Abstract
Defining the functional status of host-associated microbial ecosystems has proven challenging owing to the vast number of predicted genes within the microbiome and relatively poor understanding of community dynamics and community–host interaction. Metabolomic approaches, in which a large number of small molecule metabolites can be defined in a biological sample, offer a promising avenue to ‘fingerprint' microbiota functional status. Here, we examined the effects of the human gut microbiota on the fecal and urinary metabolome of a humanized (HUM) mouse using an optimized ultra performance liquid chromatography–mass spectrometry-based method. Differences between HUM and conventional mouse urine and fecal metabolomic profiles support host-specific aspects of the microbiota's metabolomic contribution, consistent with distinct microbial compositions. Comparison of microbiota composition and metabolome of mice humanized with different human donors revealed that the vast majority of metabolomic features observed in donor samples are produced in the corresponding HUM mice, and individual-specific features suggest ‘personalized' aspects of functionality can be reconstituted in mice. Feeding the mice a defined, custom diet resulted in modification of the metabolite signatures, illustrating that host diet provides an avenue for altering gut microbiota functionality, which in turn can be monitored via metabolomics. Using a defined model microbiota consisting of one or two species, we show that simplified communities can drive major changes in the host metabolomic profile. Our results demonstrate that metabolomics constitutes a powerful avenue for functional characterization of the intestinal microbiota and its interaction with the host.
Keywords: metabolomics, humanized mice, gut microbiota, UPLC-MS
Introduction
The metabolic capacity of the human body is a product of resident microbial communities integrated with human cells. This view has generated an increasing interest in defining the effect of the resident microbiota on the set of small molecule metabolites associated with a host. A frontier in intestinal microbiota studies is to extend our understanding of host-resident microbes beyond microbial species or genes, to link microbial community structures with microbial functions that could impact human health. Metabolomics has emerged as a technique that allows the functional status of host–microbial integration to be defined in biological fluids and tissues, such as urine, blood and feces. Metabolites originate from a complex network of chemical and biochemical pathways and offer a chemical fingerprint of microbiota functional status. Although confirming the identity of individual small molecules is currently challenging, when established, this information offers insight into the chemical messengers that mediate microbe–microbe and microbe–host interactions. Ultra performance liquid chromatography (UPLC) coupled with high-resolution mass spectrometry (MS) allows reproducible measurements of a wide range of metabolites in a complex sample.
Mice provide an experimentally tractable model to investigate how resident microbial communities impact small molecule metabolites present in host biofluids. Comparisons of conventional mice that harbor a complex microbiota with germ-free (GF) mice that lack a microbiota have established the profound microbial impact on host metabolomic profile (Claus et al., 2008; Swann et al., 2011a). Manipulation of microbiota composition via oral antibiotic treatment, cohousing mice with different microbiotas, and creation of simplified microbiotas in ex-GF (gnotobiotic) mice (Yap et al., 2008) have illustrated the dynamic nature of microbial host metabolomic integration to influence metabolomic signatures (Goodwin et al., 1994). Additional experimental manipulation of external factors known to modify the gut microbiota (for example, dietary change and probiotic treatment) constitute important first steps toward elucidating the mechanisms that drive alterations in microbe-dependent metabolomic phenotypes (Marchesi et al., 2007; Holmes et al., 2008; Kessner et al., 2008; Wikoff et al., 2009).
Although the distal gut microbiota of mice and humans harbor the same bacterial phyla, 85% of genera and microbial species found in mice are not seen in humans (Ley et al., 2005). HUM mice (ex-GF mice colonized with a human microbiota) are emerging as a powerful model for studying human-relevant microbes in a controlled experimental setting (Mallett et al., 1987; Bowey et al., 2003). Here we use HUM mice as a model for studying metabolomic changes within urine and feces. To define microbe-related signatures in the urine and feces of this mouse model, we have optimized a nontargeted metabolomics UPLC-MS method. We have applied this method to samples from HUM and conventional mice, demonstrating that the mice harboring the human microbiota express a set of metabolites distinct from that of their mouse microbiota-harboring counterparts. We have established a significant overlap in microbe-dependent metabolomes of mice humanized with different donor samples. Colonization of GF mice with one or two microbial species shows that simple models of the microbiota can drive major changes in host metabolomic profile, although distinct from the impact exerted by a complex microbiota. Our results demonstrate that the application of metabolomic analysis to HUM mice constitutes a powerful approach for characterization of intestinal microbiota functional status.
Materials and methods
Mice
GF Swiss-Webster (SW) and C57BL/6J and conventional (RF, Taconic Inc., Cambridge city, IN, USA) SW mice were maintained as previously described (Sonnenburg et al., 2010). Humanization was performed using human fecal samples obtained from healthy anonymous donors. Frozen feces were thawed by dilution in an equal volume of pre-reduced phosphate-buffered saline in anerobic conditions and 0.2 ml non-settling material was gavaged into GF-recipient mice. Mice were fed standard diet (Purina LabDiet 5K67) or polysaccharide-deficient diet (Bio-Serv, Frenchtown, NJ, USA; Sonnenburg et al., 2010). GF mice were mono- or bi-associated using oral gavage with 108 colony-forming units of Bacteroides thetaiotaomicron (Bt) VPI-5482 and/or Bifidobacterium longum (Bl) NCC2705. Bacteria were cultured under anerobic conditions at 37 °C in tryptone-yeast extract-glucose medium (Sonnenburg et al., 2010) or Reinforced Clostridial Medium (Becton Dickinson and Company, Sparks, MD, USA). All experiments were done according to the A-PLAC, the Stanford IACUC. Urine and fecal samples were collected and placed in a freezer at −70 °C within 30 min of collection until analysis.
Urine sample preparation
Twenty microlitres of urine sample was mixed with 40 μl of 10 mM ammonium formate. Five microlitres of the mixture was used in the reverse-phase liquid chromatography (RPLC)-electrospray ionization (ESI) analysis. When reverse-phase analysis was completed, samples were decapped, 10 μL aliquots of remaining samples were transferred into another set of high-performance liquid chromatography vials and resuspended with 30 μL acetonitrile for hydrophilic interaction chromatography (HILIC)-ESI analysis, using 10 μl per injection.
Fecal sample preparation
Five hundred microlitres of 1:1 methanol: acidified water (0.1% formic acid) was added to 10–30 mg of feces. Tubes were vortexed at 4 °C for 15 min. After vortexing, the tube was placed in a 4 °C centrifuge at 500 × g for 15 min. The supernatant was pipetted into a new tube, with care to minimize disruption of the pellet. The resulting fecal water samples were extracted by using solid-phase OASIS extraction cartridges (Waters, Mifford, MA, USA). Metabolites were eluted with 500 μl methanol, dried in a speedvac and resuspended in 100 μl of initial mobile phase composition corresponding to the two different chromatography methods below. Five microlitres of the 100 μl sample was used in the analysis.
RPLC conditions
Chromatographic separation was performed on a 150-mm × 2.1-mm Kinetex 1.7 μm C18 column(Phenomenex, Torrance, CA, USA) using a ACQUITY Ultra Performance Liquid Chromatography system (Waters). The flow rate was 0.25 ml min−1. Column was held at 40 °C. Solvent A was 10 mM ammonium formate in water and solvent B was 10 mM ammonium formate in methanol. The gradient started at 5% B and linearly increased to 10% B at 14 min; then linearly increased to 100% B at 22 min; and held at 100% B for 5 min. The column was equilibrated at 5% B for 3 min before starting the run. First 1.3 min of mobile phase flow were diverted from the ion source into the waste.
HILIC conditions
Chromatographic separation was performed using the 150-mm × 2.1-mm Cogent Diamond Hydride, 4 μm 100 A HILIC column (MicroSolv, Eatontown, NJ, USA). Column temperature was 40 °C. The flow rate was 0.4 ml min−1. Solvent A was 10% acetonitrile with 15 mM ammonium formate and solvent B was 90% acetonitrile with 15 mM ammonium formate. The gradient started at 100% B for 1 min, followed by a linear decrease to 30% B over 14 min, a hold at 30% B for 1 min, and a 0.6 ml min−1 hold at 100% B for 1 min. The column was equilibrated at 100% B for 5 min before starting the run.
MS conditions
MS was performed on the Exactive (Thermo Fisher, Waltham, MA, USA) orbitrap mass spectrometer operated in positive and negative electrospray mode and controlled by Xcalibur 2.1 software. The scan range was from 70 to 800 m/z, at 50 000 full width at half maximum resolution. For positive mode (ESI+), we used the following conditions: sheath gas flow rate 40 (arbitrary units), auxilary gas flow rate 8 (arbitrary units), sweep gas flow rate 1 (arbitrary units), spray voltage 3.5 kV, capillary temperature 275 °C, capillary voltage −60 V, tube lens voltage −100 V and skimmer voltage −20 V. For negative electrospray mode (ESI−), the following conditions were used: sheath gas flow rate 30 (arbitrary units), auxiliary gas flow rate 4 (arbitrary units), sweep gas flow rate 1 (arbitrary units), spray voltage 3.5 kV, capillary temperature 275 °C, capillary voltage −60 V, tube lens voltage −100 V and skimmer voltage −21 V.
Data analysis
Profile mode raw data from the individual analyses were converted to centroid mode mzXML files with msconvert and subjected to nonlinear data alignment by xcms (Smith et al., 2006; Kessner et al., 2008). Intensity data files were imported into the web-based program MetaboAnalyst for statistical univariate and multivariate analysis (http://www.metaboanalyst.ca). Unbiased metabolite identification was determined using METLIN database using a maximum error of 3 p.p.m., empirical formulas (Smith et al., 2005). When possible, compound identity validations were performed by analyzing pure compounds (Supplementary Figure S8). Tryptamine, indoxyl glucuronide, creatine, creatinine and 5-hydroxyindole were obtained from Sigma-Aldrich (St Louis, MO, USA). Features that vary significantly between the two sample sets were evaluated by univariate statistical analysis, and significance was defined as >10-fold change with P-value<0.01.
Detection of indole-3-lactic acid production by liquid chromatography-tandem mass spectrometry
Overnight culture of Bl NCC2705 was grown in Reinforced Clostridial Medium supplemented with 0.1% L-tryptophan (Sigma-Aldrich). Two hundred microlitres of culture before and after growth were collected, centrifuged (2700 × g, 10 min) and filtered. Four volumes of cold methanol were added and protein precipitate was removed by centrifugation. Supernatants were dried in a speedvac and reconstituted in 250 μl of acetonitrile:water (10:90). Ten microlitres of each samples were used for the analysis. Chromatographic separation was performed on a 150-mm × 2.1-mm Zorbac 5 μm C18 column using Agilent liquid chromatography (LC) pump/autosampler system (Agilent, Santa Clara, CA, USA). The flow rate was 0.2 ml min−1. Solvent A was 0.1% formic acid in water and solvent B was 0.1% formic acid in acetonitrile. The gradient started at 5% B, then linearly increased to 95% B at 25 min and held at 95% B for 5 min. The column was equilibrated at 5% B for 5 min before starting the run. MS was performed on the LTQ orbitrapXL (Thermo Fisher) mass spectrometer operatedin positive electrospray mode (full scan/CID,m/z=206.2). The scan range was from 120 to 1000 Da. Standards of tryptophan and indole-3-lactic acid were used in the analysis (Sigma-Aldrich).
16S ribosomal RNA sequencing
After fecal DNA isolation (MoBio fecal DNA kit, Carlsbad, CA, USA), 626 bp amplicons (including a unique 12 bp Golay barcode) spanning V3–V5 region of bacterial 16S ribosomal RNA (rRNA) were generated using barcoded forward primer (338F, 906R). Samples were sent for pyrosequencing to Duke ISGP using the Roche 454 titanium platform (Indianapolls, IN, USA). Post processing of pyrosequencing data was done using QIIME (Caporaso et al., 2010). As a first step, data from the sequencing and the metadata were combined to demultiplex the barcoded reads, and quality filtering was done using the default parameters in QIIME. Sequences were grouped into operational taxonomic units at 97% sequence similarity using uclust. Taxonomy was assigned using RDP (Ribosomal Database Project) classifier against GreenGenes database, sequences were aligned and a phylogenetic tree was built from reference sequences using FastTree. An operational taxonomic unit table showing counts of each operational taxonomic unit in each sample was produced. Beta diversity was determined using unweighted UniFrac (Lozupone et al., 2006) and is represented as principal co-ordinate plots.
Results
UPLC-MS reveals the effect of the gut microbiota on the urine and fecal metabolomes of HUM mice
We have optimized a UPLC-MS-based method that allows the examination of the metabolomic profile of murine fecal and urine samples. Our goal was to investigate the dynamics of small molecules produced by a human gut microbiota using a HUM mouse model. Urine and fecal samples were collected from adult (8 weeks old; GF SW mice, n=3), followed by colonization of these mice using a fecal sample from an adult human (‘humanization', achieved by gavage of GF mice with 200 μl of frozen fecal sample reconstituted 1:1 using pre-reduced phosphate-buffered saline). Eight weeks after humanization, urine and fecal samples were collected again (HUM samples, n=3). Throughout the experiment, mice were maintained on a standard plant polysaccharide-rich diet (RD) inside gnotobiotic isolators. An UPLC-MS-based untargeted metabolomic approach consisted of either of two types of chromatographic conditions (RPLC or HILIC), and two ionization conditions (ESI in positive mode, ESI+ or negative mode, ESI−), resulting in four possible combinations of these conditions (see Figure 1a for a schematic overview of the approach). The rationale behind employing these different conditions is to detect as many classes of small molecules (for example, hydrophobic, hydrophilic, negatively charged and positively charged) as possible.
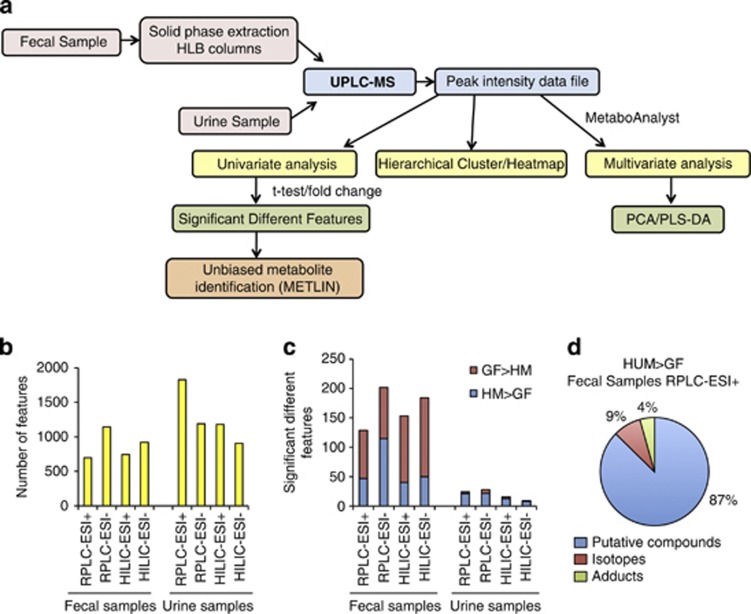
Figure 1.
Ultra performance liquid chromatography (UPLC)-MS analysis reveals microbial-related metabolites in fecal and urine samples of HUM mice. (a) Experimental workflow for metabolomic data generation and analysis. (b) Number of features detected in feces and urine of at least one of three HUM mice by different UPLC-MS methods. (c) Number of significantly different features (P<0.01, >10-fold difference) between HUM (n=3) and GF (n=3) mice found in urine or fecal metabolome. (d) Percentage of predicted isotopes and adducts in the set of features characteristic of feces from HUM mice (RPLC-ESI+).
MS was performed with a mass accuracy within 2 p.p.m. and metabolites were detected within the mass range of 70–800 m/z. The resulting peak intensity data matrix was filtered to eliminate features obtained when column conditioning and washing steps were performed (Supplementary Figure S1). Features were obtained with the following chromatography retention times: 1.5–20 min for RPLC, or 1–17 min for HILIC. RPLC-ESI+ detected the largest number of features in urine, and RPLC-ESI− detected the largest number of features in the fecal samples (Figure 1b). Despite urine having generally a larger number of features than feces, the number of significantly different features between HUM and GF mice metabolomes was higher in fecal samples than in urine, independent of the chromatographic and ionization method utilized (Figure 1c). Our analysis included the identification of clusters of masses with same retention time, which correspond to naturally occurring isotopes, clusters, fragments or adducts (for example, M+H+, M+Na+ and M+K+) of the same molecule. Although the number of possible clusters or adducts varies in each method, an example of the percentage of these features as a percentage of total features is shown in Figure 1d. Our analysis shows that these features do not affect our estimate of the number of unique metabolic features. The complete set of these features characteristic for HUM or GF mice in each of the methods can be found in Supplementary Table S1.
Fecal and urine metabolome from HUM mice is distinct from conventional mice
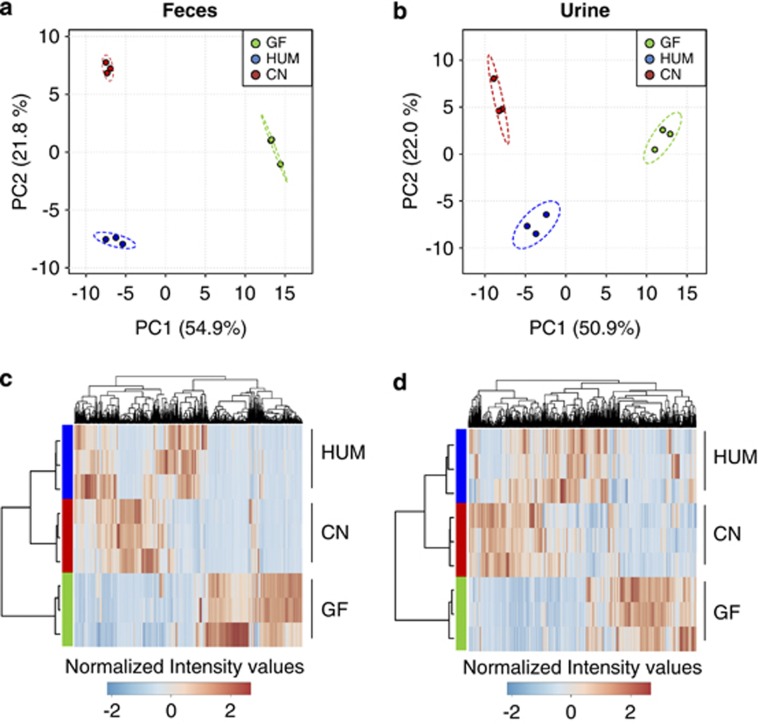
The intestinal microbiota of HUM mice reconstitutes many compositional aspects of the human microbiota and is distinct from that of a mouse (Ley et al., 2005; Turnbaugh et al., 2009b), but the extent to which these communities functionally differ remains underexplored. We assessed the fecal and urinary metabolome of mice harboring a native (murine) versus human-derived microbiota as a measure of community functionality. Fecal and urine samples from HUM, conventional and GF SW mice (n=3 mice sampled/colonization state) were analyzed using the optimized RPLC-ESI+-based method. Principal component analysis of metabolomic profiles reveal colonization-specific clustering for both fecal and urine samples (Figures 2a and b). Colonization-specific clustering was observed using all four chromatographic and ionization conditions described in Figure 1 in both fecal and urine samples, consistent with the differences between colonization states being independent of the method of data generation. The differentiation between colonization states was also observed by hierarchical clustering performed using the intensities of the detected features in each sample (Figure 2c for fecal metabolomes and Figure 2d for urine metabolomes). Ninety-nine of 821 analyzed features differentiated the fecal metabolomes of HUM and conventional mice; 45 of 2081 features differentiated the urine metabolomes of these two groups (n=3 per group; >10-fold difference in abundance and P-value<0.01, t-test). These data show that HUM mice derived from a human donor sample have a collection of metabolites in urine and feces that is distinct from those generated by this group of conventional mice.
Figure 2.
Humanized (HUM) mice fecal and urine metabolomes are distinct from those of conventional mice. (a, b) Principal component analysis (PCA) plot of fecal (a) and urine (b) metabolome from germ-free (GF, pre-humanization), conventional (CN) and HUM mice. (c, d) Hierarchical clustering of the fecal (c) and urine (d) metabolome from GF, CN and HUM mice samples.
The HUM microbiota alters levels of identifiable metabolites
A major hurdle in metabolomic studies is confirming the chemical identity of detected m/z values. This challenge is due to deficiencies in pure chemical standards and a paucity of database entries related to microbial metabolites and their host-conjugated counterparts (for example, glucuronidated). We used an unbiased metabolite identification approach on the features associated with HUM mouse samples (10-fold relative to GF samples, P<0.01, t-test). Empirical formulas were assigned to a subset of features with a maximum error of 3 p.p.m. using the METLIN database (Smith et al., 2005). When an unambiguous assignment was possible and the candidate metabolite was available, the pure metabolite standard was obtained and analyzed under the identical chromatographic and ionization conditions. The resulting spectra were compared against the experimental spectra for compound verification. The resulting verified identification data are listed in Supplementary Table S2.
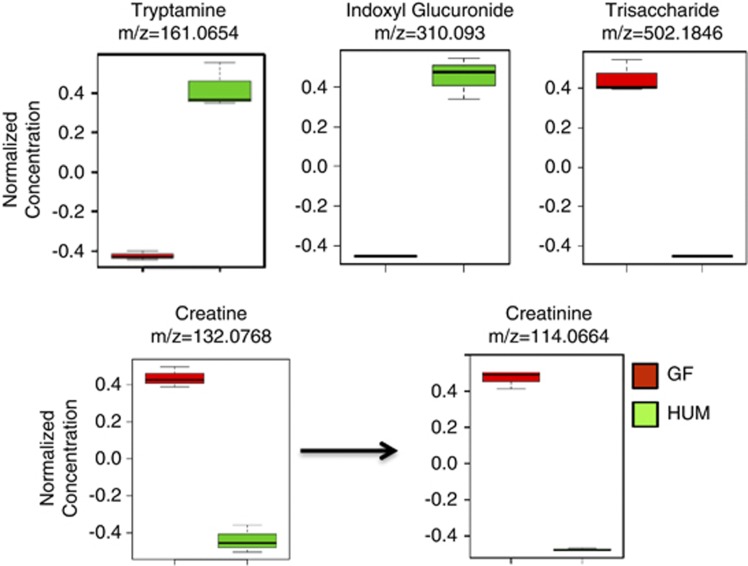
Metabolites resulting from the transformation of the aromatic amino-acid tryptophan into tryptamine and indole derivatives are consistent with previous metabolic activities assigned to the gut microbiota (Martin et al., 2008; Wikoff et al., 2009). Elevated level of fecal tryptamine (m/z 161.0654, ESI+) and urinary indoxyl glucuronide (m/z 310.0930, ESI+) were detected in HUM mice and confirmed using purified standards (Figure 3). The urine of HUM mice was also enriched for a feature with a m/z 134.0598 (ESI+), confirmed as 5-hydroxyindole, a known urinary marker of the intestinal microbiota (Supplementary Table S2; Meyer and Hostetter, 2012).
Figure 3.
Humanized (HUM) microbiota-dependent changes in fecal metabolites. Normalized mean values in GF and HUM mice±s.d. of three biological replicates are plotted for tryptamine, indoxyl glucuronide, a trisaccharide, creatine and creatinine.
As the gut microbial community consumes non-digestible plant-derived dietary polysaccharides (for example, dietary fiber), high levels of features that correspond to these carbohydrates in the feces are clear markers of the absence of gut microbial community in GF mice. Using HILIC-ESI−, a peak assigned to a trisaccharide (for example, raffinose or maltotriose, m/z 502.1846, ESI−), likely a component of the diet consumed by the mice, was present in GF fecal samples but absent in the HUM feces (Figure 3). Another marker of the microbiota is creatine and its cyclic derivative creatinine, as they are eliminated from the host by the action of intestinal microbiota (Wyss and Kaddurah-Daouk, 2000). Fecal levels of creatinine (m/z 114.066, ESI+) and creatine (m/z 132.076, ESI+) were elevated in the GF mice relative to HUM mice (Figure 3), consistent with previous studies showing an increase of these molecules in biofluids of antibiotic-treated mice (Romick-Rosendale et al., 2009; Swann et al., 2011a). These results indicate that although the available databases used for the identification of metabolite masses are incomplete and accurate mass is not sufficient to distinguish isomeric metabolites, it is still possible to identify metabolomic features consistent with microbiota colonization state in these highly complex environments. The data also suggests a large amount of novelty in the yet-to-be-described small molecules associated with the gut microbiota.
Dietary impact on the metabolome of HUM mice mirrors community compositional changes
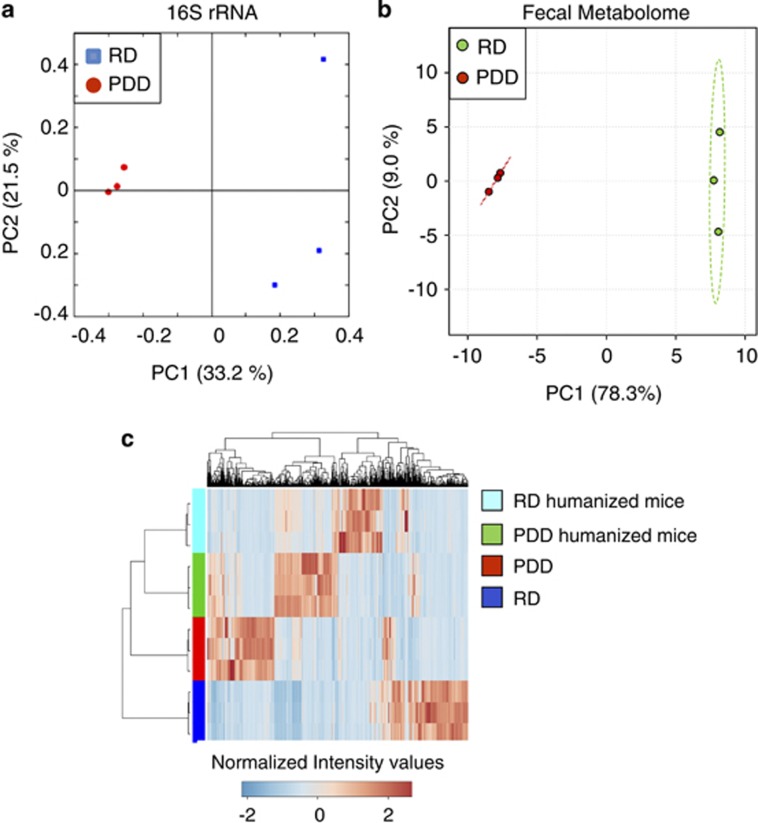
We assessed the effect of a dietary change on the C57BL/6J HUM mouse metabolome using a custom diet. HUM mice (n=3) were fed RD. After collection of feces and urine, the diet was switched to one devoid of fermentable polysaccharides (polysaccharide-deficient diet; PDD), and fecal and urine samples were collected after 4 weeks. The microbial community membership for the two diet conditions was determined by 454 pyrosequencing of the variable region V3–V5 of bacterial 16S rRNA genes present in fecal DNA. A total of 14 306 sequencing reads were obtained from the six fecal samples (2384 ± 329 sequences per sample) and analyzed using QIIME (Caporaso et al., 2010). UniFrac-based principal co-ordinate analysis plot shows that the presence or absence of polysaccharides in the diet produced a significant and reproducible effect on microbial composition as the RD mice cluster separately from the PDD mice (Figure 4a). Samples from the PDD group cluster tightly, indicating greater homogeneity within the bacterial profile for this dietary condition compared with the more complex RD. Although both dietary conditions resulted in a dominance of Firmicutes and Bacteroidetes, the Bacilli (P=0.017) and β-proteobacteria (P=0.02) were more highly represented in PDD mice than the RD-fed mice (Supplementary Figure S2).
Figure 4.
Dietary change alters composition and function of HUM mouse microbiota. (a) Principal co-ordinate analysis (PCOA) plot of 16S rRNA-based gut microbial profiling from HUM mice fed a regular diet (RD) or polysaccharide-deficient diet (PDD; n=3 mice per group). (b) PCA plot of fecal metabolite profiling from HUM mice fed with two different diets. (c) Hierarchical clustering of fecal metabolomes from HUM mice fed with two different diets versus metabolomes of the respective mouse diets.
Next, we addressed the effect of diet on the metabolomic profile of fecal and urine samples from HUM mice fed RD or PDD. Multivariate statistical analyses reveal that the fecal metabolomes of the two different dietary conditions (RPLC-ESI+) are highly distinguishable and mirror the compositional differences that we observe between samples (Figure 4b, Supplementary Table S3). The urine samples also showed a similar separation by diet (Supplementary Figure S3). To determine whether the separation between samples was due to unmodified compounds present in the diet, fecal metabolites from RD- and PDD-fed mice were compared with extracts of the two different diets. This analysis revealed that the contribution of compounds present in the diet to the differences identified between the two dietary groups is very low (7.1% of the features in feces of RD-fed mice are present in RD; 6.3% of the features in feces of PDD-fed mice are present in PDD). Hierarchical clustering of the metabolomic profiles from feces and diet illustrates that metabolites from the diet contribute only a small portion of the unique fecal metabolites associated with a specific diet condition (Figure 4c). These data reveal that diet-induced changes in microbial composition are mirrored by changes in the microbiota-dependent metabolome and suggest that metabolomics can serve as an effective read-out of diet-induced alterations in microbiota composition and function.
A simplified model microbiota drives major changes in the metabolome of GF mice
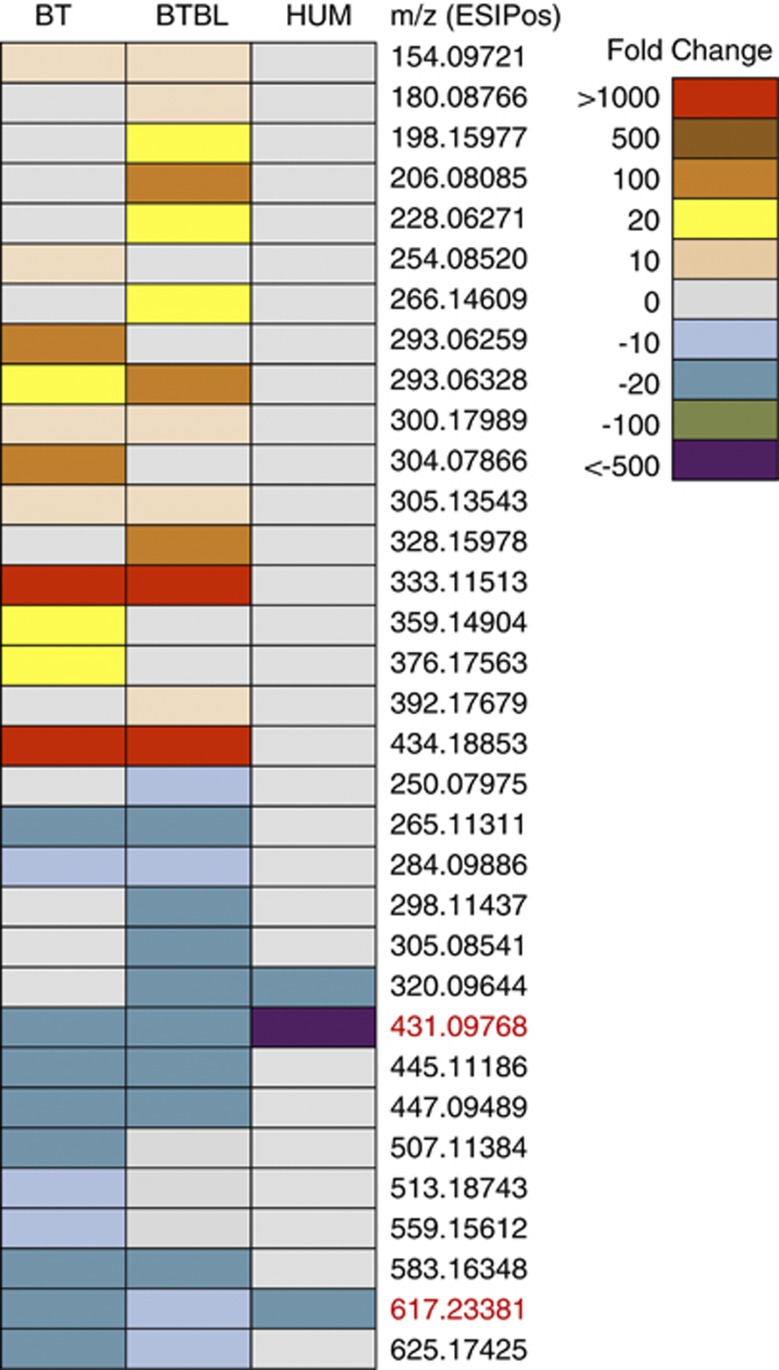
We next introduced one or two species of bacteria into GF SW mice to assess the impact of simplified communities on the fecal and urine metabolome compared with mice colonized with a complex microbial community. GF SW mice were colonized with either Bt (n=3) or with a mix of equal concentration of Bt and Bl (n=3). Ten days after colonization, urine and fecal samples were collected from both groups of mice. Relative bacterial densities within fecal samples were determined for theBt/Bl group using differential plating (see Materials and methods), revealing that Bt colonizes oneorder of magnitude higher than Bl (90.2±7.7% ofBt; 9.8±7.7% of Bl). The impact of these simplified microbiotas on the host metabolome was determined by analyzing fecal and urine samples by RPLC-ESI+. Metabolomic profiles of the gnotobiotic samples were compared with those of GF mice (10-fold, P<0.01,t-test) to identify the microbial signatures of mono and bi-colonization (Supplementary Table S4). The introduction of one or two microorganisms in the intestine significantly shifts the urine and fecal metabolomic profile of the host (Figure 5 for fecal samples; Supplementary Figure S4 for urine samples). Although the Bt mono- and Bt/Bl bi-associations produced metabolomic profiles that are similar to one another, features unique to the bi-colonized mice indicate an impact of Bl, despite the fact that it colonizes at one order of magnitude less than Bt. The bi-association-induced changes in metabolomic profile may be a result of multiple factors that are altered upon different colonization states including changes in community member abundance, interactions with the host and altered gene expression of the microbes. One feature corresponding to indolelactic acid (m/z 206.0739, ESI+) was detected in Bt/Bl fecal samples, which was not present in the GF and Bt samples. To verify that Bl is able to produce indolelactic acid, we grew Bl in culture medium supplemented with 0.1% tryptophan (a precursor of indole derivatives). The production of indolelactic by Bl was confirmed by UPLC-MS comparison with a purified standard (Supplementary Figure S5). Together, these data demonstrate that even a low level of colonization, as seen with Bl, is sufficient to impact the fecal metabolome.
Figure 5.
Simplified model microbiotas alter fecal metabolome. Heat-map of microbe-dependent metabolites from fecal samples of gnotobiotic mice colonized with B. thetaiotaomicron (BT), Bt and B. longum (BTBL) or humanized (HUM) mice. Features significantly increased or decreased in either BT or BTBL relative to GF are shown; fold change>10 and P-value<0.01 m/z values in red indicate GF-related features that disappear in all three colonization states.
To determine whether colonization with a simplified microbiota drives similar changes in the metabolomic profiles as colonization with a complete human microbiota, metabolites associated with humanization were compared with those associated with the Bt and Bt/Bl colonization states (that is, metabolites not present in GF). No features were identified that were commonly present in HUM mice and either the mono- or bi-colonized mice. However, two metabolites showed a decrease in prevalence relative to GF in all three colonization states (Figure 5). These data indicate that the metabolome of one and two member model microbiotas do not reconstitute the metabolomic complexity of a HUM microbiota (Supplementary Figure S6); however, these simple microbiotas can model some aspects of a complete microbiota, such as depletion of certain compounds. The extent to which additional other species within simplified microbiotas can represent metabolomic features of a complex community requires further investigation.
Humanization of mice reconstitutes gut microbial diversity and human fecal metabolomic profile
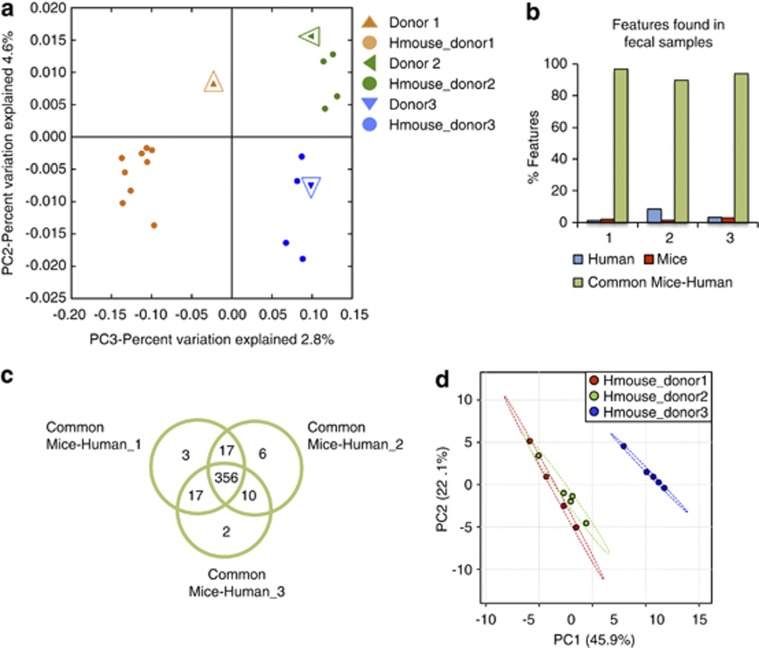
We wished to test whether the gut microbial communities in human feces retain their composition when transferred to GF mice, and whether aspects of metabolomic individuality of each human can be mirrored in the HUM animals. We collected feces from three healthy human donors: two males and one female. Four weeks following humanization of GF SW mice (n=4–8 per donor), fecal samples were collected from each mouse and submitted for 16S rRNA-based microbial community enumeration by pyrosequencing. Unweighted UniFrac-based principal co-ordinate analysis plots of the 16S rRNA microbial composition data show that the microbiota of HUM mice clusters with the corresponding human fecal inoculum (Figure 6a). These data reveal that individual-specific differences in gut microbial communities can be faithfully reconstituted in GF mice by humanization using human feces.
Figure 6.
Gut microbial diversity and human fecal metabolome are reconstituted in HUM mice. (a) Principal co-ordinate analysis (PCOA) plot of 16S rRNA-based gut microbial profiling from HUM mice from three different donors (n=4–8 mice per group) and the respective donor samples (open enlarged symbols). (b) Percent of features found in human feces, HUM mouse feces or both, for the three respective donor/recipient groups using RPLC-ESI+. (c) Venn diagram with metabolites common to the human and HUM mice samples for each donor/recipient group. (d) PCA plot of fecal metabolite profiling from HUM mice colonized with three different fecal samples.
Although a small portion of the detected features are specific to either human or mouse samples, most of the compounds are independent of the source of the feces (RPLC-ESI+, Figure 6b), suggesting that much of the microbiota functional status is recalcitrant to modification by the host species. Moreover, comparison of metabolites common to mice and human from the three different sets of samples (donor 1-, 2- or 3-derived) revealed a high degreeof overlap in the metabolomes of these groups,and a low percentage of donor-specific features (Figure 6c). These findings support the idea that many functional attributes are shared between microbiotas that differ compositionally (Turnbaugh et al., 2009a). Conversely, some of the features that differentiate donor 3 HUM mouse fecal metabolome from those of donor 1 and donor 2 were also detected in the human fecal sample of donor 3, and absent in the other human samples (Figure 6d and Supplementary Figure S7). Therefore, some individualized aspects of the human gut microbiota's metabolomic activity can be reconstructed within HUM mice.
Discussion
Sequencing-focused techniques have been used extensively to characterize the species and gene content of gut microbial communities associated with health or a variety of diseases (Qin et al., 2010; Arumugam et al., 2011; Huttenhower et al., 2012). It is apparent that common functional attributes are found in divergently composed communities and conversely, distinct functional traits can be expressed by similarly composed microbiotas (Turnbaugh et al., 2009a; Nicholson et al., 2012). Therefore, characterization of microbiota function may be more illuminating to the status of host–microbial interaction than the microbial species or gene content. Coupled to the importance of attaining the functional state of a specific microbiota under specific conditions are additional challenges spanning from data generation (for example, technical) to interpretation and integration of new types of function-focused data sets. Metabolomics offers a rapid method of defining hundreds to thousands of small molecules associated with a sample; the resulting spectrum of masses offers a chemical ‘fingerprint' of microbiota function that is informative even in the absence of definitive compound assignments. Grouping of different samples/communities based on metabolomic profile, quantifying the extent of functional change over time or based on different treatments, and characterizing the chemical complexity associated with different communities are all possible in the absence of knowing the compound to which each feature corresponds. A growing number of examples illustrate that the metabolites derived from this diverse microbial community can have a direct role in human health and disease, indicating that this universe of small molecules not only serves as an excellent proxy for functional state, but also provides a window into the compounds that are mediating microbe–microbe and microbe–host interaction (Nicholson et al., 2012).
The investigation of microbiota function is met with two related challenges. First, the sensitivity of microbiota adaptation to environmental variables (for example, diet) necessitates the need for a highly controlled experimental platform to generate reproducible, interpretable data, such as a mouse model. Second, the desire to study human-relevant microbes, which differ from those that are found in the native mouse microbiota (Ley et al., 2005), is not immediately compatible with standard mouse models. The transplantation of human gut microbiota into GF mice represents a human-relevant experimental model for pursuing microbiota functionality and its impact on host biology in healthy and diseased states (Goodman et al., 2011).
Here we present a study that applies an optimized metabolomics method on urine and fecal samples derived from mice that have been colonized with a human microbiota to permit the study of human-relevant microbes in a controlled experimental setting. Using this method, we demonstrate that the vast majority of metabolomic features that we detected in human feces are reconstituted in HUM mice. These data establish that mice harboring a human gut microbiota are a valid model for reconstructing human microbiota functionality in an experimental setting. Furthermore, the overlap in metabolomic features associated with three groups of HUM mice, each colonized with a different donor microbiota, is consistent with the implied functional conservation in divergently composed human gut communities (Turnbaugh et al., 2009a). Despite the common functional attributes, our study and others show that dietary modifications of the microbiota are a significant factor in shaping the metabolomic features associated with feces and urine (Legido-Quigley et al., 2010). The set of diet-induced metabolites provides an example of how the ability of complex external factors to interact with and influence the functionality of human gut microbes can be chemically quantified, a first step in unraveling the dynamics of microbiota function and its impact on the host.
Although humanization of GF mice provides an attractive model of how the human microbiota interacts with a host, the microbiota in this model remains a highly complex system that is resistant to mechanistic and hypothesis-based experimentation. The possibility of evaluating more simplified, defined communities on the host metabolomic profile is a promising avenue to gain mechanistic insights. Simplified and defined model systems have been used extensively to study functional properties of common gut residents in vivo (Sonnenburg et al., 2006; Samuel et al., 2007; Marco et al., 2010; Rey et al., 2010; Faith et al., 2011). Our use of one- and two-member model communities indicates that there are aspects of the host metabolome that are conserved between these simplified ecosystems and an entire human microbiota, specifically the common depletion of metabolites that are present in GF mice. An important remaining question is at what point of constructing a defined microbiota is the functionality associated with a complex HUM community sufficiently approximated. Although the answer will vary based on the scientific question being addressed, it is likely that metabolomic profiling will be an important component of assessing the functional capacity of simplified communities for comparison with that of a complete human microbiota.
Our results set the stage for discovery and elucidation of small molecule-mediated interactions between human-associated microbes and the host. Our detection of individual-specific metabolomic features suggests that personalized HUM mice could be used in a variety of applications relevant to human health, ranging from biomarker discovery to characterization and categorization of community response to microbiota-targeted treatments. In addition, the experimental approach that we have implemented to investigate the gut microbiota functional status should be straightforward to apply to microbial communities associated with other body habitats.
Acknowledgments
We thank Erica Sonnenburg for providing comments on the manuscript and Tim Meyer for helpful discussions. This work was funded in part by grants from National Institutes of Health (R01-DK085025 and DP2-OD006515).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowey E, Adlercreutz H, Rowland I. Metabolism of isoflavones and lignans by the gut microflora: a study in germ-free and human flora associated rats. Food Chem Toxicol. 2003;41:631–636. doi: 10.1016/s0278-6915(02)00324-1. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus SP, Tsang TM, Wang Y, Cloarec O, Skordi E, Martin F-P, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci USA. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin BL, Ruthven CRJ, Sandler M. Gut flora and the origin of some urinary aromatic phenolic compounds. Biochem Pharmacol. 1994;47:2294–2297. doi: 10.1016/0006-2952(94)90268-2. [DOI] [PubMed] [Google Scholar]
- Holmes E, Loo RL, Stamler J, Bictash M, Yap IKS, Chan Q, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–U350. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legido-Quigley C, Stella C, Perez-Jimenez F, Lopez-Miranda J, Ordovas J, Powell J, et al. Liquid chromatography-mass spectrometry methods for urinary biomarker detection in metabonomic studies with application to nutritional studies. Biomed Chromatogr. 2010;24:737–743. doi: 10.1002/bmc.1357. [DOI] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Nat Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac - an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallett AK, Bearne CA, Rowland IR, Farthing MJG, Cole CB. The use of rats associated with a human fecal flora as a model for studyng the effects of diet on the human gut microflora. J Appl Bacteriol. 1987;63:39–45. doi: 10.1111/j.1365-2672.1987.tb02415.x. [DOI] [PubMed] [Google Scholar]
- Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- Marco ML, de Vries MC, Wels M, Molenaar D, Mangell P, Ahrne S, et al. Convergence in probiotic Lactobacillus gut-adaptive responses in humans and mice. ISME J. 2010;4:1481–1484. doi: 10.1038/ismej.2010.61. [DOI] [PubMed] [Google Scholar]
- Martin F-PJ, Wang Y, Sprenger N, Yap IKS, Lundstedt T, Lek P, et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:1–15. doi: 10.1038/msb4100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer TW, Hostetter TH. Uremic solutes from colon microbes. Kidney Int. 2012;81:949–954. doi: 10.1038/ki.2011.504. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–U70. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FE, Faith JJ, Bain J, Muehlbauer MJ, Stevens RD, Newgard CB, et al. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem. 2010;285:22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romick-Rosendale LE, Goodpaster AM, Hanwright PJ, Patel NB, Wheeler ET, Chona DL, et al. NMR-based metabonomics analysis of mouse urine and fecal extracts following oral treatment with the broad-spectrum antibiotic enrofloxacin (Baytril) Magn Reson Chem. 2009;47:S36–S46. doi: 10.1002/mrc.2511. [DOI] [PubMed] [Google Scholar]
- Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci USA. 2007;104:10643–10648. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Maille GO, Want EJ, Qin C, Trauger SA, Brandon TR, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–U1256. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Chen CTL, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:2213–2226. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann JR, Tuohy KM, Lindfors P, Brown DT, Gibson GR, Wilson ID, et al. Variation in antibiotic-induced microbial recolonization impacts on the host metabolic phenotypes of rats. J Proteome Res. 2011a;10:3590–3603. doi: 10.1021/pr200243t. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009a;457:480–U487. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009b;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol Rev. 2000;80:1107–1213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- Yap IKS, Li JV, Saric J, Martin F-P, Davies H, Wang Y, et al. Metabonomic and microbiological analysis of the dynamic effect of vancomycin-induced gut microbiota modification in the mouse. J Proteome Res. 2008;7:3718–3728. doi: 10.1021/pr700864x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.