Abstract
The zebrafish is an important animal model for stem cell biology, cancer, and immunology research. Histocompatibility represents a key intersection of these disciplines, however histocompatibility in zebrafish remains poorly understood. We examined a set of diverse zebrafish Class I major histocompatibility complex (MHC) genes that segregate with specific haplotypes at chromosome 19, and for which donor-recipient matching has been shown to improve engraftment after hematopoietic transplantation. Using flanking gene polymorphisms we identified six distinct chromosome 19 haplotypes. We describe several novel Class I U lineage genes and characterize their sequence properties, expression, and haplotype distribution. Altogether ten full-length zebrafish Class I genes were analyzed, mhc1uba through mhc1uka. Expression data and sequence properties indicate that most are candidate classical genes. Several substitutions in putative peptide anchor residues, often shared with deduced MHC molecules from additional teleost species, suggest flexibility in antigen binding. All ten zebrafish Class I genes were uniquely assigned among the six haplotypes, with dominant or co-dominant expression of one to three genes per haplotype. Interestingly, while the divergent MHC haplotypes display variable gene copy number and content, the different genes appear to have ancient origin, with extremely high levels of sequence diversity. Furthermore, haplotype variability extends beyond the MHC genes to include divergent forms of psmb8. The many disparate haplotypes at this locus therefore represent a remarkable form of genomic region configuration polymorphism. Defining the functional MHC genes within these divergent Class I haplotypes in zebrafish will provide an important foundation for future studies in immunology and transplantation.
Keywords: zebrafish, Class I, MHC, histocompatibility, transplantation, region configuration polymorphism
Introduction
Major histocompatibility complex (MHC) gene matching between transplant donors and recipients largely determines histocompatibility, an observation that has profoundly increased human transplantation success (Chinen and Buckley 2010; Horan et al. 2012). The more ‘natural’ intrinsic roles of MHC genes in antigen presentation also remain topics of active investigation, with implications for infectious disease studies (Blackwell, Jamieson, and Burgner 2009). However, histocompatibility remains poorly understood in many organisms including zebrafish (de Jong and Zon 2012), an important preclinical animal model for immunology, stem cell biology, and cancer research (Sullivan and Kim 2008; Ignatius and Langenau 2009; Mione and Trede 2010; Renshaw and Trede 2011).
In humans the MHC includes both Class I and Class II genes that are linked on chromosome 6. Human Class I gene organization is well conserved between individuals and includes three classical human leukocyte antigen (HLA) genes (HLA-A, -B, and -C; designated Class Ia) and three non-classical genes (HLA-E, -F, and -G; representing Class Ib) (Shiina et al. 2009). The classical MHC genes can be defined, in part, as those that encode molecules presenting peptide antigens to T cell receptors. Class Ia molecules are found on the surface of nearly all cells and form a complex with β2-microglobulin to present peptide antigens derived from processed intracellular proteins to CD8+ cytotoxic T cells. Class Ib molecules perform other functions such as regulation of innate immunity through binding to natural killer cell receptors (Rodgers and Cook 2005; Parham 2005). In contrast, Class II gene expression is restricted to antigen-presenting cells that present processed extracellular antigens to CD4+ T cells involved in the adaptive immune response.
Classical human MHC genes maintain high levels of sequence polymorphism, particularly for amino acid residues associated with the peptide binding site (Bjorkman et al. 1987). Classical genes in humans have hundreds of variants (alleles) that differ mostly by single nucleotide polymorphisms (SNPs). Non-classical genes are much less polymorphic, and generally play a role in innate immunity. Alleles of each of the linked HLA genes are found as combinations that comprise distinct haplotypes on human chromosome 6. The polymorphism within many HLA genes is thought to be under strong selection thereby helping to ensure that individuals can efficiently display a wide array of pathogen-derived peptides (Hughes and Nei 1988; Nikolich-Zugich 2004). While the rate of MHC gene mutation does not appear to be significantly greater than the rest of the genome, selection can maintain high levels of sequence diversity within populations (Klein et al. 1993).
The study of zebrafish histocompatability genes is complicated by the fact that unlike mammals and other vertebrates, teleost fish do not have a single MHC locus, and instead the Class I and II genes are located on different chromosomes (Bingulac-Popovic et al. 1997). Previous studies have identified several Class I MHC genes in zebrafish that belong to distinct sequence lineages (Takeuchi et al. 1995; Graser et al. 1998; Sültmann, Murray, and Klein 2000). Prior work also showed that some of the genes are distributed among separate chromosome 19 haplotypes (Bingulac-Popovic et al. 1997; Michalová et al. 2000; de Jong et al. 2011), and identified that haplotype matching was important for improved engraftment following marrow transplantation (de Jong et al. 2011; de Jong and Zon 2012). This chromosome 19 locus maintains synteny with mammalian Class I loci including humans and mice (Matsuo et al. 2002; Kulski et al. 2002; Sambrook, Figueroa, and Beck 2005). In this report we characterize the expression, sequence properties, and haplotype distribution of Class I U lineage genes in zebrafish at the chromosome 19 locus, and provide a description of novel genes and haplotypes, each carrying distinct sets of zebrafish candidate classical genes.
Methods
Zebrafish Class I gene nomenclature
Class I genes in teleosts are labeled according to standardized nomenclature (Klein et al. 1990). Among teleost Class I MHC genes, the major lineage designation is U (for ‘uno’), which contains the genes that most consistently maintain classical characteristics. The three other lineages are L, for linkage to Class II (Dijkstra et al. 2007), S, for salmon (Lukacs et al. 2010), and Z (Kruiswijk et al. 2002). An example is Dare-uba (in this report also referred to as uba), which is the second (b) gene identified from the U lineage in the zebrafish, Danio rerio. In current zebrafish nomenclature the gene name is appended to ‘mhc1’ for Class I genes (e.g. mhc1uba). Z lineage gene nomenclature (Figure 1) was provided by personal communication from Hayley Dirscherl and Jeffrey Yoder.
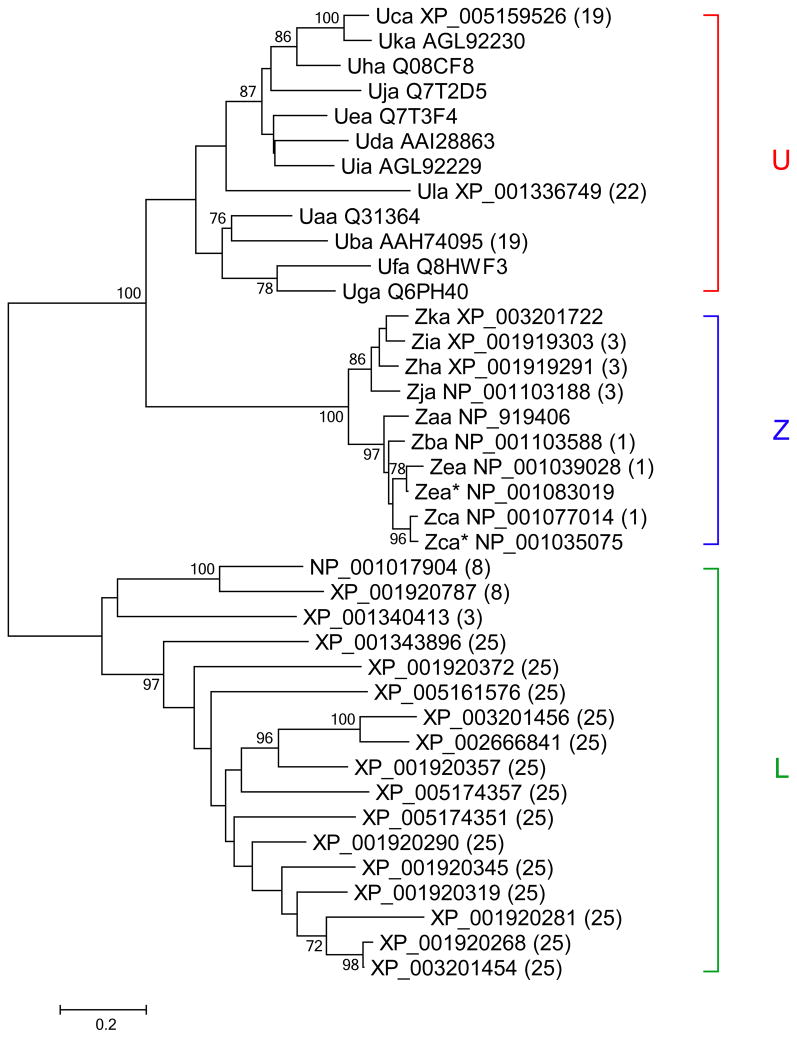
Fig. 1.
Comprehensive analysis of Class I gene relationships in zebrafish. Following alignment of predicted amino acid sequences from zebrafish Class I genes, neighbor-joining trees were constructed and bootstrap values over 70% are shown. Our analysis revealed 12 genes of the U lineage, 10 genes of the Z lineage, and 17 genes of the L lineage. Sequences assigned as allelic variants are marked with asterisks. Chromosomal location is given in parentheses for genes that map to the current reference genome (Zv9)
Genomic sequence analysis of zebrafish Class I genes on chromosome 19
The sequence for haplotype A includes PAC clones (Michalová et al. 2000) represented by AL675121, AL672185 and AL672164 as assembled on the AB chromosome. The AB chromosome contains PAC clones from the AB strain of zebrafish that were sorted out due to high variation compared with the current reference Tuebingen strain (http://vega.sanger.ac.uk/Danio_rerio/Info/Index), and manually annotated by the Zebrafish Group at the Wellcome Trust Sanger Institute as part of Zv9 (Howe et al. 2013). The sequence for haplotype B is composed of BAC clone sequences FP102097, FP085393, and FP074889 from the Tuebingen strain as assembled on chromosome 19 in the most recent reference genome version, Zv9. These sequences were analyzed using the VEGA genome browser (http://vega.sanger.ac.uk/Danio_rerio/), with sequence similarity searches (www.ncbi.nlm.nih.gov/BLAST), and with SeqBuilder (DNAstar).
Zebrafish lines
All zebrafish experiments were performed as approved by the University of Chicago Institutional Animal Care and Use Committee. AB-derived zebrafish were generously provided by Dr. Leonard Zon. The AB strain of laboratory zebrafish was originally derived by George Streisinger in the 1970’s by crossing the A and B lines that had been purchased from a pet store in Oregon (ZFIN ID: ZDB-GENO-960809-7, Zebrafish International Resource Center). These fish have been bred for >30 years to propagate the healthiest offspring. It is a genetically heterogeneous line considered to be wild type, although these fish have been subject to population bottlenecks over time and therefore would be expected to carry a limited number of MHC haplotypes within the population. AB individuals were genotyped (as described below) to examine their Class I loci on chromosome 19, and were identified as being compound heterozygous for two haplotypes or being homozygous for one haplotype. Fish were selectively bred to maintain homozygosity of certain haplotypes (e.g. haplotypes A, B, E or F).
Clonal fish were also obtained for haplotype analysis. The golden-derived clonal lines CG1 (Smith et al. 2010), carrying haplotype C, and CG2 (Mizgirev and Revskoy 2010), carrying haplotype D, were each generated through two rounds of parthenogenesis and generously provided by Dr. Sergei Revskoy. These clonal golden lines were derived on the AB background.
RNA purification
RNA was purified by a combined Trizol/column method, with modification (Lan et al. 2009). Briefly, adult zebrafish were euthanized in 1 mg/mL tricaine (MS222). Organs were removed from euthanized fish under a dissecting microscope as described (Gupta and Mullins 2010) and placed into Eppendorf tubes containing 500 μL Trizol. A pellet pestle with a rotor-stator (Kimble Chase) was used to homogenize the tissues, followed by addition of 100 μL chloroform and centrifugation at 12,000 × g for 10 min at 4°C. The aqueous phase was removed, mixed with an equal volume of 60% ethanol, and applied to an RNA binding column (Aurum Total RNA Mini Kit, Biorad). Subsequent steps were according to the manufacturer’s protocol, including DNase digestion, washes, and elution in 40 μL of nuclease-free water.
Preparation of cDNA and quantitative Polymerase Chain Reaction (qPCR)
RNA from each tissue was reverse transcribed in a 20 μL volume with random primers using the Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Fermentas) as described in the manufacturer’s instructions. qPCR was performed in 10 μL reactions that included 1 μL diluted cDNA, primers and SsoAdvanced SYBR Green Supermix supplemented with ROX (Biorad). Data were collected in triplicate reactions according to recommended settings (Biorad) on a 7900HT instrument with 384-well format (Applied Biosystems), and analyzed by the delta-delta CT method (Livak and Schmittgen 2001). β2-microglobulin (β2m) was used as a reference gene. Dissociation analysis was performed to check for non-specific products.
Primer design
Starting with the 10 full-length zebrafish Class I cDNA sequences, we designed four sets of primer pairs specific to each gene. The first qPCR primer set was used for expression analysis (supplementary Table S1) with approximately 100 bp amplicons. These primers were designed within the most polymorphic region of each gene, exons 2 and 3, spanning intron 2. Primers specific for each gene were designed using Primer3Plus (Untergasser et al. 2012) and checked for dimers using OligoAnalyzer (http://IDTDNA.com).
A second primer set for each gene (supplementary Table S2) was manually designed to amplify the full-length Class I coding sequences that are approximately 1 kb in length in order to verify the full-length sequence and evaluate for polymorphisms. Amplification of full-length genes was performed with 40 cycles consisting of 95°C for 30 seconds, 60°C for 60 seconds and 72°C for 150 seconds using 500 nM primers, cDNA, and ChoiceTaq Master Mix (Denville). These longer amplicons, in conjunction with overlapping amplicons obtained using primer sets specific for the 5′ (supplementary Table S3) and 3′ regions (supplementary Table S4) of the genes were used to provide additional sequence information.
Haplotype analysis
DNA was prepared from zebrafish tailclips as previously described (Meeker et al. 2007). 500 nM primers for either the zbtb22b gene (5′-GTTGACTATTGTCAGTCCTCGGAA-3′ and 5′-GTGCTCTGTGAGGTGATGCTTC-3′) or psmb8 gene (5′-CCTGACATTTGGTCCCAAAA-3′ and 5′-CCCTAAAGGACAACATTTATGTACA-3′) were used in a 20 μL reaction with PCR MasterMix (Fermentas) and 1 μL tail DNA. These psmb8 primers are specific for the psmb8a gene form, which is present in five of the six haplotypes, all except haplotype D. Hence no band is amplified by these psmb8 primers from haplotype D, which instead carries the divergent psmb8f gene. We amplified the specific F form of zebrafish psmb8 using previously published alternative primers (Tsukamoto, et al. 2012). In addition, PCR bias is likely to be observed using the psmb8a primer set with DNA from heterozygous haplotype E fish, displaying lower signals for the haplotype E specific SNPs compared with the other psmb8a allelic sequences. Therefore, considering lack of amplication of the psmb8f allele using psmb8a primers, together with potentially unequal amplification of select psmb8a alleles, haplotype assignment via these flanking genes may be more reliable by focusing first on the zbtb22b sequences and then using psmb8 alleles for confirmation.
Amplification was performed using touchdown PCR starting from 60°C and ending at 48°C (Korbie and Mattick 2008). Prior to sequencing, PCR reactions (5 μL aliquots) were first mixed with 3 units of Exonuclease (Affymetrix) and 0.3 units of Shrimp Alkaline Phosphatase (NEB) and incubated at 37°C for 30 minutes followed by inactivation at 80°C for 15 minutes. Sequencing was performed with forward and reverse primers. Sequences were analyzed using SeqMan (DNAstar) to identify specific SNPs and assign haplotypes (Table 2).
Table 2.
SNPs in psmb8 and zbtb22b examined for zebrafish Class I haplotype determination
| Haplotype | A | B | C | D | E | F |
|---|---|---|---|---|---|---|
|
|
||||||
| zbtb22b | 51A>G 111C>T 267C>A 445C>T |
FP102097 | 51A>G 84G>A 345C>T |
51A>G 126C>G 345C>T |
51A>G 165C>T |
51A>G |
| Class I genes |
mhc1uda mhc1uea mhc1ufa |
mhc1uba mhc1uca |
mhc1uja mhc1uka |
mhc1uga |
mhc1uia (unknown) | mhc1uha |
| psmb8a (psmb8) | 209C>G 250C>T 337C>A 364G>A |
FP074889 | 95T>C 98A>G 291G>A 364G>A |
− | 174T>A 209C>G 215T>A 250C>T |
95T>C 98A>G 324G>T 364G>A |
| psmb8f | − | − | − | + | − | − |
SNPs (arbitrary numbering) in the psmb8 and zbtb22b genes on Chromosome 19 are shown relative to reference sequences (haplotype B sequence with GenBank accession numbers). Based on genomic Southern blot data, an additional Class I U lineage gene with unknown sequence is likely to be found on haplotype E. A divergent psmb8f gene is associated with haplotype D in lieu of the psmb8a (psmb8) gene associated with all the other haplotypes. + indicates amplification, while − indicates no amplification
Sequencing and analysis of MHC genes
Fish homozygous for each haplotype were examined by amplifying the full length MHC genes expressed, followed by DNA sequencing of those MHC genes to verify sequence. At least 2 of 3 fish per haplotype that were analyzed by qPCR for expression analysis also had sequencing of the full length MHC genes. In addition we also analyzed full length MHC gene sequences from other fish in our lab stock populations. Full-length gene products and smaller qPCR amplicons were both sequenced directly after cleaning as described above.
Similarity searches against the non-redundant sequence database were performed for each domain of individual zebrafish MHC genes (www.ncbi.nlm.nih.gov/BLAST). Full-length genes and sequencing results were analyzed using SeqMan (DNAstar). Multiple sequence alignments were performed using ClustalW and adjusted by eye using MEGA version 4.0 (Tamura et al. 2007). Phylogenetic trees were constructed using the neighbor-joining method (Saitou and Nei 1987), and bootstrapped using 1000 replicates.
Haplotype linkage analysis
A fish heterozygous for haplotypes C and F was crossed with a fish heterozygous for haplotypes D and E. Offspring were analyzed by tail clip and half of the tail was isolated to prepare DNA for genotyping using lysis buffer (Zhou and Zon 2011). The other half of the tail was collected in Trizol and used to generate cDNA to analyze for MHC gene expression as described above.
Genomic Southern blot analysis
DNA was purified from whole fish as previously described (Westerfield 2000). 20 μg of DNA was digested overnight with NdeI (Fermentas) and loaded on a 0.8% agarose gel. After downward capillary transfer of DNA in alkaline buffer (Brown 2001) to a positively charged nylon membrane (Roche), the blot was probed with a PCR-amplified, digoxigenin-labeled probe specific to the α3 domain of uda (Table S5). Stringency washes were performed at 62° C following the Roche DIG High Prime DNA Labeling and Detection Starter Kit II protocol. The blot was exposed to HyBlot CL film (Denville) for chemiluminescent detection.
Results
Relationships among zebrafish Class I genes
We started by searching sequence library databases for all zebrafish Class I genes and found 39 zebrafish Class I genes, 26 of which are mapped to chromosomes within the current reference genome, Zv9. Comparisons of their amino acid sequences revealed the presence of at least three distinct lineages (Fig. 1). Previous studies have identified several Class I major histocompatibility genes (e.g. uba, uca, uda, uea and ufa) from the U lineage associated with zebrafish chromosome 19 (Takeuchi et al. 1995; Bingulac-Popovic et al. 1997; Michalová et al. 2000). The proteins encoded by the zebrafish U lineage genes cluster together in phylogenetic trees rather than with other Class I proteins from zebrafish such as those from the Z and L lineages (Fig. 1). Zebrafish U lineage genes are closely related to Class I U lineage genes considered to have classical function from better characterized teleost species such as medaka and salmon (Aoyagi et al. 2002; Kiryu et al. 2005; Tsukamoto et al. 2005; Nonaka and Nonaka 2010). The U lineage genes have also been shown to play a role in rejection of transplanted allogeneic cells in rainbow trout (Sarder et al. 2003).
We identified a total of twelve zebrafish Class I MHC genes that may be assigned to the U lineage (Table 1). Only three of these twelve U lineage genes are mapped to the current reference genome (uba and uca on chromosome 19 and ula on chromosome 22). Besides the U lineage genes, our sequence database searches for related genes yielded 23 zebrafish Class I genes mapped to additional chromosomes (Chr. 1, 3, 8, 25). These genes include members of the Z and L lineages that populate separate branches on the phylogenetic tree from the U lineage. Many likely represent non-classical genes as indicated by their sequence properties.
Table 1.
Zebrafish Class I gene information and accession numbers
| Gene | Aliases | Hap | aa | nt | GenBank Accession # |
|---|---|---|---|---|---|
| mhc1uaa | na | na | na | Z46776b, AL672185b | |
| mhc1uba | zgc:91955, ENSDART00000009689 | B | 348 | 1047 | BC074095, BC164159f, Z46777a,b, FP074889a |
| mhc1uca | DKEY-264N13.4, si:ch73-213j4.4, ENSDART00000133954 | B | 443g | 1332g | XM_005159469, AF182157b, FP085393, Z46778b |
| mhc1uda | zgc:158407 | A | 344 | 1035 | BC128862, AF182155b BC095216ac, AL672151c |
| mhc1uea | wu:fb69e07 | A | 346 | 1041 | BC053140, AF182156b, BC097061a, AL672164a |
| mhc1ufa | mhc1ubal | A | 349 | 1050 | AF137534, BC066754c, AL672151c |
| mhc1uga | mhc1uxa2, zgc:65799, zgc:77509e | D | 350 | 1053 | BC056726, BC066745e |
| mhc1uha | LOC767703, zgc:153138 | F | 341 | 1026 | BC124257, BC164146f |
| mhc1uia | mhc1uxa1, LOC550387, zgc:111997 | E | 345 | 1038 | KC626502, BC093149a, AH008873d, BC165457af |
| mhc1uja | LOC393377, zgc:64003 | C | 346 | 1041 | BC054592, BC164155f |
| mhc1uka | LOC751750, zgc:153728 | C | 356 | 1071 | KC626503, BC122401a, BC164173af |
| mhc1ula | LOC796419, ENSDART00000082034 | - | 344 | 1035 | XM_001336713, EH452054b, EH480036bc, NC_007133.5 |
Hap indicates chromosome 19 haplotype assignments, aa represents predicted amino acid length, nt is the coding sequence nucleotide length, na indicates no data available, - indicates chromosome 19 haplotype assignment not applicable (ula is instead located on chromosome 22).
partial matches with SNP differences,
partial sequences,
insertion/deletion potentially introducing frameshift,
sequence gap,
divergent variant in 3′ portion of gene,
modified stop codon,
size for one possible splice variant. GenBank accession numbers exactly matching sequences found in our fish are highlighted in bold
Several of the U lineage genes we found were uncharacterized. We proposed new names for the uncharacterized genes following accepted nomenclature, as well as renaming several other zebrafish genes in order to standardize terminology among zebrafish Class I U lineage genes (Table 1). These new names have been accepted by the Zebrafish Nomenclature Committee. Two of the genes, uaa and uia, were earlier reported as partial cDNA sequences (Takeuchi et al. 1995; Michalová et al. 2000). However, full length sequence can now be assigned for one of these genes. The uia gene, previously designated uxa1, shares nearly 100% sequence identity with a full length cDNA sequence labeled loc550387 that is unmapped to the zebrafish reference genome.
The gene uxa2 originally had two distinct sequences assigned, zgc:77509 and zgc:65799. The new name, uga, has been assigned to zgc:65799 corresponding to the sequence that retains homology with other MHC genes throughout its full length. Although the 3′ portion of zgc:77509 lacks discernible homology to other MHC genes, the 5′ portion was previously used as a probe for in situ hybridization experiments (Thisse and Thisse 2004). We have proposed that zgc:77509 retain association with the name uxa2. In addition, the gene previously labeled loc7501750 has been renamed uka, loc767703 is renamed uha, and loc393377 is renamed uja. These five genes (uga, uha, uia, uja, and uka) are all unmapped to the zebrafish reference genome.
Comparison of zebrafish Class I gene organization
While only two of these zebrafish Class I U lineage genes (uba and uca) are present on chromosome 19 in the current reference sequence (Zv9), three other Class I U lineage genes (uda, uea and ufa) are present on a separate haplotype assembly from the AB strain that still shares many of the same genes as the chromosome 19 Tuebingen reference sequence. Each of these different genomic assemblies (Fig. 2) has been defined as a unique haplotype. The genomic sequence assembled from the AB strain comprises haplotype A, while the chromosome 19 reference sequence (Zv9, assembled from the Tuebingen strain) is haplotype B.
Fig. 2.
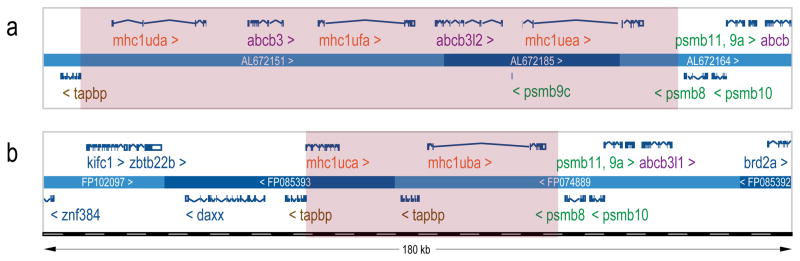
Genomic organization of zebrafish Class I genes on chromosome 19. The top panel displays gene organization for haplotype A including PAC clones AL675121, AL672185 and AL672164 as assembled on an AB chromosome containing sequence variants from the zebrafish AB strain. The bottom panel is haplotype B consisting of BAC clones FP102097, FP085393, and FP074889 from the zebrafish Tuebingen strain assembled on chromosome 19 in the most recent reference genome version, Zv9. For both haplotypes the genes that are oriented in the forward sequence direction (>) are placed above the lines labeled with the genomic sequence identifiers, while genes located in the reverse direction (<) are found below. The Class I genes are located in a highly divergent center region highlighted in pink shading, while other genes are present in an adjacent conserved framework. Haplotype A contains three Class I major histocompatibility genes, uda, uea and ufa that are separated by abcb3/Tap2 genes. Haplotype B contains two Class I major histocompatibility genes, uba and uca separated by a tapbp gene. Some of the conserved flanking genes encode proteins that interact directly with the Class I molecule, including tapbp on the 5′ side, along with a distal abcb3/Tap2 gene on the 3′ side. Others are involved in peptide processing, such as the 3′ flanking psmb8-11 genes
Detailed comparison of these genomic haplotype sequences reveals some interesting features. We were able to define the haplotypes on the basis of conserved flanking genes on either side of the Class I genes at the locus; beginning with tapbp on the 5′ end and psmb8 on the 3′ end. Additional genes including zbtb22b and abcb3/tap2 are also conserved within these flanking boundary regions. However, within this conserved genomic framework, marked differences in gene copy number and gene content are observed between the Class I haplotypes. Within the divergent central region of haplotype A, there are two copies of the abcb3/tap2 gene interspersed between the three Class I genes, uda, uea and ufa (Fig. 2a). In contrast, this region within haplotype B contains the Class I genes uba and uca, which are instead separated by a single tapbp gene (Fig. 2b). Interestingly, these two haplotypes not only lack shared Class I genes, but they also contain different interspersed genes and are divergent enough to impede direct sequence alignment.
At the individual gene level, most of the Class I genes have a conserved, seven exon structure with relatively large second introns. However, uca has a strikingly different organization compared with the other Class I genes on haplotypes A and B. While intron 2 sequences for uba, uda, and ufa exceed 10 kb in length, the uca intron 2 sequence is only about 1 kb, resulting in a much more compact genomic organization. In addition, uca appears to have additional exons spliced in its 3′ region, including one exon that is nearly identical in sequence to an adjacent exon within the uca gene. These observations may indicate functional or regulatory differences between uca and the other Class I genes.
Zebrafish Class I haplotype determination and Class I gene expression analysis
To better understand Class I histocompatibility genes in zebrafish, we focused our studies on the chromosome 19 locus. Given the existence of additional zebrafish Class I U lineage genes (e.g. uga-uka) not present on either known haplotype, we reasoned that haplotype variability at the chromosome 19 locus may extend beyond haplotypes A and B. We set out to make haplotype assignments for these remaining genes. As the genes generally share low levels of sequence identity, this limited our ability to design a common set of primers. Instead we used a haplotyping strategy that relies on conserved primer binding sites within nearby flanking genes, with primers for the zbtbt22b gene at the 5′ end of the Class I region, and the psmb8 gene at the 3′ end. SNPs within the amplified sequences of these two flanking genes (zbtb22b and psmb8, Table 2) were used for haplotype assignments. Each unique pattern of SNPs in these flanking genes is associated with one specific haplotype and given a letter designation.
Six haplotypes were identified using this method. Interestingly, psmb8 has two divergent alleles in zebrafish, which are amplified by distinct primer sets (Tsukamoto, et al. 2012). The psmb8f form was found associated with haplotype D, while several psmb8a alleles differing only by a small number of SNPs were amplified from the remaining haplotypes (Table 2). We selected zebrafish that were homozygous for each of the six different haplotypes at the Class I locus on chromosome 19 for further analysis. This enabled us to examine and compare zebrafish Class I gene expression and tissue distribution among each of the divergent haplotypes.
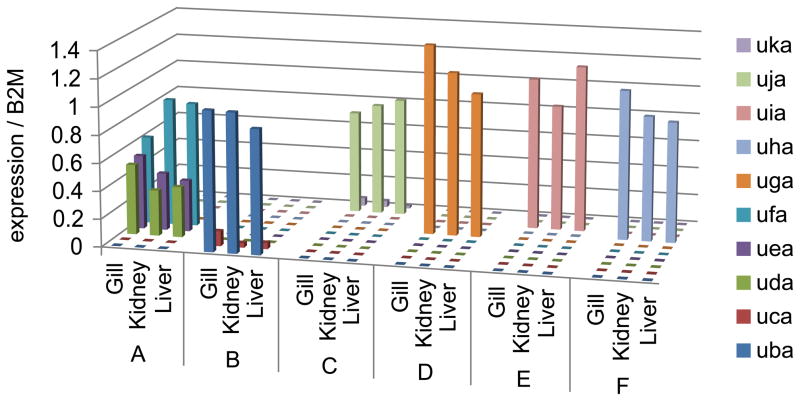
To facilitate association of Class I genes with each haplotype, and to examine tissue distribution and expression levels, separate primer sets were designed to amplify each of the Class I genes. Using qPCR, we measured gene expression for ten zebrafish Class I genes (uba-uka) in seven different tissues (spleen, liver, kidney, heart, testis, gill, intestine) from three fish per haplotype. These tissues were selected to represent both immune and non-immune related cells from zebrafish. Expression levels for each gene were highly consistent among the different fish for each haplotype examined. Furthermore, the Class I gene patterns and expression levels were similar in each of the seven tissues examined from a given fish, showing minimal variation between tissues from the same animal.
We found that expression of the Class I genes was distributed in distinctive, non-overlapping patterns among the six haplotypes, with one to three genes represented per haplotype (Fig. 3). We note that none of these genes was expressed in more than one haplotype. As expected from the genomic sequences, two Class I genes (uba and uca) were expressed from haplotype B while three genes (uda, uea and ufa) were expressed from haplotype A. Furthermore, on haplotype B, uba, was expressed at levels approximately an order of magnitude higher than uca. In contrast, expression levels of uda, uea, and ufa from haplotype A were more equivalent, with ufa expressed at the highest levels. Haplotype C expressed uja with lower levels of uka. Haplotype D expressed only uga. Haplotype E expressed uia. Haplotype F expressed only uha. We were not able to identify animals expressing the gene with a partial sequence labeled uaa, however all ten of the remaining genes (uba-uka) had expression assigned to specific chromosome 19 haplotypes in this manner.
Fig. 3.
Expression patterns of zebrafish Class I genes from six haplotypes. Tissues (gill, kidney, liver) were dissected from adult male zebrafish homozygous for a single haplotype (A through F). Following RNA extraction and cDNA synthesis, qPCR data were generated using gene specific primers and normalized to β2-microglobulin. Data represent the average of three fish for each haplotype. Similar expression levels and patterns of Class I genes for each haplotype were also observed in the spleen, heart, intestine, and testis from all fish tested (data not shown)
Haplotype linkage analysis
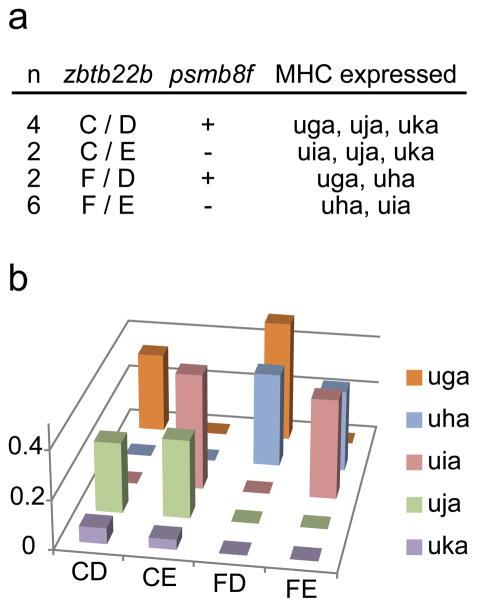
As five of our Class I genes were unmapped to the zebrafish reference genome, we performed linkage analysis to provide evidence that these genes are also linked to the psmb8 and zbtb22b alleles found on chromosome 19. After mating fish that were each heterozygous for two distinct haplotypes (haplotypes C/F and haplotypes D/E), genotyping using zbtb22b, psmb8a and psmb8f primer sets revealed that the offspring each had only two of the four parental alleles, inheriting one allele from each parent (Fig. 4a), with segregation ratios as would be expected for linked genes. Among 14 offspring examined, six inherited haplotype C and eight inherited haplotype F. Six fish inherited haplotype D and eight inherited haplotype E. We never observed co-inheritance of haplotypes C and F, or haplotypes D and E, indicating independent assortment of parental alleles that are each linked to the same chromosomal locus (Fig 4a). In addition, these specific alleles for the zbtb22b and psmb8 genes did not sort independently but segregated in every animal by coupling. That is, the SNP pattern for the psmb8 gene designating a specific haplotype was only found in fish with the zbtb22b SNP pattern for that same haplotype. These findings provide strong evidence for genetic linkage of the psmb8 and zbtb22b genes among the four additional haplotypes. Chi-square analysis of flanking gene linkage for haplotypes C through F was highly significant (p<0.001).
Fig. 4.
Linkage analysis for haplotypes C through F. A fish heterozygous for haplotypes C and F was crossed with a fish heterozygous for haplotypes D and E. Tail clips from offspring were analyzed both by genotyping for haplotype flanking allele sequences and also by examining which MHC genes were expressed by qPCR. Genotyping results for these offspring are given in the top panel. The expression levels of the different MHC genes (normalized to β2-microglobulin) are provided for one representative fish heterozygous for each of the four haplotype combinations in the bottom panel
Furthermore, analysis of Class I MHC gene expression by qPCR in these same progeny confirmed that specific Class I genes for each haplotype co-segregated only with the specific alleles of zbtb22b and psmb8 associated with that haplotype. Thus uga was only expressed in fish that inherited haplotype D, as defined by the unique SNP pattern for zbtb22b along with the divergent psmb8f allele. These heterozygous haplotype D fish also expressed additional genes depending upon the other co-inherited haplotype (C or F). Among these fish, only fish with haplotype C expressed uja and uka, which were always expressed together, while only fish with haplotype F expressed uha. Similarly, only fish inheriting haplotype E expressed uia and these heterozygous haplotype fish (E/C or E/F) also co-expressed the genes from either haplotype C or haplotype F. Thus four unique Class I gene expression patterns were observed among the heterozygous offspring (Fig. 4b). We conclude therefore that each of the unmapped Class I genes (uga-uka) is linked with a single haplotype, as defined by the SNP patterns of the psmb8 and zbtb22b genes. In support of our haplotype definitions, Chi-square analysis showed that these specific Class I gene and haplotype associations were also highly significant (p<0.001), providing strong support that all of these genes are linked to the same locus on chromosome 19.
Genomic Southern blot analysis
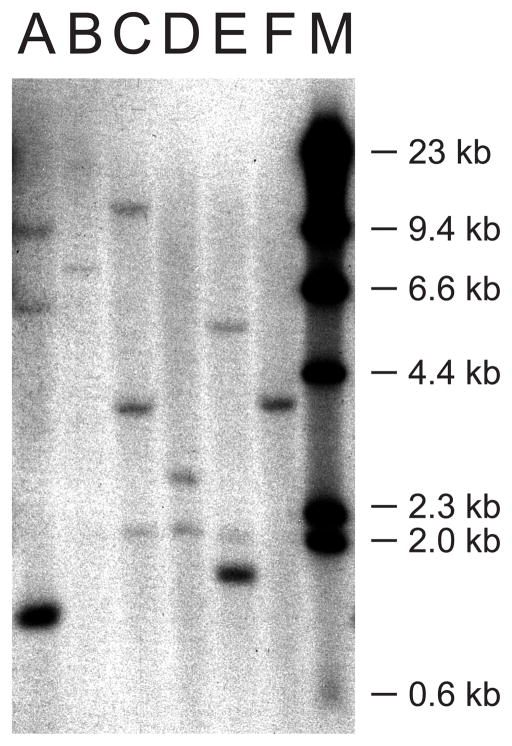
One challenge with using PCR-based methods to examine gene distributions among the haplotypes is the inability to look for genes with unknown sequence. Thus we used genomic Southern blot analysis to measure in a more unbiased manner the number of genes per haplotype and to help confirm our haplotype assignments. We designed a probe against the relatively conserved α3 domain of the uda gene, with the hope that it would cross-react broadly with other Class I genes. This probe proved to be capable of hybridizing to all known zebrafish Class I U lineage genes despite low levels of sequence identity.
We detected three genes with predicted fragment sizes corresponding to uda, uea and ufa for haplotype A (Fig. 5). For haplotype B we detected two bands with sizes corresponding to predicted fragments for uba and uca. Although restriction digest fragment sizes could not be predicted for the remaining haplotypes due to a lack of genomic sequence information, Southern blot analysis shows that each remaining haplotype is associated with a unique band pattern corresponding to one or two Class I genes per haplotype. Bands also had mostly unique sizes, with the apparent exception of one band from haplotype C (uja or uka) and another (uha) from haplotype F. Furthermore, we intermittently detected a band approximately 2kb in size in some DNA samples from haplotypes including C, D and E that corresponds in size to a predicted fragment for the ula gene located on chromosome 22, indicating that this locus may also display haplotype variation. Interestingly, in addition to the expected band for uia from haplotype E, an additional band was always detected that may correspond to an unknown gene also residing within this haplotype. Overall, genomic Southern blot analysis of these different haplotypes was consistent with our qPCR findings showing expression of one to three unique genes per chromosome 19 haplotype.
Fig. 5.
Genomic Southern blot analysis of chromosome 19 haplotypes A through F. Lane M contains DNA Molecular Weight Marker II (Roche). Lane A has genomic DNA obtained from fish homozygous for haplotype A with predicted bands for uda, uea and ufa at 10,283, 1349, and 6145 bp, respectively. Lane B (Haplotype B) displays two bands consistent with predicted sizes for uba and uca at 18,913 and 7509 bp, respectively. For the remaining haplotypes no genomic sequence information is available, thus restriction digest fragment sizes could not be predicted. For haplotype C (Lane C), bands were observed at 4 and 7 kb, corresponding to the two genes expressed by this haplotype, uja and uka. We note that an additional band at approximately 2 kb was detected in samples from haplotypes C, D, and E, likely representing hybridization with an uncharacterized U lineage gene ula found on chromosome 22, as the predicted restriction fragment of this gene is 2041 bp. Haplotype D (Lane D) had only one specific band at approximately 2.5 kb, consistent with our expression data indicating only the presence of uga. Haplotype E (Lane E) had bands at approximately 5.5 and 1.5 kb. Thus, in addition to uia, another gene with unknown sequence is likely to be found in Haplotype E. Haplotype F (Lane F) had only one specific band at approximately 4 kb, consistent with the expression of only uha
Sequence comparisons to identify Class I gene polymorphisms
The full-length sequences for the Class I genes associated with each haplotype were amplified from cDNA and analyzed for polymorphisms. Gene sequences amplified from our fish populations lacked polymorphism, likely because most of these fish are offspring of two founder fish that were inbred to obtain homozygous fish at the desired haplotypes (de Jong et al. 2011). When comparing the database sequences with cDNA sequences identified for the various genes from our fish, we noted very few or no differences, limited predominantly to one or two SNPs. For example, the uga sequence for the uga cDNA amplicons obtained from our haplotype D fish was identical to that of a previous clone found in a sequence library.
Furthermore, sequences derived from our fish showed matches to some but not all cDNA variants for each gene found within sequence libraries. Not all of the sequence variants deposited by other investigators have yet been assigned to specific lines of fish. For example, an alternative version of the ufa sequence was found in the sequence database (but not in our fish) that would introduce a frameshift in the 3′ portion of the gene and that would be predicted to impact function. These variants are summarized in Table 1.
Only two genes were found to have sequences in our fish without corresponding cDNAs in sequence databases. The uia sequence obtained from our fish had two non-synonymous SNPs relative to the loc550387 reference sequence, with R->H near the start of the α2 domain, and T->I in the transmembrane/cytoplasmic portion of the molecule. This uia sequence has been deposited as GenBank accession number KC626502. The uka sequence obtained from our fish had a single synonymous SNP and also a non-synonymous SNP resulting in an I->F amino acid change in the cytoplasmic tail region. This uka sequence has been deposited as GenBank accession number KC626503.
Since no full-length cDNA clone for uca has yet been described, we used predicted transcript ENSDART00000133954 to design primers. However, sequencing of uca amplicons obtained from our fish showed that an additional exon was spliced in the transmembrane/cytoplasmic domain region relative to the predicted transcript. Thus this uca gene product apparently contains 443 amino acids, nearly 100 amino acids longer than the other MHC genes we examined. However, it remains unclear whether this represents the only transcript from the uca gene.
Conserved sequence properties suggest classical gene function
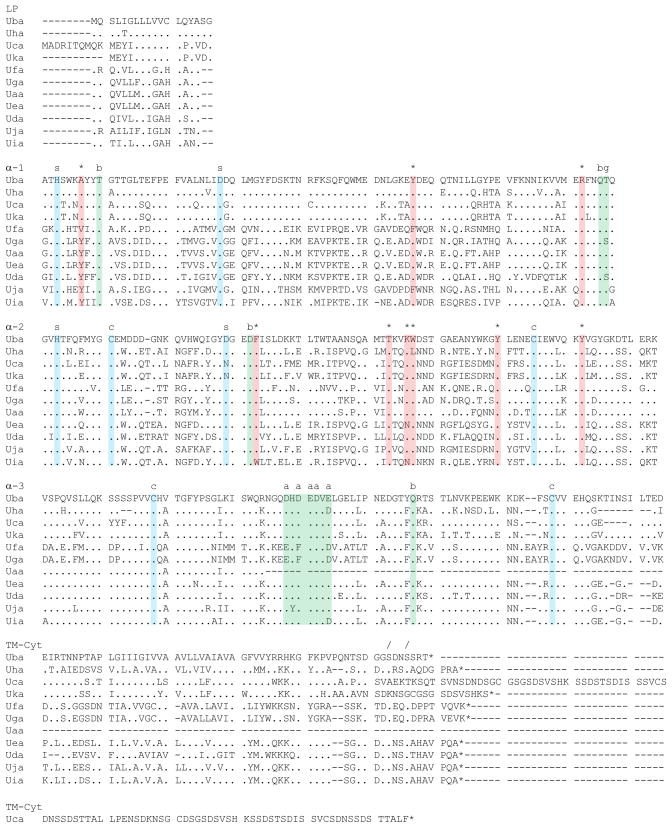
We next examined the sequences of these zebrafish Class I genes in an effort to identify those with properties most consistent with classical gene function. Several important characteristics are highlighted in the multiple sequence alignment (Fig. 6). Structural features include four cysteine residues within the α2 and α3 domains that are absolutely conserved among the eleven genes. Putative salt bridges include two in the α1 domain that are completely conserved among all eleven genes, and two within the α2 domain. Several predicted sites of contact with β2-microglobulin are completely conserved within all three domains. The CD8 co-receptor interaction region found within the α3 domain is also well-conserved, comprised of at least four acidic residues. The glycosylation motif (NQT/S) is present within all sequences near the 3′ end of the α1 domain. Finally, a putative phosphorylation motif (SXXS) is conserved within the cytoplasmic tail of the genes. Beyond these conserved sequence landmarks, this alignment demonstrates the remarkably high levels of sequence variation present between the different zebrafish Class I genes.
Fig. 6.
Multiple sequence alignment for zebrafish Class I U lineage genes. Coding sequences were aligned and translated using the MEGA 4 program. This analysis emphasizes the high levels of diversity present throughout the sequences. Conserved structural residues are highlighted in blue, predicted peptide anchor residues are highlighted in red, and predicted sites of protein-protein interactions are highlighted in green. Within the alignment, dots indicate amino acids that are identical and dashes indicate gaps in the sequence. Conserved features are also indicated with symbols above the alignment: predicted salt bridges (s), peptide anchor residues (*), phosphorylation sites (/), acidic residues for binding to CD8 (a), β2-microglobulin contacts (b), cysteine residues involved in disulfide bonds (c), and a glycosylation site (g)
Interestingly, amino acid variation is present even at the predicted peptide anchor residues (Table 3). Overall, putative peptide anchor residues (based on an alignment with other vertebrate Class I MHC genes such as human HLA-A2) are mostly conserved within Danio rerio (zebrafish) Class I genes. However, some genes have one or two changes among their nine putative peptide anchor residues relative to the consensus fish sequence. Several of these substitutions have also been observed within the genes (some classical) of other fish species (see discussion).
Table 3.
Zebrafish Class I putative peptide anchor residues
| N-terminus | C-terminus | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| 7 | 59 | 159 | 171 | 84 | 123 | 143 | 146 | 147 | |
| HLA | Y | Y | Y | Y | Y | Y | T | K | W |
| Fish | Y | Y | Y | Y | R | F | T | K | W |
| Uaa | Y | Y | Y | Y | R | F | T | K | W |
| Uba | A | Y | Y | Y | R | F | T | K | W |
| Uca | A | Y | Y | Y | R | F | T | K | W |
| Uda | Y | Y | Y | Y | R | F | T | K | W |
| Uea | Y | Y | Y | Y | R | F | T | N | W |
| Ufa | V | F | Y | Y | R | F | T | K | W |
| Uga | Y | Y | Y | Y | R | F | T | K | W |
| Uha | A | Y | Y | Y | R | F | T | K | L |
| Uia | Y | Y | Y | Y | R | W | T | N | W |
| Uja | Y | F | Y | Y | R | F | T | K | W |
| Uka | A | Y | Y | Y | R | F | T | K | W |
Putative peptide anchor residues, based on an alignment with other vertebrate Class I MHC genes. HLA represents a consensus of classical human Class I sequences (HLA-A, B, C). Fish represents a consensus sequence from teleost Class I MHC genes thought to be classical. Anchor residue numbering is based on the human HLA-A2 gene
Discussion
Our findings provide new insights into the immunogenetics of histocompatibility in zebrafish. We characterize novel zebrafish Class I U lineage genes, and present data suggesting that most may be considered candidate classical MHC genes. A total of ten full-length Class I genes were found within zebrafish cDNA libraries (uba-uka, Table 1). We confirmed expression for all ten of these genes among assorted fish with a total of six unique haplotypes at chromosome 19, thus permitting assignment of each of these genes among the different haplotypes (Fig. 7).
Fig. 7.
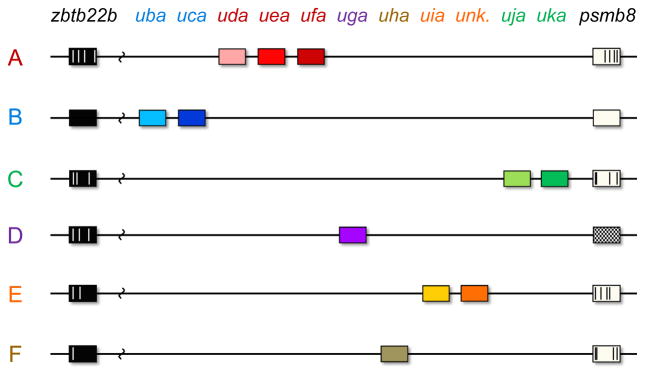
Model of zebrafish Class I gene distribution among six haplotypes. The specific order of the genes for haplotypes C through F is presently unknown, but MHC genes associated with each haplotype are presented in the context of known linked gene polymorphisms in psmb8 and zbtb22b that distinguish these haplotypes from one another and from haplotypes A and B. Specific polymorphisms are illustrated within each allele relative to the haplotype B reference sequence. For haplotype D the divergent psmb8f allele is depicted with cross-hatches. An unknown Class I gene associated with haplotype E is also indicated
Our data show that disparate Class I MHC haplotypes in zebrafish express distinct sets of genes, such that varying numbers of genes and altogether different genes are present within each haplotype of this species. The absence of gene overlap for these haplotypes indicates that genomic large-scale structural variation may represent an important mechanism of maintaining diversity of the antigen-binding repertoire in zebrafish. In contrast, previous studies in salmonids (Aoyagi et al. 2002) have identified a single classical Class I gene on each haplotype, while medaka have two genes per haplotype (Nonaka and Nonaka 2010). The Class I genes are highly polymorphic in these fish species with a set number of classical genes per haplotype. Extensive haplotype variation in gene copy number exemplified by region configuration polymorphism (also called large-scale structural variation) has been identified at MHC loci in other species such as rhesus macaques and cattle (Doxiadis et al. 2011; Codner et al. 2012). However, in these studies a single classical gene is typically found in the majority of haplotypes examined, with variability most apparent by the presence or absence of additional Class I genes, many of which are shared among several haplotypes. Other studies with rhesus macaques have examined genomic regions with extreme variation in gene copy number (Otting 2005; Blokhuis 2010). Although distributed in distinctive patterns throughout disparate haplotypes, in general these genes appear to be relatively closely related. In contrast, the haplotype diversity that we have described here in zebrafish is remarkable in that each haplotype expresses a unique set of one to three genes out of a variety of distantly related Class I genes, and also includes diversity in additional Class I pathway genes. This is even more striking when considering that most of these divergent Class I genes are also likely to have classical function.
Classical MHC gene definition is informed by several factors including expression levels, polymorphism, evidence of selection, and ultimately function through immune response to pathogens or allograft rejection. For MHC gene studies, substitutions within peptide anchor positions have previously been used to help differentiate between classical and non-classical gene in many species (Kaufman, Salomonsen, and Flajnik 1994; Kruiswijk et al. 2002). In mammals, substitutions at one or two of these anchor residues have only rarely been found within classical genes, and accumulation of these substitutions indicates non-classical gene function in general. Although peptide anchor residue substitutions may be considered an indication that these zebrafish genes lack classical function, several of the substitutions have been described among Class I genes from other teleost species (Aoyagi et al. 2002; Kiryu et al. 2005; Miller et al. 2006; Lukacs et al. 2010; Chen et al. 2010; Urabe et al. 2011; Nonaka et al. 2011) including some that are considered to have classical gene function. For example, the Y7A substitution found in zebrafish Class I genes such as uba has also been found among salmonid classical alleles. While one simple explanation is shared ancestry, such widespread conservation of these unique substitutions across great phylogenetic distances (and despite high divergence of the individual genes) instead indicates that the substitutions may have functional roles with respect to antigen presentation by the Class I molecules. Therefore these substitutions may not necessarily disqualify these genes from classical function.
Considering that we identified two haplotypes (D and F) that each carry only a single Class I gene, both of these isolated genes (uga and uha) would be expected to maintain classical function, as loss of function for the sole classical gene within a given haplotype would likely be strongly selected against. Similarly, we characterized two additional haplotypes (B and C) in which only a single gene each was expressed in each at levels comparable to β2-microglobulin, again indicating that these two highly expressed genes (uba and uja) may be considered classical. In contrast, the three expressed Class I genes from haplotype A (uda, uea and ufa) may all be considered candidate classical genes. While the status of a possible second gene associated with haplotype E remains unknown, the known gene (uia) with an expression level comparable to β2-microglobulin may at present be considered a candidate classical gene.
In zebrafish, these genes have evidently been maintained separately for long periods of time, based on their extremely high levels of sequence diversity, similar to what has been found in alleles of uba in salmonids (Shum et al. 2001). Furthermore, divergent alleles of psmb8 are also distributed among these haplotypes, which may also confer additional Class I pathway diversity. Thus, the multiple disparate haplotypes that we characterize in zebrafish may contribute to the functional pathway diversity within this species by isolating distinct sets of MHC genes and flanking genes that may each contribute their unique functional properties within the context of specific haplotypes.
Genomic sequences are available for haplotypes A and B, but genomic sequence data for other zebrafish Class I haplotypes are lacking, and would help to shed further light on their diversity. Interestingly, it has been suggested that the genomic sequence for haplotype A might actually be assembled from sequences derived from distinct haplotypes (Sambrook, Figueroa, and Beck 2005), possibly with a contig sequence breakpoint within intron 2 that joins the closely related uaa and uea genes. This composite zebrafish haplotype A sequence would thus be analogous to the first composite human MHC sequence (The MHC Sequencing Consortium 1999). Nevertheless, the MHC gene sequences and flanking gene sequences that we obtained from our fish with haplotypes A and B closely match sequences from their respective reference genomic assemblies. In addition, we obtained the expected band sizes for each MHC gene via genomic Southern blot analysis, adding to our confidence in the reliability of these assemblies.
The zbtb22b and psmb8 genes that we use to define our haplotypes have highly conserved linkage to the Class I genes within zebrafish. Moreover, zbtb22b (BING1) and psmb8 (LMP7) are linked to Class I genes found within other vertebrate species including medaka and humans (Matsuo 2002). In zebrafish, zbtb22b and psmb8a sequences from separate haplotypes differ at only a small number of SNPs, with the exception of the divergent psmb8f allele. This unique psmb8f allele has been preserved within only a subset of zebrafish haplotypes, despite being apparently lost from mammals and many other teleosts (Tsukamoto et al. 2012).
In contrast to selective pressure to maintain relative stability in the flanking regions, a different form of selective pressure appears to be operating on the Class I genes themselves. Extreme sequence diversity has been found within individual exons of teleost Class I genes (Shum 2001 and Fig. 6). These remarkably high levels of diversity indicate trans-species polymorphism, which is also supported by specific polymorphisms found shared among the sequence lineages that have been identified in species as divergent as zebrafish and salmon (Grimholt et al. 2002, Kiryu et al. 2005). Therefore selection appears to have helped maintain many ancient Class I sequences within the core of these divergent haplotypes, while simultaneously conserving largely similar flanking sequences. In order to estimate the age of these haplotypes, large numbers of zebrafish from diverse populations are needed to search for evidence of recombination within and between the MHC genes from different haplotypes. Although we have found limited evidence for recombination between MHC genes (data not shown), further examination of haplotypes in other zebrafish populations and genomic sequencing of haplotypes C-F is needed.
While this study reports the most comprehensive analysis of zebrafish Class I genes on chromosome 19 to date, the list of haplotypes studied is unlikely to be exhaustive. Future studies are needed to reveal the full repertoire of gene arrangements at this locus within the zebrafish species. The experiments reported here include a subset of laboratory strains, limited to two clonal golden lines and related AB-derived fish. However, we also detected many of the haplotypes described in this study in additional zebrafish strains including TL and WIK (data not shown), suggesting that these haplotypes may be more widespread among laboratory zebrafish. Much less is known about zebrafish populations in the wild, which may be expected to have even more diverse MHC genes.
We searched zebrafish cDNA libraries in an attempt to identify as many MHC genes as possible, especially those with full-length cDNA sequences or those containing sequences for multiple exons. Several of these genes were found in common among previously published studies (Takeuchi et al. 1995; Bingulac-Popovic et al. 1997; Graser et al. 1998; Michalová et al. 2000), in addition to Zv9, sequence libraries, and our study. However, our current list of zebrafish Class I genes is presumably not comprehensive. Importantly, pseudogenes and genes with low levels of expression may be particularly under-represented within sequence libraries, so that identification and assignment of this type of gene may require genomic sequence information. For example, genomic sequencing will be required to identify the additional Class I gene in haplotype E seen on Southern blot analysis (Fig. 5). Nevertheless, genes with the highest expression levels, including those with properties consistent with being Class Ia (classical) genes, are the gene type most likely to be included in our study.
Antibodies against zebrafish MHC complexes are not yet available, impeding measurement of zebrafish Class I molecules at the cell surface. For this reason our study focused on mRNA expression levels. However, measurement of mRNA for each Class I gene relative to β2-microglobulin within the samples provided a reasonable first assessment of which genes are expressed at physiologically-relevant levels. Interestingly, total Class I expression levels from each of the six haplotypes were similar overall to β2-microglobulin expression. The expression levels and tissue distribution are thus consistent with many of these genes functioning as classical genes, but confirmation via functional studies is still needed in order to exclude the possibility that they could be non-classical MHC genes.
In summary, we provide data regarding the expression and sequence properties of zebrafish Class I genes from divergent haplotypes. While our findings for these genes may be significant from an evolutionary perspective, they may also have practical value as a foundation for future studies. Polymorphic Class I genes form haplotypes that can significantly affect immune responses to pathogens (The International HIV Controllers Study 2010; Grimholt et al. 2003). Knowing the specific chromosome 19 haplotype present in an immune response or transplantation experiment may influence the interpretation of the results. We anticipate other histocompatibility loci will also play a significant role. Realizing the full potential of immune matching will depend in part on uncovering the functional roles of the zebrafish Class I molecules, particularly for antigen presentation and initiation of an immune response, studies that are now underway in our laboratory.
Supplementary Material
Acknowledgments
We thank Leonard Zon for AB-derived zebrafish and Sergei Revskoy for clonal zebrafish. We are grateful to Aye Chen and Yi Zhou for sharing primers for haplotype determination. We thank Michael Peters, Wilfredo Marin, Jissy Cyriac, William Tian, William Buikema and the University of Chicago sequencing core for technical assistance. We thank Xinan (Holly) Yang for help with statistical analyses. We also thank Hayley Dirscherl, Jeffrey Yoder, Erin Adams, Anita Chong, Derek Stemple, Amy Singer and members of the de Jong laboratory for helpful discussion. This work was supported by grants from the NIDDK (K08DK074595 and R03DK091497), as well as funds from the University of Chicago Cancer Research Foundation Auxiliary Board.
References
- Aoyagi K, Dijkstra JM, Xia C, Denda I, Ototake M, Hashimoto K, Nakanishi T. Classical MHC Class I genes composed of highly divergent sequence lineages share a single locus in rainbow trout (Oncorhynchus mykiss) The Journal of Immunology. 2002;168:260–273. doi: 10.4049/jimmunol.168.1.260. [DOI] [PubMed] [Google Scholar]
- Bingulac-Popovic J, Figueroa F, Sato A, Talbot WS, Johnson SL, Gates M, Postlethwait JH, Klein J. Mapping of MHC Class I and Class II regions to different linkage groups in the zebrafish, Danio rerio. Immunogenetics. 1997;46:129–134. doi: 10.1007/s002510050251. [DOI] [PubMed] [Google Scholar]
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A 2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. Clinical Microbiology Reviews. 2009;22:370–385. doi: 10.1128/CMR.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhuis JH, van der Wiel MK, Doxiadis GG, Bontrop RE. The mosaic of KIR haplotypes in rhesus macaques. Immunogenetics. 2010;62:295–306. doi: 10.1007/s00251-010-0434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. Current Protocols in Protein Science A.4G.1–8. 2001. Southern blotting. [DOI] [PubMed] [Google Scholar]
- Chen W, Jia Z, Zhang T, et al. MHC Class I presentation and regulation by IFN in bony fish determined by molecular analysis of the Class I locus in grass carp. The Journal of Immunology. 2010;185:2209–2221. doi: 10.4049/jimmunol.1000347. [DOI] [PubMed] [Google Scholar]
- Chinen J, Buckley RH. Transplantation immunology: Solid organ and bone marrow. Journal of Allergy and Clinical Immunology. 2010;125:S324–S335. doi: 10.1016/j.jaci.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codner GF, Birch J, Hammond JA, Ellis SA. Constraints on haplotype structure and variable gene frequencies suggest a functional hierarchy within cattle MHC class I. Immunogenetics. 2012;64:435–445. doi: 10.1007/s00251-012-0612-6. [DOI] [PubMed] [Google Scholar]
- de Jong JL, Burns CE, Chen AT, Pugach E, Mayhall EA, Smith AC, Feldman HA, Zhou Y, Zon LI. Characterization of immune-matched hematopoietic transplantation in zebrafish. Blood. 2011;117:4234–4242. doi: 10.1182/blood-2010-09-307488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong JL, Zon LI. Histocompatibility and hematopoietic transplantation in the zebrafish. Advances in Hematology. 2012;2012:1–8. doi: 10.1155/2012/282318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra JM, Katagiri T, Hosomichi K, Yanagiya K, Inoko H, Ototake M, Aoki T, Hashimoto K, Shiina T. A third broad lineage of major histocompatibility complex (MHC) class I in teleost fish; MHC class II linkage and processed genes. Immunogenetics. 2007;59:305–321. doi: 10.1007/s00251-007-0198-6. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, Groot N, Otting N, Blokhuis JH, Bontrop RE. Genomic plasticity of the MHC Class I A region in rhesus macaques: extensive haplotype diversity at the population level as revealed by microsatellites. Immunogenetics. 2011;63:73–83. doi: 10.1007/s00251-010-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser R, Vincek V, Takami K, Klein J. Analysis of zebrafish MHC using BAC clones. Immunogenetics. 1998;47:318–325. doi: 10.1007/s002510050364. [DOI] [PubMed] [Google Scholar]
- Grimholt U, Drabløs F, Jørgensen SM, Høyheim B, Stet RJ. The major histocompatibility class I locus in Atlantic salmon (Salmo salar L.): polymorphism, linkage analysis and protein modelling. Immunogenetics. 2002;54:570–581. doi: 10.1007/s00251-002-0499-8. [DOI] [PubMed] [Google Scholar]
- Grimholt U, Larsen S, Nordmo R, Midtlyng P, Kjoeglum S, Storset A, Saebø S, Stet RJ. MHC polymorphism and disease resistance in Atlantic salmon (Salmo salar); facing pathogens with single expressed major histocompatibility Class I and Class II loci. Immunogenetics. 2003;55:210–219. doi: 10.1007/s00251-003-0567-8. [DOI] [PubMed] [Google Scholar]
- Gupta T, Mullins MC. Dissection of organs from the adult zebrafish. Journal of Visualized Experiments. 2010 doi: 10.3791/1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan J, Wang T, Haagenson M, et al. Evaluation of HLA matching in unrelated hematopoietic stem cell transplantation for nonmalignant disorders. Blood. 2012;120:2918–2924. doi: 10.1182/blood-2012-03-417758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S, Nei M. Pattern of nucleotide substitution at Major Histocompatibility Complex Class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Ignatius MS, Langenau DM. Zebrafish as a model for cancer self-renewal. Zebrafish. 2009;6:377–387. doi: 10.1089/zeb.2009.0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman J, Salomonsen J, Flajnik MF. Evolutionary conservation of MHC Class I and Class II molecules: different yet the same. Seminars in Immunology. 1994;6:411–424. doi: 10.1006/smim.1994.1050. [DOI] [PubMed] [Google Scholar]
- Kiryu I, Dijkstra JM, Sarder RI, Fujiwara A, Yoshiura Y, Ototake M. New MHC class Ia domain lineages in rainbow trout (Oncorhynchus mykiss) which are shared with other fish species. Fish & Shellfish Immunology. 2005;18:243–254. doi: 10.1016/j.fsi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, Jones PP, Parham P, Wakeland EK, Watkins DI. Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics. 1990;31:217–219. doi: 10.1007/BF00204890. [DOI] [PubMed] [Google Scholar]
- Klein J, Satta Y, O’hUigin C, Takahata N. The molecular descent of the major histocompatibility complex. Annual Review of Immunology. 1993;11:269–295. doi: 10.1146/annurev.iy.11.040193.001413. [DOI] [PubMed] [Google Scholar]
- Korbie DJ, Mattick JS. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nature Protocols. 2008;3:1452–1456. doi: 10.1038/nprot.2008.133. [DOI] [PubMed] [Google Scholar]
- Kruiswijk CP, Hermsen TT, Westphal AH, Savelkoul HF, Stet RJ. A novel functional Class I lineage in zebrafish (Danio rerio), carp (Cyprinus carpio), and large barbus (Barbus intermedius) showing an unusual conservation of the peptide binding domains. The Journal of Immunology. 2002;169:1936–1947. doi: 10.4049/jimmunol.169.4.1936. [DOI] [PubMed] [Google Scholar]
- Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H. Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunological reviews. 2002;190:95–122. doi: 10.1034/j.1600-065x.2002.19008.x. [DOI] [PubMed] [Google Scholar]
- Lan C, Tang R, Un San Leong I, Love D. Quantitative Real-Time RT-PCR (qRT-PCR) of zebrafish transcripts: Optimization of RNA extraction, quality control considerations, and data analysis. Cold Spring Harbor Protocols. 2009;10:1–12. doi: 10.1101/pdb.prot5314. [DOI] [PubMed] [Google Scholar]
- Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lukacs M, Harstad H, Bakke H, Beetz-Sargent M, McKinnel L, Lubieniecki K, Koop B, Grimholt U. Comprehensive analysis of MHC Class I genes from the U-, S-, and Z-lineages in Atlantic salmon. BMC Genomics. 2010;11:154–170. doi: 10.1186/1471-2164-11-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo MY, Asakawa S, Nobuyoshi S, Kimura H, Nonaka M. Nucleotide sequence of the MHC Class I genomic region of a teleost, the medaka (Oryzias latipes) Immunogenetics. 2002;53:930–940. doi: 10.1007/s00251-001-0427-3. [DOI] [PubMed] [Google Scholar]
- Meeker ND, Hutchinson SA, Ho L, Trede NS. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Bio Techniques. 2007;43:610–613. doi: 10.2144/000112619. [DOI] [PubMed] [Google Scholar]
- Michalová V, Murray BW, Sültmann H, Klein J. A contig map of the MHC Class I genomic region in the zebrafish reveals ancient synteny. The Journal of Immunology. 2000;164:5296–5305. doi: 10.4049/jimmunol.164.10.5296. [DOI] [PubMed] [Google Scholar]
- Miller KM, Li S, Ming TJ, Kaukinen KH, Schulze AD. The salmonid MHC Class I: more ancient loci uncovered. Immunogenetics. 2006;58:571–589. doi: 10.1007/s00251-006-0125-2. [DOI] [PubMed] [Google Scholar]
- Mione MC, Trede NS. The zebrafish as a model for cancer. Disease Models & Mechanisms. 2010;3:517–523. doi: 10.1242/dmm.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizgirev IV, Revskoy S. A new zebrafish model for experimental leukemia therapy. Cancer biology & therapy. 2010;9:895–902. doi: 10.4161/cbt.9.11.11667. [DOI] [PubMed] [Google Scholar]
- Nikolich-Zugich J. The role of MHC polymorphism in anti-microbial resistance. Microbes and Infection. 2004;6:501–512. doi: 10.1016/j.micinf.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Nonaka MI, Aizawa K, Mitani H, Bannai HP, Nonaka M. Retained Orthologous Relationships of the MHC Class I Genes during Euteleost Evolution. Molecular Biology and Evolution. 2011;28:3099–3112. doi: 10.1093/molbev/msr139. [DOI] [PubMed] [Google Scholar]
- Nonaka MI, Nonaka M. Evolutionary analysis of two classical MHC Class I loci of the medaka fish, Oryzias latipes: haplotype-specific genomic diversity, locus-specific polymorphisms, and interlocus homogenization. Immunogenetics. 2010;62:319–332. doi: 10.1007/s00251-010-0426-3. [DOI] [PubMed] [Google Scholar]
- Otting N, Heijmans CM, Noort RC, de Groot NG, Doxiadis GG, van Rood JJ, Watkins DI, Bontrop RE. Unparalleled complexity of the MHC class I region in rhesus macaques. PNAS. 2005;102:1626–1631. doi: 10.1073/pnas.0409084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P. MHC Class I molecules and KIRs in human history, health and survival. Nature Reviews Immunology. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- Renshaw SA, Trede NS. A model 450 million years in the making: zebrafish and vertebrate immunity. Disease Models and Mechanisms. 2011;5:38–47. doi: 10.1242/dmm.007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JR, Cook RG. MHC Class Ib molecules bridge innate and acquired immunity. Nature Reviews Immunology. 2005;5:459–471. doi: 10.1038/nri1635. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Figueroa F, Beck S. A genome-wide survey of Major Histocompatibility Complex (MHC) genes and their paralogues in zebrafish. BMC Genomics. 2005;6:152–161. doi: 10.1186/1471-2164-6-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarder MR, Fischer U, Dijkstra JM, Kiryu I, Yoshiura Y, Azuma T, Kollner B, Ototake M. The MHC class I linkage group is a mojor determinant in the in vivo rejection of allogeneic erythrocytes in rainbow trout (Oncorhynchus mykiss) Immunogenetics. 2003;55:315–324. doi: 10.1007/s00251-003-0587-4. [DOI] [PubMed] [Google Scholar]
- Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. Journal of Human Genetics. 2009;54:15–39. doi: 10.1038/jhg.2008.5. [DOI] [PubMed] [Google Scholar]
- Shum BP, Guethlein L, Flodin LR, Adkison MA, Hedrick RP, Nehring B, Stet RJ, Secombes C, Parham P. Modes of Salmonid MHC Class I and II Evolution Differ from the Primate Paradigm. Journal of Immunology. 2001;166:3297–3308. doi: 10.4049/jimmunol.166.5.3297. [DOI] [PubMed] [Google Scholar]
- Smith AC, Raimondi AR, Salthouse CD, Ignatius MS, Blackburn JS, Mizgirev IV, Storer NY, de Jong JL, Chen AT, Zhou Y. High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood. 2010;115:3296–3303. doi: 10.1182/blood-2009-10-246488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan C, Kim CH. Innate immune system of the zebrafish, Danio rerio. Innate Immunity of Plants, Animals, and Humans. 2008:113–133. [Google Scholar]
- Sültmann H, Murray BW, Klein J. Identification of seven genes in the Major Histocompatibility Complex Class I region of the zebrafish. Scandinavian Journal of Immunology. 2000;51:577–585. doi: 10.1046/j.1365-3083.2000.00729.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Figueroa F, O’hUigin C, Klein J. Cloning and characterization of Class I MHC genes of the zebrafish, Brachydanio rerio. Immunogenetics. 1995;42:77–84. doi: 10.1007/BF00178581. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- The International HIV Controllers Study. The major genetic determinants of HIV-1 control affect HLA Class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The MHC Sequencing Consortium. Complete sequence and gene map of a human major histocompatibility complex. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Fast release clones: A high throughput expression analysis. 2004. [Google Scholar]
- Tsukamoto K, Hayashi S, Matsuo MY, Nonaka MI, Kondo M, Shima A, Asakawa S, Shimizu N, Nonaka M. Unprecedented intraspecific diversity of the MHC class I region of a teleost medaka, Oryzias latipes. Immunogenetics. 2005;57:420–431. doi: 10.1007/s00251-005-0009-x. [DOI] [PubMed] [Google Scholar]
- Tsukamoto K, Miura F, Fujito N, Yoshizaki G, Nonaka M. Long-lived dichotomous lineages of the proteasome subunit beta type 8 (PSMB8) gene surviving more than 500 million years as alleles or paralogs. Molecular Biology and Evolution. 2012;29:3071–3079. doi: 10.1093/molbev/mss113. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3—new capabilities and interfaces. Nucleic Acids Research. 2012;40:e115–e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urabe S, Sonamoto T, Sameshima S, Unoki-Kato Y, Nakanichi T, Nakao M. Molecular characterization of MHC Class I and beta-2 microglobulin in a clonal strain of ginbuna crucian carp, Carassius auratus langsdorfii. Fish and Shellfish Immunology. 2011;31:469–474. doi: 10.1016/j.fsi.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Westerfield M. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 4. University of Oregon Press; Eugene, OR: 2000. The Zebrafish Book. [Google Scholar]
- Zhou Y, Zon LI. Methods in Cell Biology. Elsevier; 2011. The Zon Laboratory Guide to Positional Cloning in Zebrafish; pp. 287–309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.