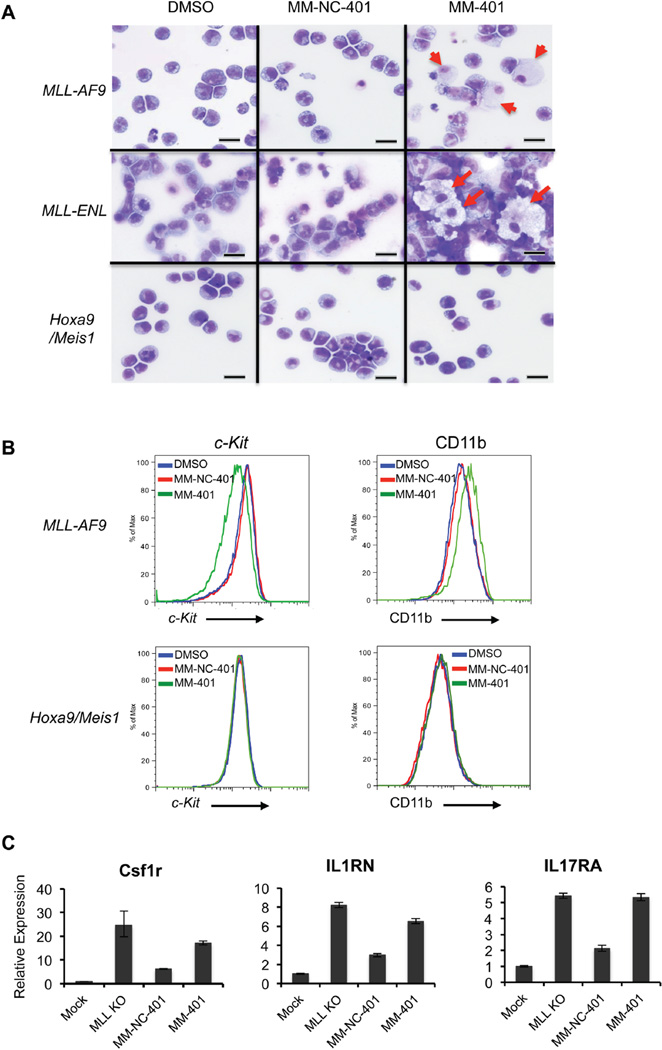
In addition to growth inhibition and apoptosis, MM-401 specifically induced differentiation of MLL-AF9 and MLL-ENL leukemic blasts towards more mature lineages (e.g. macrophage). Upon 20 µM MM-401 treatment for 7 days, significant number of cells underwent differentiation with characteristic small nuclei and large foamy cytoplasm as shown by Wright-Giemsa staining (Figure 6A, right panels). No differentiation was observed for MLL-AF9 and MLL-ENL leukemic blasts treated with the control MM-NC-401 (Figure 6A, middle panels). Consistent with the selectivity of MM-401, Hoxa9/Meis1 leukemic blasts remain undifferentiated under the same condition. Hematopoietic differentiation towards more mature myeloid lineage was further confirmed by fluorescence-activated cell sorting (FACS) analyses. MM-401 treatment led to a marked reduction of the progenitor cell surface marker c-Kit and an attendant increase of the myeloid marker CD11b in MLL-AF9 cells (Figure 6B, top panels). As a control, no change in c-Kit and CD11b surface markers was observed after MM-NC-401 treatment or in MM-401 treated Hoxa9/Meis1 cells (Figure 6B, bottom panels). Concomitant with morphological and immunophenotypical changes, several genes expressed in mature myeloid cells such as Csf1r, IL1rn and IL17ra were up regulated in MM-401 treated MLL-AF9 cells (Figure 6C). Up regulation of these genes were also found after MLL1 deletion (Supplemental Table 2).
Figure 6. MM-401 specifically induces myeloid differentiation in MLL leukemia cells.
A. Wright-Giemsa-stained cytospins for MLL-AF9 (top), MLL-ENL (middle) and Hoxa9/Meis1 (bottom) transduced leukemia cells treated with DMSO, 40 µM MM-NC-401 or 20 µM MM-401. Arrowhead, differentiated macrophages. Scale bars are 10µm. B. FACS analyses for c-kit (left) or CD11b (right) of MLL-AF9 (top) or Hoxa9/Meis1 (bottom) cells treated with DMSO, 40 µM of MM-NC-401 or MM-401. Y-axis is normalized % events defined by Flow-Jo on overlay histogram. X-axis is signal intensity in arbitrary unit. C. Q-PCR for MLL1 targets after MM-401 treatment or MLL1 deletion. Gene expression was normalized against Gapdh and presented as fold change against mock treated cells, which is arbitrarily set at 1. Data are presented as mean ± s.d. from three experiments. See also Figure S6.
MM-401 shows efficacy in human MLL leukemia
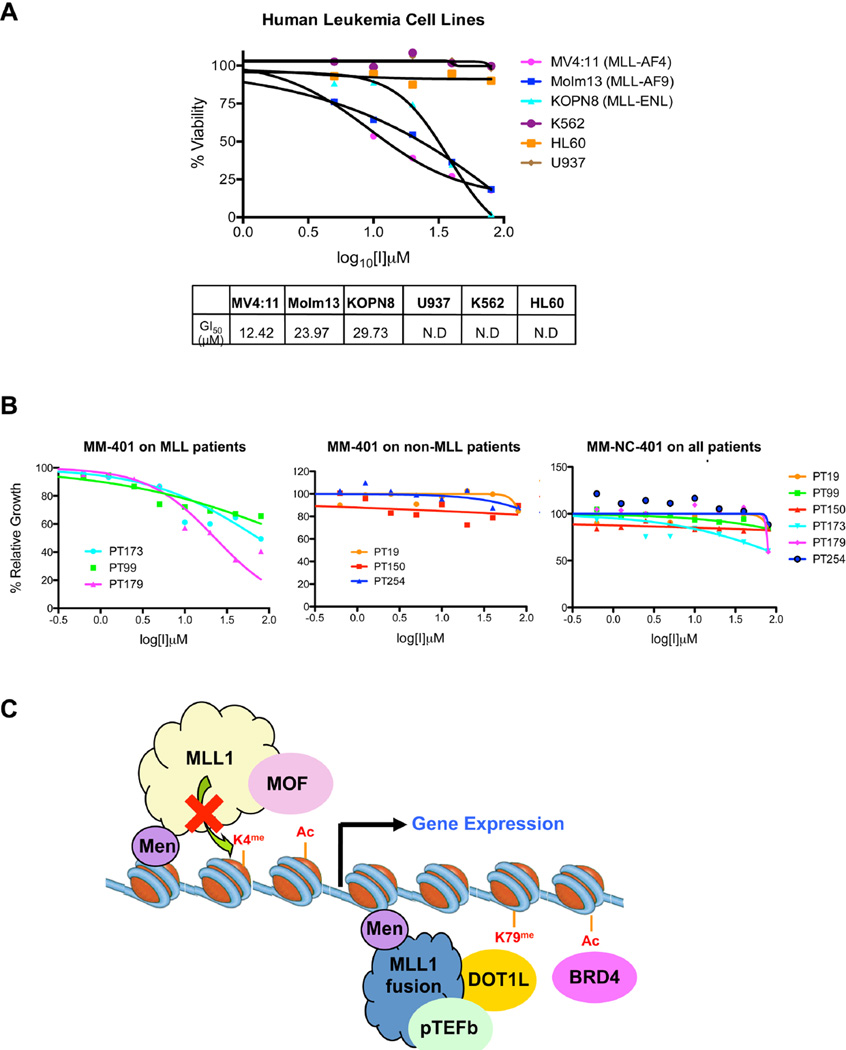
In addition to murine cell lines, MM-401 showed efficacy against human MLL leukemia cell lines as well. We tested three leukemia cell lines with single allele MLL1 translocation including MV4:11 (MLL-AF4, AML), MOLM13 (MLL-AF9, AML), and KOPN8 (MLL-ENL, ALL) as well as three control cell lines that carry no MLL1 related genetic lesions (i.e. K562, HL60 and U937) for cell growth. As shown in Figure 7A, MM-401 inhibited growth of MV4:11, MOLM13 and KOPN8 leukemia cells whereas it had no effects on leukemia driven by other oncogenic mutations. The GI50 of MM-401 for MLL leukemia cells was summarized in Figure 7A. Despite small differences in GI50 for different MLL leukemia cells, a phenomenon also described for other epigenetic inhibitors (Daigle et al., 2011a; Dawson et al., 2011b), MM-401 demonstrates high specificity for MLL leukemia, supporting the mechanism based on-target effects.
Figure 7. MM-401 specifically inhibits human MLL leukemia cells.
A. human leukemia cell lines were tested for growth inhibition in the presence of MM-401 or MM-NC-401 for three days. MLL1 translocation was indicated in parenthesis. K562, HL60 and U937 cells were used as non- MLL leukemia control cell lines. GI50 was summarized in the table with data presented as mean from three experiments. For non-MLL leukemia cell lines, GI50 was not determined due to the lack of inhibition. B. MM-401 inhibits MLL leukemia blasts isolated from human patients. PT, patient case number. Patient cytogenetic and molecular diagnosis data see Supplemental Table 6. For all panels, Y-axis, percent of relative growth of MM-401 vs. mock treated cells; X-axis, compound concentration (log10). C. Schematic for the potential mechanism of action of MM-401, details see text. See also Figure S7 and Table S6.
Increased apoptosis, cell cycle arrest as well as cell differentiation upon MM-401 treatment were also observed in human leukemia cells MV4:11 (MLL-AF4). We found that MM-401 induced apoptosis as well as dose-dependent G1/S arrest after 48-hour treatment in MV4:11 cells (Supplemental Figure 7A and 7B). A prominent dose-dependent increase of the myeloid surface marker CD11b was also detected in MV4:11 after MM-401 treatment (Supplemental Figure 7C), indicative of shift of leukemic blasts towards more differentiated myeloid cells. In contrast, MM-401 had minimal impact on cell cycle index, apoptosis and myeloid differentiation of K562 (BCR-ABL) leukemia cells (Supplemental Figure 7), confirming selectivity of MM-401 in human MLL leukemia.
Finally, we sought to establish the potential applicability and selectivity of MM-401 to human diseases. To this end, we tested efficacy of MM-401 on human leukemia blasts isolated from human AML patients with or without MLL1 rearrangements. As shown in Figure 7B, while control MM-NC-401 had no effects on leukemia blasts from AML patients, MM-401 was able to inhibit blast cells isolated from MLL leukemia patients, but not those isolated from AML patients carrying no MLL1 abnormality (cytogenetic information see Supplemental Table 6). Taken together, these results suggest that MM-401 has therapeutic potential for MLL leukemia in patients.
Discussion
Here we describe the development of a highly specific inhibitor (MM-401) for H3 K4 methyltransferase MLL1. Our results show, for the first time, the critical role of MLL1 catalytic activity in MLL1 rearranged leukemia and the potential of the MLL1-WDR5 interaction interface as a therapeutic target. Furthermore, we demonstrate a novel strategy for selective targeting MLL1 methyltransferase activity. Instead of directly targeting the catalytic SET domain, we exploit the unique regulation of the MLL1 complex by WDR5 and target MLL1 complex assembly without affecting other MLL family HMTs. As a result, our compound shows no inhibition for global H3K4me and little toxicity for normal BM cells. Transcriptome analyses also confirmed the remarkable selectivity of MM-401, which induces changes in gene expression that are highly correlative with MLL1 gene deletion. The specificity of MM-401 for MLL1 HMT activity ensures its broad applications in basic research (e.g. to probe functions of H3K4 methylation by MLL1 in different biological context) and, with further optimization, in targeted epigenetic therapies.
Targeting MLL1 activity for treatment of MLL leukemia
MLL leukemia arises from combined cell proliferation abnormality and differentiation block as a result of balanced translocation of one MLL1 allele (Ayton and Cleary, 2001; Krivtsov and Armstrong, 2007). MLL1 translocation generates over 70 in-frame MLL1 fusion proteins, which lead to aberrant expression of hematopoietic transcription factors (e.g. Hoxa9) in BMPCs whose down regulation are essential for normal myeloid differentiation. Mechanistic studies have shown that these MLL1 fusion proteins recruit cofactors such as the EAP complex (Monroe et al., 2011; Mueller et al., 2007), PAF1 (Milne et al., 2010; Muntean et al., 2010), DOT1L (Jo et al., 2011; Krivtsov et al., 2007; Okada et al., 2005) and pTEF-b (Lin et al., 2010; Yokoyama et al., 2010), which facilitate transcription elongation of MLL1 target genes (Fig 7B). Recently, small molecular inhibitors for DOT1L, BRD4 and MENIN that target MLL1 fusion dependent gene pathways have shown efficacies in treatment of MLL leukemia and some of them are quickly moving to clinical trials (Bernt et al., 2011; Daigle et al., 2011a; Dawson et al., 2011b; Grembecka et al., 2012). With rare exception, most MLL1 rearranged MLL leukemia maintains one functional wild type MLL1 allele. MLL1 tandem duplication and MLL1 amplification have also been reported in subpopulations of AML and ALL patients (Ayton and Cleary, 2001). Here we show that targeting wild type MLL1 methyltransferase activity selectively inhibits MLL leukemia by reducing expression of Hoxa9 and other genes important for leukemogenesis, thus serving as an alternative therapeutic approach for treatment of MLL leukemia.
One previous concern for targeting MLL1 methyltransferase activity is its similarity to other MLL family HMTs. It is conceivable that general inhibition of the MLL family HMTs will lead to high toxicity in normal cells given the importance of H3K4 methylation in many biological pathways. Our study here shows that targeting the MLL/SET1-WDR5 interaction interface selectively inhibits MLL1 activity but not that of other MLL family HMTs. This selectivity is important for potential therapeutic applications since it will limit general toxicity of the inhibitor. Similar concerns can also be raised for targeting the highly conserved WDR5 protein. WDR5 resides in multiple protein complexes (e.g. the MOF-MSL1v1 complex (Li et al., 2009)) and forms conserved interactions with other proteins as well as nucleic acids (Trievel and Shilatifard, 2009). However, in most reported interactions, WDR5 does not directly contribute to enzymatic activities of the residing complexes with the exception of the MLL1 complex. Furthermore, the arginine binding central cavity has only been reported to interact with the WIN motif of MLLs or H3 tail in vitro (Trievel and Shilatifard, 2009), suggesting limited effects by MM-401 on MLL1-independent WDR5 functions. Genome-wide transcriptome analyses confirm the specificity of this targeting strategy. About 60% genes that show more than 2 fold changes in gene expression upon MM-401 treatment are also affected by MLL1 deletion including key targets (e.g. Myc and Bcl2) for leukemogenesis (p< 10−10, Fisher’s exact test). We also fail to detect any global changes in histone modifications (Figure 3B). These results suggest that blocking WDR5 arginine-binding pocket mainly affects MLL1 dependent transcription regulation in the context of MLL leukemia. The on-target effects of MM-401 on MLL-AF9 transcriptome and the lack of general toxicity for normal BM cells strongly argue that targeting MLL1-WDR5 interaction is a viable therapeutic approach for MLL leukemia. Future characterizations of MM-401 in different biological context are needed to fully assess its overall impact in vivo.
Distinct regulation of the MLL1 complex
Mammals have six MLL family H3 K4 methyltransferases in response to increased complexity of the organism. These MLL family HMTs are ubiquitously expressed and share a common core complex configuration that is essential for their methyltransferase activities (Dou et al., 2006). However, both mouse genetics and whole-exome sequencing studies for mutations of the MLL family members lead to conclusions supporting more specialized functions for each of the MLL family HMTs (Dou and Hess, 2008). Functional and regulatory specification of MLL family HMTs are likely due to 1) recruitment to different genomic loci by interacting with distinct chromatin binding factors (Cho et al., 2007; Hughes et al., 2004; Patel et al., 2007) and 2) intrinsic differences in their biochemical characteristics (Wu et al., 2013). We have recently reported that MLL3 is a H3 K4 mono-methyltransferase and is not regulated by H2Bub-mediated trans-tail regulation. In contrast, MLL1 is able to mono-, di- and tri-methylate H3 K4 and is subject to regulation by H2Bub (Wu et al., 2013). Our study here reveals a distinct feature of the MLL1 complex. To our surprise, we find that MLL1-WDR5 interaction is uniquely required for the overall activity of the MLL1 complex, but not other MLL family HMTs (Figure 2C). This is due to essential function of MLL1-WDR5 interaction in stabilizing interactions between MLL1 SET domain and the ASH2L SPRY domain (Cao et al., 2010). Since MLL3 directly interacts with RbBP5/ASH2L heterodimer, disrupting MLL3-WDR5 interaction does not have net effects on the catalysis of the MLL3 complex. It also does not alter MLL3 methylation state specificity (Supplemental Figure 2E). Of note, it has been reported that WDR5 is essential for global H3K4 methylation and knockdown WDR5 in cells leads to reduced global H3K4 di- and tri-methylation (Wysocka et al., 2005). These results are not in conflict with our conclusion here since WDR5 is involved in multiple protein complexes and its depletion may have indirect effects on H3K4 methylation machineries. On the contrary, MM-401 only blocks a single protein interaction interface while leaving other WDR5 intercom intact. In sum, we show that MLL family HMTs have distinct biochemical properties that allow for differential inhibition of their respective activities. It would be interesting to further analyze the distinct features of the MLL1 complex as well as their structural basis in future.
MLL1 methyltransferase activity is essential for MLL leukemia
Extensive studies have demonstrated the importance of wild type MLL1 gene in both fetal and adult hematopoiesis (Hess et al., 1997; Jude et al., 2007; McMahon et al., 2007; Yu et al., 1995). However, deletion of the MLL1 SET domain in mice does not lead to embryonic lethality or gross defects in hematopoiesis (Terranova et al., 2006). The MLL1ΔSET mice are relatively normal, raising the question on whether the MLL1 H3K4 methyltransferase activity is obligatory in MLL1 functions. The function of MLL1 methyltransferase activity has not been directly tested in leukemogenesis. Our study here firmly establishes the importance of MLL1 activity in the maintenance of MLL leukemia. ChIP assays show that MM-401 treatment leads to reduction of H3K4me2 and H3K4me3 without affecting chromatin binding of WDR5 and RbBP5 at Hox A loci (Supplemental Figure 3G), allowing us to causally attribute changes in MLL-AF9 cells to MLL1-dependent methylation. Our results highlight the importance and specific requirement of H3K4me by MLL1 in MLL leukemia: 1) MM-401 alters global gene expression in the same way as MLL1 deletion (Figure 4); 2) MM-401 selectively inhibits MLL-AF9 cell growth and induces apoptosis, cell cycle arrest and myeloid differentiation of MLL leukemic blasts while has no effects on non-MLL leukemia (Figure 5 and 6); and 3) consistent with mouse genetic studies using MLL1ΔSET mice (Terranova et al., 2006), MM-401 has little impact on proliferation and colony formation of normal BM cells (Supplemental Figure 6). These results raise an interesting possibility that MLL1 methyltransferase activity is essential for MLL leukemia but redundant for normal hematopoiesis. The redundancy can be that other chromatin modifying enzymes may regulate expression of MLL1 target genes or that MLL1 target genes (e.g. Hoxa9) are dispensable for normal hematopoietic processes. The detailed mechanism warrants future studies.
Perspective
In summary, the highly selective MLL1 inhibitor we reported here will have broad applications in basic research and lay the foundation for future development of effective therapeutics in clinical arenas. We envision several lines of research may arise in future: First, with development of chemical probe MM-401, the requisite function of H3K4 methylation in various biological contexts can be examined. Compare to genetic knockout models, on-target inhibition by pharmacological compounds such as MM-401 introduces small but specific perturbation to the MLL1 complex, allowing functional attribution to its methyltransferase activity. Second, given that MLL1 and H3K4me regulate several key targets (e.g. Hoxa9, Myc and Bcl2) in MLL leukemia (Figure 7C), it will be important to compare MLL1 and MLL fusion protein dependent gene pathways to see if they largely overlap with each other or they belong to distinct pathways that are important for progression of MLL leukemia. Third, it will be important to test whether MLL1 inhibitor has therapeutic potential for a broad spectrum of diseases beyond MLL leukemia. Specifically, inhibiting MLL1 activity may be useful for treatment of AMLs and myelodysplastic syndrome (MDS) that have wild type MLL1 alleles and Hoxa9 overexpression (Ayton and Cleary, 2001; Dou and Hess, 2008). It may also be applicable for AMLs with MLL1 amplification and tandem duplication. Testing MM-401 or its derivatives on broad spectrum of human diseases will provide greater insights in future.
Experimental Procedures
Protein expression and affinity purifications
Recombinant fragments of MLL family members, MLL13762-C’, MLL25062-C’, MLL34707-C’ and MLL42508-C’, as well as WDR523-C’, RbBP5 and Ash2L were expressed from the pET28A-SUMO as SUMO fusions. Protein purification details see Supplemental information.
Histone Methytransferase Assays
The HMT assay was performed as described previously (Dou et al., 2005). For inhibitor studies, compounds at various concentrations were incubated first with the pre-assembled complex and reactions were initiated by addition of substrates. For kinetic analyses, the reaction progression curve was established to determine the linear range of the reaction at room temperature. For Lineweaver-Burk curve, reactions (0.5µM enzyme complex and 50µM substrates) were initiated and quenched after 4 minutes by addition of β-mercaptoethanol (Fisher) at a final concentration of 178µM.
Crystal Structures
WDR5/MM-401 or WDR5/MM-NC-401 binary complex was obtained by mixing WDR5 and compounds at molar ratio 1: 2. The complex was crystallized by hanging-drop-vapor-diffusion at 22°C. Details see supplemental information.
Cell Culture Conditions
MLL-AF9-transduced mouse bone marrow was cultured in IMDM + 15% FBS and supplemented with 0.01ng/ml IL-3 at each passage, every other day. Human leukemia cell lines MV4:11, K562, MOLM-13, U937 and KOPN8 were maintained in RPMI1460 (GIBCO) supplemented with 10% FBS. Cells were passaged to 1-×105/ml every 2 days.
Assays for Cell Viability, Wright-Giemsa staining, apoptosis, cell cycle and cell differentiation
Inhibitors were diluted from stock to culture media containing 0.1% DMSO final concentration. For viability assays, cells were cultured at 1×105/ml and passaged every 2 days. Viability was determined using the CellTitreGlo® Kit (Promega) according to the manufacturer’s directions. Luminescence was monitored on a Molecular Dynamics plate reader. For staining, cells treated with 10, 20 and 40µM MM-401, or 40µM MM-NC-401 or DMSO vehicle for 4 days were diluted to 2.5×105/ml in 1× PBS and fixed to glass slides by cytospin followed by Wright-Giemsa staining (Fisher Scientific Inc). Cell images were taken at 40× magnification by light microscopy (Olympus BX41). Apoptosis, cell cycle and cell differentiation analyses were performed using standard protocols (see supplemental information).
Real Time-PCR, RNA-seq and CHIP assays
MLL1-AF9 cells were cultured for 2 days in the presence of MM-401 or MM-NC-401. At the end of treatment, cells were harvested by centrifugation at 300×g and washed with 1xPBS. RNAs for duplicated biological samples were extracted by a standard protocol. 10ng RNAs were used for Illumina sequencing library. Four RNA seq samples were multiplexed and loaded into one lane in Hi-seq sequencer. RNA-seq analyses were described in supplemental information. CHIP assays were performed as previously described (Dou et al., 2005).
Inhibition of Human Leukemia blasts
Patient leukemia Blasts were purified by negative selection using Miltenyi Biotec (Auburn, CA) LS columns (cat# 130-042-401), and a combination of the following four Miltenyi Biotec antihuman microbeads: CD3 (130-050-101), CD19 (130-050-301), CD14 (130-050-201), CD235a (130-050-501). Available pathology data were first studied; bead types were omitted as necessary if blasts were reported positive for CD3, CD14, or CD19. Wright-Giemsa staining and CellTiter–Glo (Promega, G7558) were performed as described above.
Supplementary Material
Highlights.
MM-401 inhibits MLL1 H3K4 methylation without affecting other MLL family members
MM-401 inhibits MLL leukemia cells but not normal BM or non-MLL leukemia cells
RNA-seq analyses show correlative changes upon MM-401 treatment and MLL1 deletion
Targeting of MLL1 activity has therapeutic potential for MLL leukemia
ACKNOWLEDGEMENTS
We thank Drs. Hugh Brady for Mllf/f mice, Andrew Muntean for MLL1 fusion gene vectors and Dr. Brian Shay at Biomedical Mass Spectrometry Facility at University of Michigan for MALDI-mass spec analyses. This work is supported by funding from Stand Up to Cancer (SU2C), Leukemia and Lymphoma Society (LLS), Novartis Research Institute and National Institute of Health (NIH) to Y.D., from LLS to S.W., from Howard Hughes Medical Institute (HHMI) to M. L., and from NIH to S.M., Q.S and J.L.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number
The structures was deposited in the Protein Data Bank with PDB number 4GM9 for crystal structure of human WD repeat domain 5 with compound MM-401 and PDB number 4GMB for crystal structure of human WD repeat domain 5 with compound MM-NC-401. The RNA-seq data was deposited in the SRA database with the accession number SRP033036.
REFERENCE
- Artinger EL, Mishra BP, Zaffuto KM, Li BE, Chung EK, Moore AW, Chen Y, Cheng C, Ernst P. An MLL-dependent network sustains hematopoiesis. Proc Natl Acad Sci U S A. 2013a;110:12000–12005. doi: 10.1073/pnas.1301278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artinger EL, Mishra BP, Zaffuto KM, Li BE, Chung EK, Moore AW, Chen Y, Cheng C, Ernst P. An MLL-dependent network sustains hematopoiesis. Proc Natl Acad Sci U S A. 2013b doi: 10.1073/pnas.1301278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- Bernt KM, Armstrong SA. Targeting epigenetic programs in MLL-rearranged leukemias. Hematology Am Soc Hematol Educ Program. 2011;2011:354–360. doi: 10.1182/asheducation-2011.1.354. [DOI] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Chen Y, Cierpicki T, Liu Y, Basrur V, Lei M, Dou Y. An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain. PLoS One. 2010;5:e14102. doi: 10.1371/journal.pone.0014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove MS, Patel A. Mixed lineage leukemia: a structure-function perspective of the MLL1 protein. Febs J. 2010;277:1832–1842. doi: 10.1111/j.1742-4658.2010.07609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011a;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao YH, et al. Selective Killing of Mixed Lineage Leukemia Cells by a Potent Small-Molecule DOT1L Inhibitor. Cancer Cell. 2011b;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha R, Dittman A, Giotopoulos G, Bantscheff M, Chan WI, Robson S, Chung CW, Hopf C, Savitski M, et al. Inhibition of BET Recruitment to Chromatin As An Effective Treatment for MLL-Fusion Leukaemia. Blood. 2011a;118:27–28. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011b;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Hess JL. Mechanisms of transcriptional regulation by MLL and its disruption in acute leukemia. Int J Hematol. 2008;87:10–18. doi: 10.1007/s12185-007-0009-8. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, Showalter HD, Murai MJ, Belcher AM, Hartley T, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8:277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JL, Yu BD, Li B, Hanson R, Korsmeyer SJ. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1997;90:1799–1806. [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117:4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatas H, Townsend EC, Cao F, Chen Y, Bernard D, Liu L, Lei M, Dou Y, Wang S. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J Am Chem Soc. 2013;135:669–682. doi: 10.1021/ja306028q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis A, Look AT. Distinct and dynamic requirements for mTOR signaling in hematopoiesis and leukemogenesis. Cell Stem Cell. 2012;11:281–282. doi: 10.1016/j.stem.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux M, Faber J, Xia X, Kung AL, Armstrong SA. Global increase in H3K79 dimethylation in murine and human MLL-AF4 lymphoblastic leukemias. Blood. 2007;110:108A–108A. [Google Scholar]
- Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu L, Corsa CA, Kunkel S, Dou Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell. 2009;36:290–301. doi: 10.1016/j.molcel.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Takeda S, Cheng EH, Hsieh JJ. Biphasic MLL takes helm at cell cycle control: implications in human mixed lineage leukemia. Cell Cycle. 2008;7:428–435. doi: 10.4161/cc.7.4.5426. [DOI] [PubMed] [Google Scholar]
- McMahon KA, Hiew SYL, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams O, Brady HJM. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1:338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, Whitcomb SJ, Wang Z, Ruthenburg AJ, Elenitoba-Johnson KS, Roeder RG, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38:853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe SC, Jo SY, Sanders DS, Basrur V, Elenitoba-Johnson KS, Slany RK, Hess JL. MLL-AF9 and MLL-ENL alter the dynamic association of transcriptional regulators with genes critical for leukemia. Exp Hematol. 2011;39:77–86. e71–e75. doi: 10.1016/j.exphem.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, Basrur V, Elenitoba-Johnson KS, Hess JL. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM, et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294:2563–2566. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- Ohyashiki K, Kocova M, Ryan DH, Rowe JM, Sandberg AA. Secondary acute myeloblastic leukemia with a Ph translocation in a treated Wegener's granulomatosis. Cancer Genet Cytogenet. 1986;19:331–333. doi: 10.1016/0165-4608(86)90062-2. [DOI] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Patel A, Dharmarajan V, Cosgrove MS. Structure of WDR5 bound to mixed lineage Leukemia protein-1 peptide. J Biol Chem. 2008 doi: 10.1074/jbc.C800164200. [DOI] [PubMed] [Google Scholar]
- Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senisterra G, Wu H, Allali-Hassani A, Wasney GA, Barsyte-Lovejoy D, Dombrovski L, Dong A, Nguyen KT, Smil D, Bolshan Y, et al. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem J. 2013;449:151–159. doi: 10.1042/BJ20121280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Kingston RE. WDR5 interacts with mixed-lineage Leukemia (MLL) protein via the histone H3 binding pocket. J Biol Chem. 2008 doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci U S A. 2006;103:6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky GA, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel RC, Shilatifard A. WDR5, a complexed protein. Nat Struct Mol Biol. 2009;16:678–680. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- Wu L, Lee SY, Zhou B, Nguyen UT, Muir TW, Tan S, Dou Y. ASH2L regulates ubiquitylation signaling to MLL: trans-regulation of H3 K4 methylation in higher eukaryotes. Mol Cell. 2013;49:1108–1120. doi: 10.1016/j.molcel.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.