Abstract
PDZ domains represent one group of the major structural units that mediate protein interactions in intercellular contact, signal transduction and assembly of biological machineries. TIP-1 protein is composed of a single PDZ domain that distinguishes TIP-1 from other PDZ domain proteins that more often contain multiple protein domains and function as scaffolds for protein complex assembly. However, the biological functions of TIP-1, especially in cell transformation and tumor progression, are still controversial as observed in a variety of cell types. In this study, we have identified ARHGEF7, a guanine nucleotide exchange factor (GEF) for Rho GTPases, as one novel TIP-1 interacting protein in human glioblastoma cells. We found that the presence of TIP-1 protein is essential to the intracellular redistribution of ARHGEF7 and rhotekin, one Rho effector, and the spatiotemporally coordinated activation of Rho GTPases (RhoA, Cdc42 and Rac1) in migrating glioblastoma cells. TIP-1 knockdown resulted in both aberrant localization of ARHGEF7 and rhotekin, as well as abnormal activation of Rho GTPases that was accompanied with impaired motility of glioblastoma cells. Furthermore, TIP-1 knockdown suppressed tumor cell dispersal in orthotopic glioblastoma murine models. We also observed high levels of TIP-1 expression in human glioblastoma specimens, and the elevated TIP-1 levels are associated with advanced staging and poor prognosis in glioma patients. Although more studies are needed to further dissect the mechanism(s) by which TIP-1 modulates the intracellular redistribution and activation of Rho GTPases, this study suggests that TIP-1 holds potential as both a prognostic biomarker and a therapeutic target of malignant gliomas.
Keywords: Tax-interacting protein-1, ARHGEF7, rhotekin, Rho GTPases, glioblastoma, cell migration
Introduction
PDZ (PSD-95/DlgA/ZO-1) domain is one of the major structural units that mediate protein interactions in intercellular contact, signal transduction and complex assembly of biological machineries (1). Composed of around 80-90 amino acids, this structurally conserved protein domain specifically recognizes a short region of C-terminal or internal sequence of its target protein. More than 180 proteins have been identified with PDZ domain in human proteome, which more often are composed of more than one PDZ or other protein domains and these combinations allow them to carry out their specific functions, such as protein complex assembly for tumor suppression (2, 3). Tax-interacting protein 1 (TIP-1, also known as Tax1bp3 or glutaminase-interacting protein, GIP) is unique in its simple structure, a single PDZ domain is the only functional and structural unit identified so far in this small PDZ protein (124 amino acids in human and mouse). This unique structure suggests that TIP-1 might function differently from other PDZ domain proteins. Previous studies indicated that TIP-1 is highly conservative in amino acids sequence across species (4), its expression has been documented in a variety of cell types. For example, high level of TIP-1 expression is limited to the central nervous system in the late stage of embryo development of zebrafish (4). In mammals, TIP-1 mRNA was detected in most regions of human brain but was abundant in spinal cord, particularly in astrocytes and neurons. In those cells, TIP-1 has been located in cytosol, mitochondria, nuclear envelope and plasma membrane (5). High level of TIP-1 expression was detected in human invasive breast cancer cells and that is important for tumor cell adhesion, migration and pulmonary metastasis (6). However, the precise biological effects of TIP-1 and its functional mechanisms have largely remained elusive.
Studies on the small PDZ protein TIP-1 have been largely conducted through identification of its interacting proteins (7-11). Among around a dozen of TIP-1 interacting proteins that have been identified so far, it has been reported that TIP-1, together with ARHGEF16, mediates the HPV16 E6 oncoprotein-induced Cdc42 activation and cell migration (8, 12). The interaction between TIP-1 and the potassium channel Kir2.3 blocks recruiting Kir2.3 to the basolateral membrane-associated Lin-7 protein and affects the polarity of the mammalian epithelial cell line MDCK (7). Other than these functions facilitating tumor formation or progression, TIP-1 was also reported with inhibitory activities towards the beta-catenin regulated gene expression and proliferation of colon cancer cells (9).
The Rho GTPases are a family of small (~21 kDa) signaling proteins that act as molecular switches, cycling between a GTP-bound active form and a GDP-bound inactive form. Once activated by guanine nucleotide exchange factors (GEFs), Rho GTPases bind different effectors and regulate various cellular processes, including cytoskeleton reorganization, cell adhesion, cell motility, vesicle trafficking, and phagocytosis (13). Among all Rho GTPases, Rac1, Cdc42 and RhoA are the most studied in cytoskeleton reorganization and cell migration. The functionality of Rho GTPases is finely coordinated at multiple levels including activation by GEFs, subcellular localization and effector association. Alterations in Rho GTPases and GEFs -- Rac1, trio, Ect2, and Vav3 -- have been reported in multiple human malignancies including gliomas (14, 15).
Malignant gliomas are the most prevalent primary brain tumors and are essentially universally fatal (16). Despite maximal treatment, patients with grade IV malignant gliomas (glioblastomas) survive only 12-15 months (17). Glioblastomas are highly invasive, and irradiation and surgical removal of these tumors are challenging due to the diffuse infiltration of individual tumor cells into adjacent brain tissue. In addition, invading tumor cells may be more resistant to conventional therapies, suggesting that studying the mechanisms of tumor invasion might lead to discoveries of novel therapeutic targets or prognostic biomarkers.
In the present study, we have identified guanine nucleotide exchange factor 7 (ARHGEF7, also known as beta-PIX), one Rho GEF, as one novel TIP-1 interacting protein in human glioblastoma cells. It was demonstrated that TIP-1 coordinates the intracellular redistribution of ARHGEF7 and rhotekin, one Rho effector for the spatiotemporally regulated activation of Rho GTPases (RhoA, Cdc42, and Rac1) in migrating glioblastoma cells. TIP-1 knockdown reduced the motility of human glioblastoma cells in vitro and significantly impaired the infiltrative growth of intracranial human glioblastoma xenografts in mouse models. Correlation of high TIP-1 expression levels in human malignant gliomas with poor prognosis of the patients further suggests that TIP-1 could be a putative prognostic biomarker and therapeutic target of human glioblastoma.
Results
TIP-1 interacts with ARHGEF7 and rhotekin
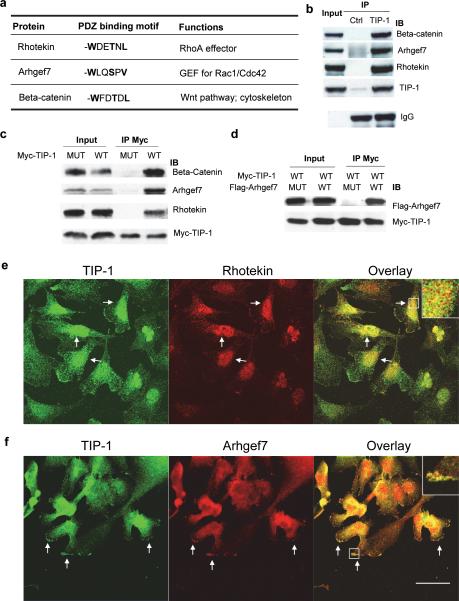
TIP-1 is composed of a single type I classic PDZ domain which selectively recognize a C-terminal S/T-X-V/L-COOH (where X represents any amino acid) motif of its interacting partners (7, 9, 10, 18, 19). In addition to the highly conserved signature motif, recent structural studies of the protein complex formed with TIP-1 and its interacting partners showed that the high affinity and selectivity of TIP-1 also requires a tryptophan residue at the −5 position to the C-terminus of the interacting proteins (20, 21). Based on this information, we searched a PDZ binding protein database (1) and discovered three proteins that contain this unique sequence signature (Figure 1a). In addition to beta-catenin (9) and rhotekin (10), which have been reported with selective binding to the TIP-1 PDZ domain, ARHGEF7 was identified as a novel TIP-1 interacting protein. The interactions between these proteins were validated by immunoprecipitation and co-immunostaining with human glioblastoma cells. In the immunoprecipitation assays, protein-protein interactions were detected with both of the endogenous (Figure 1b) and the ectopically expressed proteins (Figures 1c, d). It was also revealed that all of the three proteins were associated only with the wild type TIP-1 protein, but not with a TIP-1 mutant containing a dysfunctional PDZ domain (7) (Figure 1c). Mutations within the PDZ binding motif of ARHGEF7 from −WLQSPV to –ALQAPV (mutations are underlined) abolished its interaction with TIP-1 (Figure 1d). Immunofluorescent staining of human glioblastoma T98G cells indicated that rhotekin and TIP-1 are co-localized mainly in the cell body and the trailing edge (Figure 1e), whereas a significant amount of ARHGEF7 and TIP-1 are co-localized at the leading edge of the migrating T98G cells (Figure 1f).
Figure 1.
TIP-1 interacts with ARHGEF7 and rhotekin. (a) PDZ binding motif within the TIP-1-interacting proteins. The critical residues for TIP-1 binding are highlighted in bold. (b) Interactions of the endogenous proteins. TIP-1-specific or a control antibody was used for immunoprecipitation of proteins from T98G cell lysates. (c) Validation of the protein interactions with T98G cells transfected with either Myc-tagged TIP-1 wild type (WT) or a mutant (MUT) with a dysfunctional PDZ domain. Myc antibody was used in the immunoprecipitation. Beta-catenin, Rhotekin and ARHGEF7 were blotted with specific antibody, respectively. (d) Immunoprecipitation of Myc-TIP-1 in cells co-transfected with Myc-TIP-1 (wild type, WT) and FLAG-tagged ARHGEF7 (wild type, WT) or a mutant (MUT) with mutations in the C-terminal PDZ binding motif. (e) Immunofluorescent staining of T98G cells with TIP-1 antibody (green) and Rhotekin antibody (red). (f) Immunofluorescent staining of T98G cells with TIP-1 antibody (green) and ARHGEF7 antibody (red). Arrows indicate the colocalized proteins. Colocalized TIP-1 with Rhotekin or ARHGEF7 in the cell body or leading edge of migrating cells was illustrated as the inserts, respectively. Scale bars: 40 μm.
TIP-1 regulates the intracellular redistribution of ARHGEF7 and rhotekin in migrating glioblastoma cells
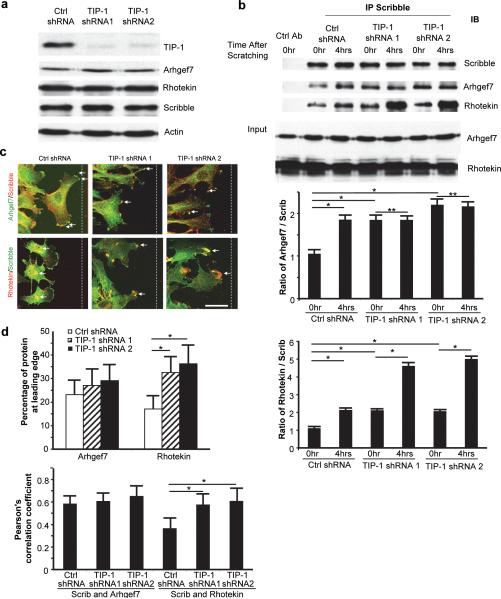
To study the biological relevance of these TIP-1 mediated protein interactions, stable T98G cell lines with TIP-1 knockdown were developed by using two independent TIP-1 targeting shRNA sequences. Western blot analyses of total cell lysates showed that TIP-1 knockdown did not significantly change the overall protein levels of ARHGEF7 or rhotekin in T98G cells (Figure 2a). However, TIP-1 knockdown significantly affected the intracellular redistribution of these two proteins in migrating T98G cells, as indicated with the colocalization studies of these two proteins with Scribble. Scribble is one of the major proteins that regulate the intracellular distributions of ARHGEF7 and rhotekin in the establishment of cell polarity (22, 23) and the directional cell migration (24). Coinciding with the published discoveries (24), co-immunoprecipitation studies indicated that dynamic interactions with Scribble exist for both of ARHGEF7 and rhotekin in the migrating T98G cells, the protein interactions was elevated by around 2-fold at 4 hours post the scratch wounding (Figure 2b). Interestingly, TIP-1 knockdown increased the basal levels of both protein interactions in the stationary T98G cells. However, ARHGEF7 lose the dynamics of the Scribble interaction with the absence of TIP-1 protein, the protein interaction between Scribble and ARHGEF7 did not change after scratch wounding with TIP-1 knockdown (Figure 2b). The interaction between rhotekin and Scribble kept a similar dynamics pattern in the migrating T98G cells after TIP-1 knockdown even the basal levels of protein interaction were increased.
Figure 2.
TIP-1 regulates intracellular redistribution of ARHGEF7 and rhotekin in migrating glioblastoma cells. (a) Western blot analysis of ARHGEF7, rhotekin and Scribble protein expression in total cell lysates of T98G cells with or without TIP-1 knockdown. (b) Interactions of Scribble with ARHGEF7 or rhotekin in T98G cells with or without TIP-1 knockdown. Protein interactions assessed with Co-IP. Cells were lysed at the indicated time points after scratch wounding. Proteins immunoprecipitated with anti-Scribble (Scrib) or irrelevant (Ctrl Ab) antibodies were analyzed by SDS-PAGE and blotted with antibodies against ARHGEF7, rhotekin or Scribble, respectively. Shown are representative blots of three independent experiments. The bar graphs represent semi-quantitative measurements from three independent experiments. (* p<0.001; ** p>0.05. (c) Intracellular distribution of ARHGEF7 and rhotekin in the T98G cells with or without TIP-1 knockdown. Four hours after the scratch wounding (toward the dashed lines), cells were fixed and co-stained with indicated antibodies. The colocalized proteins were pointed with arrows. Scale bar: 40 μm. (d) Quantitative measurement of the proteins in migrating T98G with ImageJ. Proteins were quantified upon the total fluorescence intensity at the leading edge and in the cell body. At least 50 cells were included in each measurement. Shown are percentages of each protein located at the leading edge (upper panel) and the Pearson correlation coefficients (lower panel). * p<0.05, ANOVA.
The altered intracellular redistribution of ARHGEF7 and rhotekin caused by TIP-1 knockdown was further validated with immunofluorescent staining of migrating T98G cells. In addition to confirming the observations in co-immunoprecipitation studies, the results also revealed that TIP-1 knockdown leads to a remarkable change in cellular morphology of T98G cells (Figure 2c). Compared to the even distribution alongside the lamellipodia in control cells, ARHGEF7 was only located at some isolated tips of the highly branched pseudopodia in the TIP-1 knockdown cells (Figure 2c). Rhotekin was primarily located in the cell body and trailing edge in the control cells, but was associated with the leading edge after TIP-1 knockdown (Figure 2c). Fluorescence intensity for each protein at the leading edge of migrating T98G cells was semi-quantitatively measured to determine the Pearson correlation coefficients which have been utilized to measure the colocalization of two proteins (25). It was found that TIP-1 knockdown increased (by ~2-fold) the colocalization of rhotekin with the Scribble at the leading edge of migrating T98G cells, but had a moderate impact on ARHGEF7 (Figure 2d).
TIP-1 regulates the spatiotemporal activation of Rho GTPases in migrating glioblastoma cells
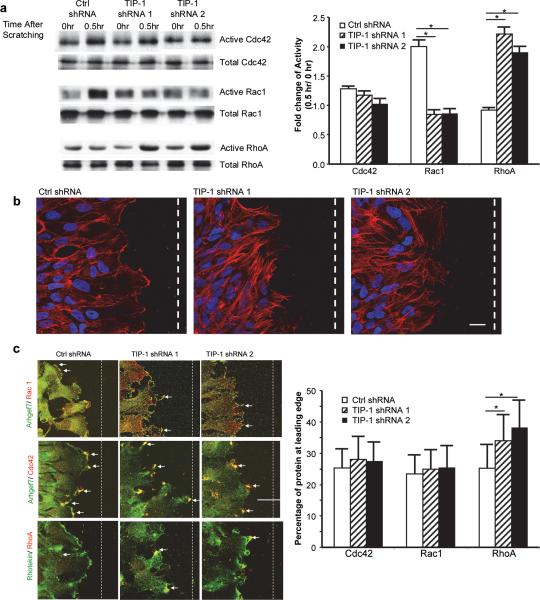
Activation of Rho GTPases, such as RhoA, Rac1 and Cdc42Rho is spatially and temporally orchestrated by selective association with effectors and GEFs in cytoskeleton reorganization, cell morphogenesis, and directional cell migration (13, 26, 27). We investigated whether the TIP-1-modulated redistribution of rhotekin (a RhoA effector) and ARHGEF7 (a GEF for Rac1/Cdc42) leads to abnormal activation of Rho GTPases within the migrating glioblastoma cells. In the control T98G cells expressing TIP-1 protein, scratch wounding on the cell monolayer induced a robust activation of Rac1 (~2-fold increase), and to a lesser extent activation of Cdc42. RhoA activity did not show a significant change after the scratch wounding at the time points surveyed in this study (Figure 3a). However, in the T98G cells with TIP-1 knockdown, Rac1 activity was slightly decreased, compared to a robust increase (>2-fold) of RhoA activity after the scratch wounding (Figure 3a).The increased RhoA activity also resulted in the formation of strong stress fibers in TIP-1 knockdown cells (Figure 3b). Immunofluorescent staining of the cells indicated that Rac1 and Cdc42 were predominantly colocalized with ARHGEF7, whereas RhoA was primarily colocalized with rhotekin. As a consequence of the aberrant intracellular redistribution of ARHGEF7 and rhotekin by TIP-1 knockdown (Figure 2), Rac1 and Cdc42 were located to some isolated tips of the branched pseudopodia whereas the cells expressing TIP-1 protein had an even distribution of the Rac1 and Cdc42 along the lamellipodia (Figure 3c). A great amount of RhoA and rhotekin were found at the leading edge in the TIP-1 knockdown cells, instead of the cell body where these proteins were primarily localized in the control cells.
Figure 3.
TIP-1 regulates the spatiotemporal activation of Rho GTPases in migrating glioblastoma cells. (a) Quantitative measurements of Rho GTPases. The active and total Rac1/Cdc42/RhoA in T98G cells were analyzed as described in Materials and methods. Results represent 3 independent experiments. The bar graph shows the densitometric quantifications of the blot results. (* p<0.002). (b) Staining of actin (red) with phalloidin to visualize stress filament formation along the leading edge of migrating T98G cells. (Scale bar: 20 μm). (c) Intracellular localization of Rho GTPases determined with immunofluorescent staining of migrating T98G cells. Four hours after cell scratching, cells were fixed and stained with antibodies as indicated. Arrows indicate the colocalized proteins. Scale bar= 40 μm. The fluorescence intensity of each protein was measured with ImageJ, and percentages of individual proteins located at the leading edge were calculated. (*p<0.001).
TIP-1 knockdown reduces glioblastoma cell migration and invasion in vitro
To study the biological relevance of the TIP-1 knockdown-caused aberrant activation of Rho GTPases, migration and invasion capabilities of glioblastoma cells, with or without TIP-1 knockdown (Figure 4a), were studied in vitro by using a Boyden chamber-based system. In those assays, all the three human glioblastoma cell lines (T98G, D54 and T4302) demonstrated a significant reduction in cell migration after TIP-1 knockdown (Figure 4b). Similarly but to a greater extent, cell invasion through matrigel was inhibited in all the three cell lines with TIP-1 knockdown (Figure 4c). Combining with studies with other human glioblastoma cell lines (supplementary Figure s1), these results indicated that TIP-1 expression is generally required for the motility of human glioblastoma cells in vitro.
Figure 4.
TIP-1 knockdown inhibits migration and invasion of human glioblastoma cells in vitro. (a) Western blot showing knockdown of TIP-1 expression in three human glioma cell lines. (b) Migration and (c) invasion of the glioma cells with or without TIP-1 knockdown were studied using Boyden chamber-based assays as described in Materials and Methods. Bar graphs show the number of cells that have migrated or invaded across the membrane. Results shown are the mean ± SD of three independent experiments (* p<0.01).
TIP-1 knockdown in glioblastoma cells reduces infiltrative tumor cell dispersal in murine xenograft models
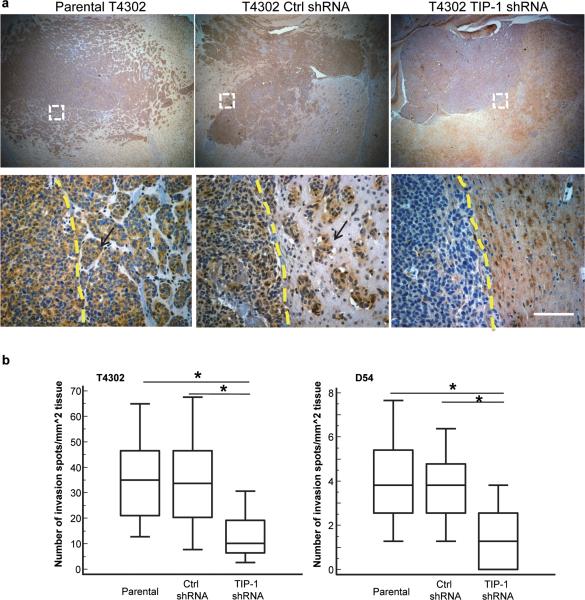
The biological significance of TIP-1 expression on the motility of human glioblastoma cells was further studied with murine orthotopic xenograft models. Two human glioblastoma cell lines (T4302 and D54), with or without TIP-1 knockdown, were implanted intracranially in athymic nude mice. Brain tissues were recovered three weeks thereafter for histological examinations. In contrast to the extensive infiltration of tumor cells into brain tissues in the control groups (non-transduced or transduced with control shRNA cells), TIP-1 knockdown tumors had a reduced invasion and infiltration into the surrounding brain tissue. Much less tumor cells (1/3 of the controls) were spotted beyond the tumor border in TIP-1 knockdown glioblastomas (Figures 5 & s2).
Figure 5.
TIP-1 knockdown inhibits the infiltrative growth of human glioblastoma cells in mice. (a) Histological examination of human glioblastoma xenografts formed in athymic nude mice. 104 of parental or transduced T4302 glioma cells with either non-targeting or TIP-1-specific shRNAs were implanted intracranially. Tumors were resected three weeks thereafter and stained with hematoxylin to visualize the invasive growth (indicated with arrows) of the tumor. TIP-1 was stained with a specific antibody (brown) to verify the efficiency of TIP-1 knockdown. Main tumor mass was highlighted with dashed line. Scale bar: 100 μm. (b) Quantification of the tumor invasion spots. An invasion spot was defined as a site of multiple (≥3) tumor cells that were not directly connected to the main tumor mass. Invasion spots beyond borders were counted at four locations on the border and on five sections of each tumor (3 tumors in each group). Data distribution was presented as box plot. * p<0.001, the Student T-test.
TIP-1 knockdown had minor impact on the beta-catenin controlled gene expression and cell proliferation in human glioblastoma cells
It was reported that TIP-1 interaction inhibits the transcriptional activity of beta-catenin and cell proliferation in colon cancer cells (9). However, microarray and western blot analyses indicated that TIP-1 had a minor or moderate impact on the expression levels of a panel of beta-catenin-regulated genes such as cyclin D1 and c-Myc in D54 cells (Figures 6a, b). This observation correlated to the results of in vitro and in vivo cell proliferation assays conducted with D54 and T4302 glioblastoma cell lines. In vitro cell doubling time assays showed that there was no significant difference in the cell proliferation rate between the control cells and the cells with TIP-1 knockdown (Figure 6c). Cell proliferation in the tumor sections recovered from the animal models was detected with immunostaining of Ki67 antigen. Quantitative measurements indicated that TIP-1 did not change the proliferation rate of glioblastoma cells in vivo (Figure 6d). These data suggested that the reduced tumor cell dispersal was not resulted from the disadvantage in cell proliferation.
Figure 6.
TIP-1 knockdown has minor impact on the beta-catenin-controlled gene transcription and cell proliferation in glioblastoma cells. (a) Microarray profiling of gene expression in D54 glioblastoma cells with or without TIP-1 knockdown. Expression levels of a panel of beta-catenin-regulated genes are presented in a heat map. TIP-1 (Tax1bp3) was included as a quality control. (b) Western blot and semi-quantification of c-MYC and Cyclin D1 protein levels in D54 glioblastoma cells with or without TIP-1 knockdown. Data were respectively normalized to the levels of actin. (c) In vitro cell proliferation rates were measured by microscopic cell counting every 24 hours. (d) Quantitative measurement of cell proliferation rate in vivo. Intracranial glioma xenografts were resected 3 weeks after the tumor implantation. Quantification of Ki67 positive cells was based on at least 7 microscopic fields on more than 5 sections for each group. Data distribution was shown as box plot. p>0.05, ANOVA.
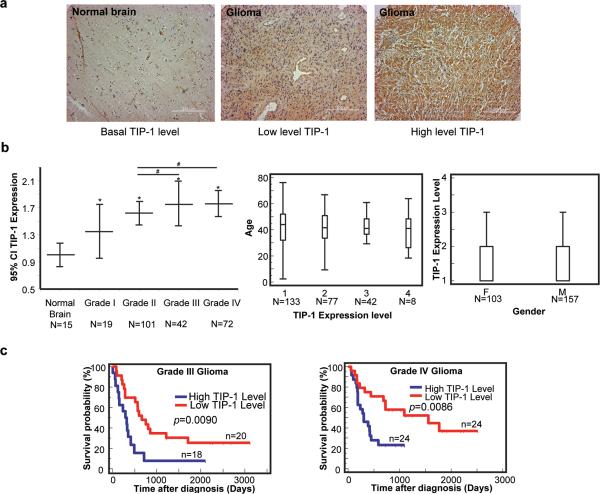
High levels of TIP-1 expression correlate with advanced stage and poor prognosis of human malignant gliomas
To determine TIP-1 expression in human malignant gliomas, a TIP-1 specific antibody (28) was used for immunohistochemical (IHC) staining of human specimens. Within the 234 cases of pathologically verified clinical specimens (supplementary table s1), high TIP-1 protein levels were only detected in high grade gliomas whereas TIP-1 protein was barely detectable in 15 cases of normal brain (Figure 7a and Supplementary table s1). IHC staining indicated that TIP-1 is predominantly located in the cytosol with some plasma membrane localization (Supplementary Figure s3). Semi-quantitative measurements revealed a correlation between TIP-1 protein levels and the stage of malignant gliomas (Figure 7b). However, TIP-1 levels were independent of patient's age or gender. A retrospective study of gene expression profiles (29, 30) from 86 patients with grade III or grade IV glioblastomas whose tumors had been surgically resected revealed that TIP-1 expression is a negative prognostic indicator after the surgical removal of the tumor. For both of grade III and IV glioma patients, those with tumors of high level of TIP-1 expression experienced a shorter survival time after surgical removal of the primary tumors (Figure 7c). The correlation between TIP-1 expression and patient survival time was further supported by analyzing a large group of glioma patients in which overexpression of TIP-1 (more than 2-fold up-regulation) was detected in over 50% of glioma patients (Supplementary Figure s4).
Figure 7.
High levels of TIP-1 expression correlate with advanced staging and poor prognosis of human malignant gliomas. (a) TIP-1 expression in human gliomas. Representative tissue core images from a tissue microarray stained with a TIP-1 antibody (brown) demonstrate differential TIP-1 protein levels in normal brain and gliomas. Scale bar: 200 μm. (b) Semi-quantitative histological evaluation of TIP-1 expression within human glioma tissues. TIP-1 level was scored from 1 (lowest) to 4 (highest), mean expression scores multiplied by the percentage of positive cells in the field (Quick's combined score system) are presented for normal brain, grade I, grade II, grade III and grade IV gliomas, respectively, in a graphical format using error bars with 95% confidence intervals (CI). Statistically significant differences of TIP-1 expression (ANOVA) were noted between normal brain tissue and gliomas of varying stages of malignancy (*, p<0.001) and between the advanced (III or IV) and low grade tumors (Student T-test, #, p<0.005). Number of samples in each group was shown under the bar graph. Box plots show no correlation of TIP-1 expression levels with patient's age or gender (F, female; M, male). (c) Kaplan-Meier plot of patient survival time upon relative TIP-1 expression levels. Patients within the top 40% of TIP-1 expression were defined as the high expression group, and those in the bottom 40% were defined as the low expression group. A log rank test was used to evaluate differences between groups.
Discussion
Several proteins have been identified as TIP-1 interacting partners in mammalian cells. Through these protein interactions, TIP-1 demonstrates a variety of biological functions, such as mediating the cellular response to serum starvation (10), inhibiting the beta-catenin-regulated gene transcription and cell proliferation (9), establishing polarity of epithelial cell (7), and protecting tumor cells from ionizing radiation-induced cell death (31). In this study, we have identified ARHGEF7 as a novel TIP-1 interacting protein, and demonstrated that this protein interaction is biologically relevant to the infiltrative growth of glioblastoma cells. TIP-1 knockdown resulted in aberrant localization of ARHGEF7 and rhotekin, as well as abnormal activation of Rho GTPases including Rac1, Cdc42, and RhoA, that was accompanied with a reduced motility of glioblastoma cells in vitro and impaired infiltrative tumor growth in vivo.
Roles of TIP-1 in the Rho GTPases activation and cell motility had been reported previously in cervical cancer cells (8, 12, 32). In those reports, data indicated that TIP-1, probably through cooperating with Arhgef16 or other mechanisms, facilitates the HPV-16 E6 oncoprotein-induced activation of RhoA, Cdc 42 and ROCK for cell migration. Knockdown of TIP-1 or inhibition of Rho kinase resulted in a reduced cell motility in the HPV16 E6-expresing cervical cancer cells (8). Since high levels of TIP-1 expression were also documented in invasive human breast tumors, and TIP-1 knockdown reduced cell migration in vitro and pulmonary metastasis in vivo (6). All these studies suggested that TIP-1 might play a general role in the Rho GTPases activation and cell migration in a variety of tumor cells.
Spatiotemporally regulated activation of Rho GTPases is finely orchestrated in cell migration, which starts with protrusion at the leading edge and ends with rear retraction of the distal cell body (27). In the past few years, accumulating evidences indicate that the dynamic interactions with scaffold proteins are essential for Rho GEFs and effectors to direct the Rho GTPase signaling outcomes in polarized migrating cells (13, 33). One polarized scaffold protein, Scribble, was reported to interact with ARHGEF7 through its PDZ domain and to translocate the activated Cdc42 and Rac1 to the migrating cell leading edge (22, 23, 34). However, little is known about the roles of rhotekin in these processes and how cells coordinately activate RhoA, Rac1 and Cdc42 during cell migration and invasion.
Previous reports (8, 12) suggested that TIP-1, probably through the PDZ domain-mediated protein interactions, modulates a delicate balance of interactions between Scribble, HPV16 E6 oncoprotein, Arhgef16, Cdc42, or other proteins that are involved in the spatiotemporal activation of Rho GTPases. Data from this study support that hypothesis. In this study, both rhotekin and ARHGEF7 interact with Scribble through the PDZ domain, the interaction dynamics and the intracellular distributions of those proteins are modulated by TIP-1 in migrating glioblastoma cells. It was noted that TIP-1 knockdown enhanced the protein interaction between ARHGEF7 and Scribble, but caused a loss of the dynamic regulation in the protein interaction after scratch wounding. TIP-1 knockdown also remarkably changed the cellular morphology and the distribution pattern of ARHGEF7 at the leading edge. On the other hand, even the basal level of Scribble and rhotekin interaction was elevated with the TIP-1 knockdown, the protein interaction was still dynamically regulated in the migrating glioma cells. The data suggested that a competition might exist between Scribble and TIP-1 in the selective binding to ARHGEF7 or rhotekin. This question was further answered by experiments with the use of T98G cells with an ectopic expression of TIP-1. The results indicated that an ectopic expression of functional TIP-1 reduced the overall levels of these protein interactions but maintained the dynamic interactions of Scribble/Arhgef7 and Scribble/Rhotekin in the migrating T98G cells, whereas ectopic expression of a TIP-1 mutant containing a dysfunctional PDZ domain had a similar but less significant effect on the protein interactions as the TIP-1 knockdown did (Supplementary Figure s5). These data suggested the importance of PDZ domain in these competitive and dynamic protein interactions. Therefore, we hypothesize that the high level expression of TIP-1 provides the flexibility of protein interactions that is required for the well-orchestrated intracellular redistribution of ARHGEF7 and rhotekin in the migrating glioblastoma cells. Since TIP-1 protein contains several hypothetical phosphorylation sites, it would be interesting to further investigate whether protein modifications, such as phosphorylation or dephosphorylation, are involved in the dynamic regulation of protein interactions between Scribble, rhotekin, ARHGEF7 and TIP-1.
In migrating cells, Rac1 induces the formation of lamellipodia and membrane ruffles, Cdc42 induces the formation of filapodia, and RhoA triggers actin stress fiber formation and retraction of the cell body (35, 36). We found that TIP-1 knockdown resulted in less activated Rac1 and more activated RhoA in T98G cells after scratch wounding. The activity reduction of Rac1 by TIP-1 knockdown is much greater than that of Cdc42. Consistent with the abnormal activation of these Rho GTPases, glioblastoma cells with TIP-1 knockdown adopted a highly branched morphology that might jointly reduce the motility of the TIP-1-depleted glioblastoma cells. We also noted that other report showed that TIP-1, acting together with ARHGEF16, activated Cdc42 and to a lesser extent, Rac1 in HPV16 E6 protein-expressing cervical cancer cells (12). These differences might be initiated through independent pathways, caused by the counteraction between RhoA and Rac1 (24), or resulted from the differential cellular context between cervical cancer cells and glioblastoma cells. Identifying proteins involved in the complex formation of Scribble, Arhgef7, rhotekin or TIP-1 would help to understand this mystery.
TIP-1 also binds to beta-catenin in the glioblastoma cells (Figure 1c). Beta-catenin associates with actins through alpha-catenin and stabilizes the cytoskeleton (37). Transactivation of nucleus-located beta-catenin also drives cell migration and invasion (38). Although our study showed that TIP-1 expression within glioblastoma cells does not significantly affect the beta-catenin-regulated gene expression and glioblastoma cell proliferation in vitro and in vivo, we cannot rule out the possibility that TIP-1 regulates glioma cell motility, cell morphology, or other cellular properties through interacting with beta-catenin. The different impacts of TIP-1 on the cell proliferation between colon cancer cells (9) and glioblastoma cells might suggest that some other cellular context would be required for the TIP-1-regulated transcriptional activity of beta-catenin or cell proliferation.
Malignant gliomas are highly invasive. The infiltrative dispersal of malignant glioma cells into normal brain impairs most of the currently available treatments. The vast majority (80-90%) of tumor recurrences arise within 2-3 cm of the resection margin (39) but tumor cells can also be found at far distant sites so that even hemispherectomies are not curative. Therefore, identifying genes regulating glioma cell migration and invasion is crucial to developing new therapeutic modalities for this fatal disease. This study demonstrated that the elevated expression of TIP-1 correlates with invasive progression and poor prognosis of human glioblastoma. Animal and cell-based studies indicated that TIP-1 expression is required for the invasive growth of human glioblastoma cells in vitro and in vivo. In fact, we found that TIP-1 knockdown in D54 glioblastoma cells greatly extended mice survival in intracranial xenograft models (Supplementary Figure s6). Other than the reduced invasion, TIP-1 knockdown also impaired the tumor-driven angiogenesis (40) and sensitized the glioblastoma cells to cytotoxic agents (31). Taken together, we speculate that TIP-1 holds potential as both a prognostic marker and a therapeutic target of malignant gliomas.
Materials and Methods
Cell culture and Stable shRNA transfection
Human glioblastoma cancer cells lines D54 and T98G were obtained from Dr. Yancie Gillespie (University of Alabama-Birmingham, Birmingham, AL) and purchased from ATCC (Manassa, VA), respectively. Both lines were maintained in DMEM/F12 media (Invitrogen, Carlsbad, CA) and stably transfected with TIP-1 targeting shRNAs (TRCN0000159034 and TRCN0000162886. Sigma-Aldrich, St. Louis, MO) using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Positive clones were selected with puromycin (Sigma-Aldrich). TIP-1 downregulation was assessed by western blot as described previously (28). Primary glioblastoma cell line T4302 was originally from Dr. Jeremy Rich (Cleveland Clinic, Cleveland, OH). Downregulating TIP-1 expression in the primary glioblastoma cell lines was achieved with recombinant lentivirus-mediated delivery of TIP-1 shRNAs (Sigma). Cell maintenance and lentivirus infection were conducted as described (41).
Antibodies and reagents
TIP-1 monoclonal (mouse) and polyclonal (rabbit) antibodies were produced in our lab and characterized as described (28). ARHGEF7 antibody (SH3 domain) was obtained from Millipore (Billerica, MA). Antibodies against Rac1and beta-catenin were purchased from BD Biosciences (Rockville, MD). Anti-Ki67 and RhoA antibodies were obtained from Abcam (Cambridge, MA). Anti-flag (M2) and beta-actin antibodies were purchased from Sigma. Anti-Scribble (H300), Cdc42 and rhotekin antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Alexa Fluor 594-labeled phalloidin and all other Alexa Fluor dye-labeled secondary antibodies were obtained from Invitrogen.
Plasmid constructs
A construct encoding a cMyc-tagged TIP-1 mutant with a dysfunctional PDZ domain (H90A) (7) in a pcDNA3.1 plasmid was a generous gift from Dr. Paul A. Welling at the University of Maryland (Baltimore, MD). The plasmid expressing flag-tagged ARHGEF7 (42) was obtained from Addgene (Cambridge, MA). To create mutations in the putative C-terminal PDZ binding motif (from −WLQSPV to –ALQAPV, mutations were underlined) of ARHGEF7, we used the QuikChange™ Site-Directed Mutagenesis Kit (Stratagene, Santa Cruz, CA) with primers 5’-atatgaacgaccctgccgcggatgaggccaatctatagctcgag-3’, and 5’-ctcgagctatagattggcctcatccgcggcagggtcgttcatat-3’.
Immunoprecipitations and western blotting
For immunoprecipitation, cells were lysed in RIPA buffer without SDS (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate). The cell lysates were centrifuged at 16,000× g for 5 minutes and the supernatant, after addition of protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO), was incubated for 2 hours at 4°C with corresponding antibody and protein A-agarose beads (Thermo Fisher Scientific, Rockford, IL). The amount of proteins loaded on SDS-PAGE gels was measured by the Bradford assay (BioRad, Hercules, CA).
Rho GTPase activation assay
Rho GTPase activation was determined using the RhoA Activation Assay Kit or Rac1/Cdc42 Activation Assay Kit (Cell Biolabs, San Diego, CA). Briefly, the cells were lysed in lysis buffer and centrifuged at 13,000× g for 10 minutes. The supernatant was respectively mixed with rhotekin RBD-agarose beads (for activated RhoA) or PAK1 PBD-agarose beads (for activated Rac1/Cdc42) for 1 hour at 4°C. The activated Rho GTPases were analyzed by western blot.
Cell staining and imaging
Cells were fixed and permeabilized in 4% formaldehyde solution before staining with specific antibodies. The images were acquired using a Zeiss LSM 510 inverted confocal microscope. To quantify the percentage of individual proteins located at the cell leading edge (those parts of cell membranes oriented toward the scratched wound), the fluorescence intensity of each protein was measured using Image J software (National Institutes of Health). The Pearson's correlation coefficients (25) were also produced with ImageJ to measure the colocalization of two proteins.
Migration and invasion assays
Cell migration and invasion assays were performed using 8-μm porous Boyden chambers (Corning Life Science, Lowell, MA) coated with or without Matrigel (BD Biosciences) according to the manufacturer's recommendations. Prior to the migration and invasion assays, cells were maintained in serum free media overnight. The next day, 10,000 cells (for migration) or 50,000 cells (for invasion) were seeded onto the upper chambers with serum free media. Full media with 10% serum were added to the bottom chamber. Twelve hours later, the cells that stayed on top of the membrane were removed with cotton swabs, and the cells migrated to the bottom of the membrane were stained with DAPI and counted under a fluorescent microscope. Cell migration was also measured using the scratch wound healing assay (43). Briefly, cells were allowed to grow to confluence on glass cover slips. After starving the cells overnight with serum free media, cells were scratched with a 200 μL pipette tip, and allowed to grow in fresh media for the indicated time before fixation and staining.
In vivo xenograft tumor model of glioblastoma
The intracranial tumor model was described previously (44). Briefly, 1×104 D54 or T4302 cells with or without TIP-1 knockdown in 10 μL of phosphate-buffered saline (PBS) were injected 3 mm deep (from the skull surface) into the right burr hole (2mm posterior to the bregma and 3mm to the right of the midline) of the brains of FoxN1-null nude mice (Harlan Laboratories, Prattville, AL). The mice were monitored daily for hypomotility, absence of grooming behavior, and weight loss. Tumor progression was verified by histological assessment. All the animal studies were conducted as approved by the Institutional Animal Care and Use Committee (IACUC) at Vanderbilt University.
Immunohistochemistry of tumor tissues
To study histological changes in the intracranial tumor model, tumors were retrieved 3 weeks after tumor cell implantation, fixed in formaldehyde and embedded in paraffin. Ten μm thick tumor sections were de-paraffinized in graded alcohols, rehydrated and heated in 10 mM citrate buffer (pH 6.0) for antigen retrieval using a pressure cooker. The tissue slides were subsequently stained with Ki67 antibody or TIP-1 antibody and counterstained with hematoxylin. Tumor invasion was quantified upon 200x microscopic images of tumor sections across main tumor mass as described (45). For the study of TIP-1 expression within human specimens, tissue microarrays containing pathologically verified astrocytoma, anaplastic astrocytoma and glioblastoma multiform (GBM) of WHO grade I to IV (46) and normal tissue were purchased from US Biomax (Rockville, MD). The tissue slides were stained with TIP-1 antibody and counterstained with hematoxylin as described above. TIP-1 expression intensity was scored using a semi-quantitative four-point scale based on light-microscopic observations as follows: 1 = background; 2 = scattered weak staining; 3 = moderately intense and more widespread staining; 4 = heavy staining.
Statistics
All numerical data are expressed as mean ± standard deviation (SD). Statistical comparisons between groups were performed by ANOVA. For correlation studies of TIP-1 expression and patient survival probability, datasets from Freije et. al. (29) and Phillips et. al. (30) that contain 86 clinically annotated brain tumor specimens were reanalyzed. All these brain tumors are WHO grade III or grade IV glioblastomas. The data from grade III or grade IV gliomas patients were analyzed separately. Patients within the top 40% of TIP-1 expression were defined as the high expression group, and those in the bottom 40% were defined as the low expression group. Kaplan-Meier survival analysis was used, and the log rank test was performed using MedCalc to determine differences between the two groups of patients. For all analyses, a p value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Drs. Barbara Fingleton, Steven K. Hanks and Mike Freeman (Vanderbilt) for critical reading and discussions of this manuscript. We are grateful to Dr. Paul A. Welling at University of Maryland (Baltimore, MD) for the kind gift of TIP-1-expressing constructs. We appreciate the technical assistance and advices from Dr. Ling Geng (Pathology, Vanderbilt University) in the animal studies, Dr. Heping Yan (Washington University at St. Louis) in the antibody development, Dr. Bret Mobley (Pathology, Vanderbilt University) in the microscopic examination of tissue blocks, and the technical staffs at the Cell Imaging, Histology, and Genomics Cores of the Vanderbilt-Ingram Cancer Center.
This study was supported in part by NIH grants R01CA127482 (Z Han), P50 CA128323 (Gore), and R01CA112385 (DE Hallahan).
Footnotes
Conflict of interest: No
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Beuming T, Skrabanek L, Niv MY, Mukherjee P, Weinstein H. PDZBase: a protein-protein interaction database for PDZ-domains. Bioinformatics (Oxford, England) 2005;21(6):827–8. doi: 10.1093/bioinformatics/bti098. [DOI] [PubMed] [Google Scholar]
- 2.Subbaiah VK, Kranjec C, Thomas M, Banks L. PDZ domains: the building blocks regulating tumorigenesis. Biochem J. 2011;439(2):195–205. doi: 10.1042/BJ20110903. [DOI] [PubMed] [Google Scholar]
- 3.Molina JR, Agarwal NK, Morales FC, Hayashi Y, Aldape KD, Cote G, et al. PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene. 2012;31(10):1264–74. doi: 10.1038/onc.2011.324. Epub 2011/08/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besser J, Leito JT, van der Meer DL, Bagowski CP. Tip-1 induces filopodia growth and is important for gastrulation movements during zebrafish development. Dev Growth Differ. 2007;49(3):205–14. doi: 10.1111/j.1440-169X.2007.00921.x. [DOI] [PubMed] [Google Scholar]
- 5.Olalla L, Gutierrez A, Jimenez AJ, Lopez-Tellez JF, Khan ZU, Perez J, et al. Expression of the scaffolding PDZ protein glutaminase-interacting protein in mammalian brain. Journal of neuroscience research. 2008;86(2):281–92. doi: 10.1002/jnr.21505. [DOI] [PubMed] [Google Scholar]
- 6.Han M, Wang H, Zhang HT, Han Z. The PDZ protein TIP-1 facilitates cell migration and pulmonary metastasis of human invasive breast cancer cells in athymic mice. Biochem Biophys Res Commun. 2012;422(1):139–45. doi: 10.1016/j.bbrc.2012.04.123. Epub 2012/05/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alewine C, Olsen O, Wade JB, Welling PA. TIP-1 Has PDZ Scaffold Antagonist Activity. Mol Biol Cell. 2006;17(10):4200–11. doi: 10.1091/mbc.E06-02-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hampson L, Li C, Oliver AW, Kitchener HC, Hampson IN. The PDZ protein Tip-1 is a gain of function target of the HPV16 E6 oncoprotein. Int J Oncol. 2004;25(5):1249–56. [PubMed] [Google Scholar]
- 9.Kanamori M, Sandy P, Marzinotto S, Benetti R, Kai C, Hayashizaki Y, et al. The PDZ protein tax-interacting protein-1 inhibits beta-catenin transcriptional activity and growth of colorectal cancer cells. J Biol Chem. 2003;278(40):38758–64. doi: 10.1074/jbc.M306324200. Epub 2003/07/23. [DOI] [PubMed] [Google Scholar]
- 10.Reynaud C, Fabre S, Jalinot P. The PDZ protein TIP-1 interacts with the Rho effector rhotekin and is involved in Rho signaling to the serum response element. J Biol Chem. 2000;275(43):33962–8. doi: 10.1074/jbc.M000465200. [DOI] [PubMed] [Google Scholar]
- 11.Zencir S, Ovee M, Dobson MJ, Banerjee M, Topcu Z, Mohanty S. Identification of brain-specific angiogenesis inhibitor 2 as an interaction partner of glutaminase interacting protein. Biochem Biophys Res Commun. 2011;411(4):792–7. doi: 10.1016/j.bbrc.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver AW, He X, Borthwick K, Donne AJ, Hampson L, Hampson IN. The HPV16 E6 binding protein Tip-1 interacts with ARHGEF16, which activates Cdc42. Br J Cancer. 2011;104(2):324–31. doi: 10.1038/sj.bjc.6606026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 14.Salhia B, Tran NL, Chan A, Wolf A, Nakada M, Rutka F, et al. The guanine nucleotide exchange factors trio, Ect2, and Vav3 mediate the invasive behavior of glioblastoma. The American journal of pathology. 2008;173(6):1828–38. doi: 10.2353/ajpath.2008.080043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang SL, Hong YR, Sy WD, Lieu AS, Lin CL, Lee KS, et al. Rac1 gene mutations in human brain tumours. Eur J Surg Oncol. 2004;30(1):68–72. doi: 10.1016/j.ejso.2003.10.018. Epub 2004/01/23. [DOI] [PubMed] [Google Scholar]
- 16.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes & development. 2007;21(21):2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 17.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The lancet oncology. 2009;10(5):459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 18.Olalla L, Aledo JC, Bannenberg G, Marquez J. The C-terminus of human glutaminase L mediates association with PDZ domain-containing proteins. FEBS Lett. 2001;488(3):116–22. doi: 10.1016/s0014-5793(00)02373-5. [DOI] [PubMed] [Google Scholar]
- 19.Rousset R, Fabre S, Desbois C, Bantignies F, Jalinot P. The C-terminus of the HTLV-1 Tax oncoprotein mediates interaction with the PDZ domain of cellular proteins. Oncogene. 1998;16(5):643–54. doi: 10.1038/sj.onc.1201567. [DOI] [PubMed] [Google Scholar]
- 20.Durney MA, Birrane G, Anklin C, Soni A, Ladias JA. Solution structure of the human Tax-interacting protein-1. Journal of biomolecular NMR. 2009;45(3):329–34. doi: 10.1007/s10858-009-9361-8. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Yan X, Shi C, Yang X, Guo Y, Tian C, et al. Structural Basis of [beta]-Catenin Recognition by Tax-interacting Protein-1. Journal of Molecular Biology. 2008;384(1):255–63. doi: 10.1016/j.jmb.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 22.Osmani N, Vitale N, Borg JP, Etienne-Manneville S. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr Biol. 2006;16(24):2395–405. doi: 10.1016/j.cub.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 23.Audebert S, Navarro C, Nourry C, Chasserot-Golaz S, Lecine P, Bellaiche Y, et al. Mammalian Scribble forms a tight complex with the betaPIX exchange factor. Curr Biol. 2004;14(11):987–95. doi: 10.1016/j.cub.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 24.Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol. 2008;9(11):846–59. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- 25.Rincon E, Saez de Guinoa J, Gharbi SI, Sorzano CO, Carrasco YR, Merida I. Translocation dynamics of sorting nexin 27 in activated T cells. J Cell Sci. 2011;124(Pt 5):776–88. doi: 10.1242/jcs.072447. Epub 2011/02/10. [DOI] [PubMed] [Google Scholar]
- 26.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461(7260):99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440(7087):1069–72. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Yan H, Fu A, Han M, Hallahan D, Han Z. TIP-1 translocation onto the cell plasma membrane is a molecular biomarker of tumor response to ionizing radiation. PloS one. 2010;5(8):e12051. doi: 10.1371/journal.pone.0012051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64(18):6503–10. doi: 10.1158/0008-5472.CAN-04-0452. Epub 2004/09/18. [DOI] [PubMed] [Google Scholar]
- 30.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Han M, Wang H, Zhang HT, Han Z. Expression of TIP-1 confers radioresistance of malignant glioma cells. PloS one. 2012;7(9):e45402. doi: 10.1371/journal.pone.0045402. Epub 2012/10/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hampson L, He XT, Oliver AW, Hadfield JA, Kemp T, Butler J, et al. Analogues of Y27632 increase gap junction communication and suppress the formation of transformed NIH3T3 colonies. Br J Cancer. 2009;101(5):829–39. doi: 10.1038/sj.bjc.6605208. Epub 2009/08/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Mata R, Burridge K. Catching a GEF by its tail. Trends in cell biology. 2007;17(1):36–43. doi: 10.1016/j.tcb.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 34.ten Klooster JP, Jaffer ZM, Chernoff J, Hordijk PL. Targeting and activation of Rac1 are mediated by the exchange factor {beta}-Pix. J Cell Biol. 2006;172(5):759–69. doi: 10.1083/jcb.200509096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279(5350):509–14. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 36.Amano M, Chihara K, Kimura K, Fukata Y, Nakamura N, Matsuura Y, et al. Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science. 1997;275(5304):1308–11. doi: 10.1126/science.275.5304.1308. [DOI] [PubMed] [Google Scholar]
- 37.Dickinson DJ, Nelson WJ, Weis WI. A polarized epithelium organized by beta- and alpha-catenin predates cadherin and metazoan origins. Science. 2011;331(6022):1336–9. doi: 10.1126/science.1199633. Epub 2011/03/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji H, Wang J, Nika H, Hawke D, Keezer S, Ge Q, et al. EGF-induced ERK activation promotes CK2-mediated disassociation of alpha-Catenin from beta-Catenin and transactivation of beta-Catenin. Mol Cell. 2009;36(4):547–59. doi: 10.1016/j.molcel.2009.09.034. Epub 2009/11/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burger PC, Dubois PJ, Schold SC, Jr., Smith KR, Jr., Odom GL, Crafts DC, et al. Computerized tomographic and pathologic studies of the untreated, quiescent, and recurrent glioblastoma multiforme. J Neurosurg. 1983;58(2):159–69. doi: 10.3171/jns.1983.58.2.0159. [DOI] [PubMed] [Google Scholar]
- 40.Han M, Wang H, Zhang HT, Han Z. Expression of Tax-interacting protein 1 (TIP-1) facilitates angiogenesis and tumor formation of human glioblastoma cells in nude mice. Cancer Lett. 2013;328(1):55–64. doi: 10.1016/j.canlet.2012.09.011. Epub 2012/09/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. doi: 10.1038/nature05236. Epub 2006/10/20. [DOI] [PubMed] [Google Scholar]
- 42.Mayhew MW, Webb DJ, Kovalenko M, Whitmore L, Fox JW, Horwitz AF. Identification of protein networks associated with the PAK1-betaPIX-GIT1-paxillin signaling complex by mass spectrometry. Journal of proteome research. 2006;5(9):2417–23. doi: 10.1021/pr060140t. [DOI] [PubMed] [Google Scholar]
- 43.Etienne-Manneville S. In vitro assay of primary astrocyte migration as a tool to study Rho GTPase function in cell polarization. Methods in enzymology. 2006;406:565–78. doi: 10.1016/S0076-6879(06)06044-7. [DOI] [PubMed] [Google Scholar]
- 44.Geng L, Shinohara ET, Kim D, Tan J, Osusky K, Shyr Y, et al. STI571 (Gleevec) improves tumor growth delay and survival in irradiated mouse models of glioblastoma. Int J Radiat Oncol Biol Phys. 2006;64(1):263–71. doi: 10.1016/j.ijrobp.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 45.Akella NS, Ding Q, Menegazzo I, Wang W, Gillespie GY, Grammer JR, et al. A novel technique to quantify glioma tumor invasion using serial microscopy sections. J Neurosci Methods. 2006;153(2):183–9. doi: 10.1016/j.jneumeth.2005.10.026. Epub 2006/01/13. [DOI] [PubMed] [Google Scholar]
- 46.Cavenee WK, Furnari FB, Nagane M, Huang H-JS, Newcomb E,W, Bigner DD, et al. In: Tumors of the Nervous System. Kleihues P, Cavenee WK, editors. IARC Press; Lyon: 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.