Abstract
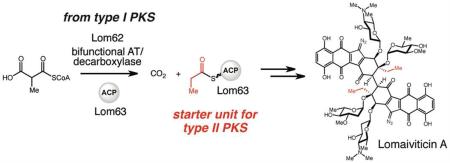
Lomaiviticin biosynthesis is thought to utilize a propionyl starter unit for a type II polyketide synthase (PKS). Discovery of the lomaiviticin (lom) biosynthetic gene cluster suggested an unusual method for starter unit generation involving a bifunctional acyltransferase/decarboxylase (AT/DC) thus far observed only in type I PKS pathways. In vitro biochemical characterization of AT/DC Lom62 confirmed its ability to generate a propionyl-acyl carrier protein (ACP), revealing a new role for this enzymatic activity within natural product biosynthesis.
Aromatic polyketides are an important family of natural products that includes clinically used antibiotics and antitumor agents.1 These secondary metabolites are constructed from malonate building blocks using iterative decarboxylative Claisen condensations carried out by a type II polyketide synthase (PKS). An important point for introducing structural diversity in these pathways is the initial priming of the PKS with a starter unit.3 Understanding the logic underlying starter unit synthesis is therefore a critical component of efforts to engineer the biosynthesis of ‘non-natural’ analogs of aromatic polyketides.4 Here we detail evidence supporting the involvement of a new strategy for starter unit generation in lomaiviticin biosynthesis that employs an enzyme previously associated only with type I modular PKSs.
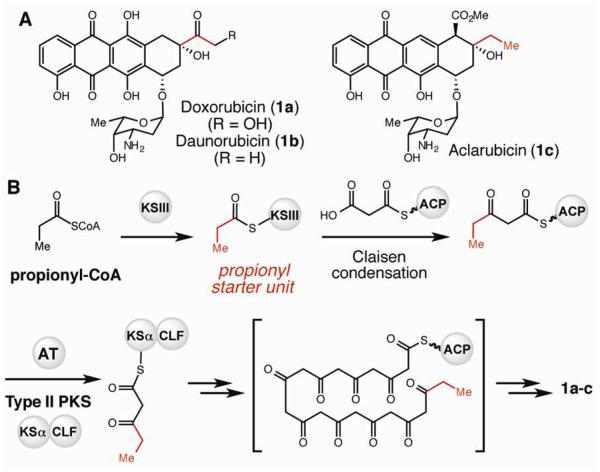
Type II PKS systems consist of a ketosynthase-chain length factor (KSα-CLF) complex, which performs the C–C bond-forming chemistry, and an acyl carrier protein (ACP) that tethers an incoming malonyl extender unit. The acyltransferase (AT) that selects and loads malonyl-coenzyme A (CoA) is shared with fatty acid synthesis.2 The majority of secondary metabolites made by type II PKS systems utilize an acetate starter unit generated by decarboxylation of a malonyl-ACP thioester substrate derived from malonyl-CoA.5 Propionyl starter units are employed less frequently, but are involved in the biosynthesis of the clinically important anti-tumor drugs doxorubicin (1a) and daunorubicin (1b) (Figure 1A).6 To date, all characterized mechanisms for incorporating propionyl starter units into type II polyketides utilize propionyl-CoA as the source of this three carbon building block. In doxorubicin/daunorubicin biosynthesis, a separate priming KS (KSIII), homologous to the E. coli FabH enzyme, catalyzes an initial Claisen condensation using propionyl-CoA and ACP-bound malonyl-CoA (Figure 1B).6 The diketide product is then transferred to the main KSα-CLF for further rounds of elongation. Other type II polyketide biosynthetic pathways that utilize longer alkyl chain starter units similarly employ this KSIII loading module strategy,7 and most of the corresponding gene clusters also encode a second ACP that is dedicated to starter unit production.7c
Figure 1.
Propionyl starter units are employed in aromatic polyketide biosynthesis. A) Natural products made by type II polyketide synthases (PKSs) that utilize a propionyl starter unit. B) Characterized pathway for type II PKS priming with a propionyl starter unit.
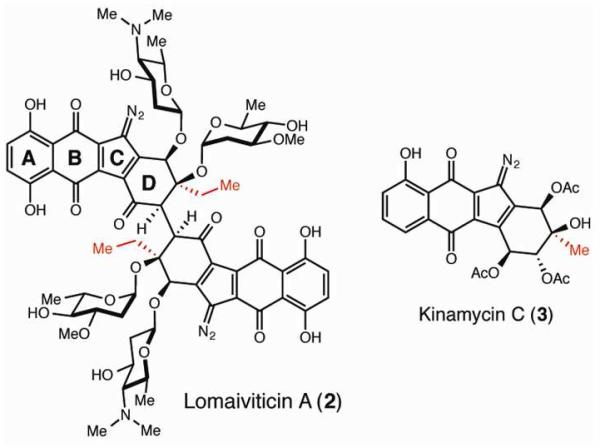
The lomaiviticins (2) and kinamycins (3) are two families of diazo-containing aromatic polyketides that have attracted attention from both synthetic and biological chemists (Figure 2).8 Though these metabolites possess considerable structural similarity, a key difference between them is the D-ring alkyl substituent, which likely arises from use of distinct type II PKS starter units. The kinamycin PKS is predicted to use a canonical acetate starter unit, resulting in a D-ring methyl group. In contrast, substitution of an ethyl group at the corresponding position of the lomaiviticin scaffold suggests use of a propionyl starter unit. Recently, Moore and co-workers discovered a lomaiviticin (lom) biosynthetic gene cluster in the marine actinomycete Salinispora tropica CNB-440.9 We have also identified a homologous biosynthetic gene cluster in the lomaiviticin producer Salinispora pacifica DPJ-0019.10 Unexpectedly, neither cluster encoded an enzyme homologous to the priming KSIII typically employed in propionyl starter unit loading.9 The absence of this enzymatic function suggested that an alternate method of starter unit synthesis could be used in lomaiviticin biosynthesis.
Figure 2.
The lomaiviticin and kinamycin families of aromatic polyketides likely use different starter units in biosynthesis.
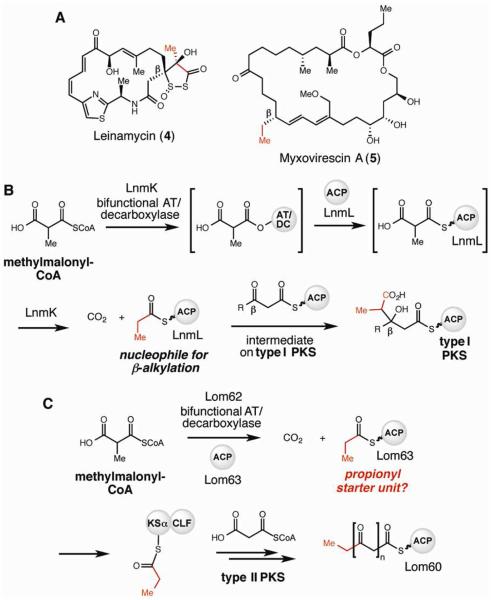
In our cluster annotation we identified several genes that we hypothesized could play roles in propionyl starter unit generation: a homolog of the bifunctional acyltransferase/decarboxylase (AT/DC) LnmK (lom62) and two ACPs (lom60 and lom63). LnmK is involved in the biosynthesis of leinamycin (3), a natural product constructed by a type I modular PKS-nonribosomal synthetase (NRPS) assembly line (Figure 3A). Biochemical characterization of LnmK has revealed its ability to catalyze both transfer of methylmalonyl-CoA to an ACP and subsequent decarboxylation to provide a propionyl-ACP thioester.11 This intermediate is ultimately utilized to install a β-propionyl substituent onto the polyketide backbone of leinamycin during chain elongation (Figure 3B).12-14 Recently, the crystal structure of LnmK was solved.15 Co-crystallization with methylmalonyl-CoA revealed putative active site residues, which led to the identification of an essential tyrosine (Tyr62) as the key functionality necessary for AT activity. Homologs of LnmK have been noted in other gene clusters encoding type I PKS-NRPS pathways, including the myxovirescin A (5) gene cluster.11
Figure 3.
An enzyme involved in type I PKS biosynthesis may be utilized in lomaiviticin starter unit generation. A) Natural products synthesized by type I PKS-nonribosomal peptide synthetase assembly lines contain β-propionyl and -ethyl branching. B) Role of acyltransferase/decarboxylase (AT/DC) LnmK in leinamycin biosynthesis. C) Hypothesized role of LnmK homolog Lom62 in lomaiviticin biosynthesis.
The presence of an LnmK homolog (44% amino acid identity, 60% similarity) in the lom gene cluster led us to hypothesize that Lom62 might catalyze an analogous reaction, transferring methylmalonyl-CoA to an ACP and then decarboxylating the resulting methylmalonyl-ACP thioester intermediate to generate a propionyl-ACP thioester (Figure 3C). Transfer of this propionyl starter unit to the KS domain of the KSα-CLF could then occur directly from the ACP or via the action of predicted AT Lom61. The presence of two ACPs in the lom cluster (Lom 60 and Lom 63) further suggests use of an alternate starter unit, with one of the ACPs likely dedicated to propionyl-ACP generation. Importantly, homologs of these enzymes (Lom62 and a second ACP) are not found in the kinamycin biosynthetic gene cluster,16 providing additional support for this hypothesis. If operative, this co-opting of type I PKS tailoring enzymes would constitute a completely new mechanism for type II PKS starter unit generation. However, the dramatically different pathway contexts of Lom62 and LnmK (type II vs. type I modular PKS systems) precluded the assumption that the two enzymes would exhibit the same biochemical activity.
To verify that the sequence similarity of Lom62 and LnmK was likely to reflect a shared biochemical function, we performed additional bioinformatic analyses. Alignment of the two amino acid sequences showed conservation of the essential tyrosine residue that is hypothesized to function as the nucleophile in catalysis (Figure S9). Key residues that contact the CoA backbone are also conserved in Lom62. These similarities were reinforced by construction of a Lom62 homology model (Figure S11). Overall, these analyses confirmed that Lom62 and LnmK might possess related functions.
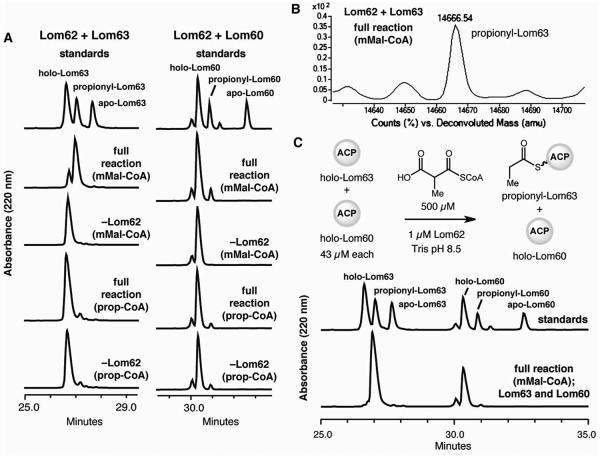
We tested the hypothesis that Lom62 plays a role in propionyl starter unit generation by characterizing its activity in vitro. We cloned lom62 and the two ACPs lom60 and lom63 from S. pacifica DPJ-0019 and expressed them individually in E. coli as His6-tagged fusions. Analysis of LC-MS data confirmed the identities of both apo-ACPs and their ability to undergo conversion to their corresponding holo-forms in the presence of CoA and the phosphopantethienyltransferase Sfp (Figures S2-S8, Tables S2-S4).17 We examined the activity of Lom62 toward each holo-ACP in the presence of potential substrates methylmalonyl-CoA and propionyl-CoA using an HPLC assay (Figure 4A). LC-MS analysis verified the identities of the major products (Tables S5 and S6). Robust, Lom62-dependent formation of the expected propionyl-ACP thioester product was observed only in the presence of methylmalonyl-CoA and Lom63. In comparison, reactions of Lom62 with methylmalonyl-CoA and alternate ACP Lom60 were sluggish, providing <10% conversion after 1 h. Lom62 was not able to generate propionyl-ACP product from propionyl-CoA in combination with either ACP partner, confirming that decarboxylation takes place after loading.
Figure 4.
Lom62 generates a propionyl-ACP intermediate from methylmalonyl-CoA for potential use as starter unit in lomaiviticin biosynthesis. A) HPLC assays of Lom62 with holo-acyl carrier proteins Lom63 and Lom60 (1 h time point) demonstrate loading and decarboxylation of methylmalonyl-CoA to generate propionyl-ACPs and a preference for Lom63. UV detection (220 nm). B) Deconvoluted mass spectrum of the Lom62 + Lom63 mMal-CoA full reaction assay. C) HPLC competition assay with equal amounts of ACPs (1 h time point) reveals selective generation of propionyl-Lom63. UV detection (220 nm). Abbreviations: ACP = acyl carrier protein, AT = acyltransferase, mMal-CoA = methylmalonyl-CoA, prop-CoA = propionyl-CoA.
To further explore the specificity of Lom62 for the two ACPs, we performed a competition experiment, incubating equal concentrations (43 μM) of the two ACPs with methylmalonyl-CoA and Lom62 for 1 h (Figure 4C). Under these conditions we observed almost exclusive formation of propionyl-Lom63 by HPLC analysis. This result suggests that LnmK preferentially interacts with only one of the two ACPs encoded in the lomaiviticin cluster. The use of a dedicated ACP for starter unit generation is also observed for other priming mechanisms that use non-acetate type II PKS starter units.7c Overall, these biochemical experiments support our hypothesis that Lom62 is involved in generating the propionyl starter unit required for lomaiviticin biosynthesis and reveal Lom63 as its partner ACP. Additional biochemical experiments are needed to elucidate the precise mechanism of propionyl starter unit transfer to the type II PKS.
The potential distribution of this alternate propionyl starter unit synthesis machinery across other type II PKS biosynthetic pathways was evaluated by searching for homologs of Lom62 in sequenced bacterial genomes (Table S8). The majority of the most closely related enzymes were part of other lom clusters (32 hits). However, we found 11 Lom62/LnmK homologs that were were co-localized with genes encoding type I PKS assembly lines and other enzymes involved in β-alkyl installation,14 and an additional two homologs located near type II PKS biosynthetic machinery that was distinct from the lom gene cluster. This analysis suggests that the decarboxylative route to propionyl starter unit generation is utilized by other type II PKS systems. We hypothesize that this priming strategy could have evolved in response to differences in the availability of methylmalonyl-CoA and propionyl-CoA precursors in producing organisms.
In summary, we have discovered that a new mechanism for type II PKS propionyl starter unit generation may be operative in lomaiviticin biosynthesis. Intriguingly, this process co-opts enzymes previously associated only with type I PKS pathways. These experiments are the first step towards understanding the logic underlying this unique priming strategy, which may represent a new target for metabolic engineering efforts targeting aromatic polyketide biosynthesis.
Supplementary Material
Acknowledgment
We acknowledge Dr. Sunia Trauger and Jennifer Wang (Harvard University Small Molecule Mass Spectrometry Core Facility) for assistance with LC-MS analyses. We received financial support from the National Institutes of Health (NIH) (DP2 GM105434 and GM095450), the Searle Scholars Program, and Harvard University.
Footnotes
Supporting Information Available Experimental details. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Nat. Prod. Rep. 2007;24:162. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]; (b) Das A, Khosla K. Acc. Chem. Res. 2009;42:631. doi: 10.1021/ar8002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Revill WP, Bibb MJ, Hopwood DA. J. Bacteriol. 1995;177:3946. doi: 10.1128/jb.177.14.3946-3952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Summers RG, Ali A, Shen B, Wessel WA, Hutchinson CR. Biochemistry. 1995;34:9389. doi: 10.1021/bi00029a015. [DOI] [PubMed] [Google Scholar]; (c) Carreras CW, Khosla C. Biochemistry. 1998;37:2084. doi: 10.1021/bi972919+. [DOI] [PubMed] [Google Scholar]
- 3.Moore BS, Hertweck C. Nat. Prod. Rep. 2002;19:70. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 4.(a) Tang Y, Lee TS, Khosla C. PLOS Biol. 2004;2:227. doi: 10.1371/journal.pbio.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lee TS, Khosla C, Tang Y. J. Am. Chem. Soc. 2005;127:12254. doi: 10.1021/ja051429z. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Xu X, Schenk A, Hertweck C. J. Am. Chem. Soc. 2007;129:6022. doi: 10.1021/ja069045b. [DOI] [PubMed] [Google Scholar]; (d) Das A, Szu PH, Fitzgerald JT, Khosla C. J. Am. Chem. Soc. 2010;132:8831. doi: 10.1021/ja102517q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Bisang C, Long PF, Cortes J, Westcott J, Crosby J, Matharu A-L, Cox RJ, Simpson TJ, Staunton J, Leadlay PF. Nature. 1999;401:502. doi: 10.1038/46829. [DOI] [PubMed] [Google Scholar]; (b) Drier J, Khosla C. Biochemistry. 2000;39:2088. doi: 10.1021/bi992121l. [DOI] [PubMed] [Google Scholar]
- 6.(a) Rajgarhia VB, Priestley ND, Strohl WR. Metab. Eng. 2001;3:49. doi: 10.1006/mben.2000.0173. [DOI] [PubMed] [Google Scholar]; (b) Bao WL, Sheldon PJ, Hutchinson CR. Biochemistry. 1999;38:9752. doi: 10.1021/bi990751h. [DOI] [PubMed] [Google Scholar]; (c) Bao W, Sheldon PJ, Wendt-Pienkowski E, Hutchinson CR. J. Bacteriol. 1999;181:4690. doi: 10.1128/jb.181.15.4690-4695.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Bibb MJ, Sherman DH, Omura S, Hopwood DA. Gene. 1994;142:31. doi: 10.1016/0378-1119(94)90351-4. [DOI] [PubMed] [Google Scholar]; (b) Meadows ES, Khosla C. Biochemistry. 2001;40:14855. doi: 10.1021/bi0113723. [DOI] [PubMed] [Google Scholar]; (c) Tang Y, Lee TS, Kobayashi S, Khosla C. Biochemistry. 2003;42:6588. doi: 10.1021/bi0341962. [DOI] [PubMed] [Google Scholar]
- 8.Woo CM, Herzon SB. Nat. Prod. Rep. 2012;29:87. doi: 10.1039/c1np00052g. For a review of the lomaiviticin and kinamycin families of natural products see. [DOI] [PubMed] [Google Scholar]
- 9.(a) Kersten RD, Lane AL, Nett M, Richter TKS, Duggan BM, Dorrestein PC, Moore BS. ChemBioChem. 2013;14:955. doi: 10.1002/cbic.201300147. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) The lom gene cluster disclosed in this work also encodes a LnmK homolog (Strop_2227; 91% amino acid identity, 95% amino acid similarity to Lom62) that was annotated as a KSIII. Our bioinformatic analyses of Strop_2227 and Lom62 indicate that these proteins are more closely related to LnmK-like AT/DCs than to KSIIIs (see Figures S9 and S10)
- 10. Details will be published elsewhere.
- 11.Liu T, Huang Y, Shen B. J. Am. Chem. Soc. 2009;131:6900. doi: 10.1021/ja9012134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Huang S-X, Ju J, Tang G, Liu T, Shen B. Org. Lett. 2011;13:498. doi: 10.1021/ol102838y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simunovic V, Müller R. ChemBioChem. 2007;8:1273. doi: 10.1002/cbic.200700153. [DOI] [PubMed] [Google Scholar]
- 14.Calderone CT. Nat. Prod. Rep. 2008;25:845. doi: 10.1039/b807243d. [DOI] [PubMed] [Google Scholar]
- 15.Lohman JR, Bingman CA, Philips GN, Jr., Shen B. Biochemistry. 2013;52:92. doi: 10.1021/bi301652y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Gould SJ, Hong ST, Carney JR. J. Antibiot. 1998;51:50. doi: 10.7164/antibiotics.51.50. [DOI] [PubMed] [Google Scholar]; (b) Bunet R, Song L, Mendes MV, Corre C, Hotel L, Rouhier N, Framboisier X, Leblond P, Challis GL, Aigle B. J. Bacteriol. 2011;193:1142. doi: 10.1128/JB.01269-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Quadri LEN, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Biochemistry. 1998;37:1585. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]; (b) Yin J, Lin AJ, Golan DE, Walsh CT. Nat. Protoc. 2006;1:280. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.