Abstract
Solving the crystal structure of Cbl(TKB) in complex with a pentapeptide, pYTPEP, revealed that the PEP region adopted a poly-L-proline type II (PPII) helix. An unnatural amino acid termed a proline-templated glutamic acid (ptE) that constrained both the backbone and sidechain to the bound conformation was synthesized and incorporated into the pYTPXP peptide. We estimated imposing structural constraints onto the backbone and sidechain of the peptide and preorganize it to the bound conformation in solution will yield nearly an order of magnitude improvement in activity. NMR studies confirmed that the ptE-containing peptide adopts the PPII conformation, however, competitive binding studies showed an order of magnitude loss of activity. Given the emphasis that is placed on imposing structural constraints, we provide an example to support the contrary. These results point to conformational flexibility at the interface, which have implications in the design of potent Cbl(TKB)-binding peptides.
Modern chemistry programs have witnessed an increased demand for inhibitors of protein-protein interactions (i-PPI). This demand is appropriate, given the recognized role of protein-protein interactions (PPI) in various diseases. Inhibition of PPI with small organic molecules has been achieved with some success1,2,3. Targeting PPI with small molecules, however, has been considered a high-risk endeavor due to the generally ill-defined nature of the PPI interface4,5. To overcome these difficulties, peptides that bind the PPI interface have been reliably used as lead compounds for the design of potent inhibitors of PPI6,7,8. In the case of well-defined pockets, it is generally accepted in the medicinal chemistry community that addition of structural constraints to bias them to the bound conformation will result in improved potency. It is rationalized that, given comparable enthalpies, a preorganized ligand to the bound conformation will have a more favorable binding entropy and thus higher binding affinity, relative to the flexible ligand. Indeed, this practice has dramatically improved binding affinity for a select number of ligands. The practice of imposing structural constraints onto ligands to improve the binding affinity has been known as “prepaying” the entropic cost associated with the binding event. By reducing the ligand's ability to sample various conformations, it remains locked into a single conformation to readily bind to the cognate partner.
These lessons from small molecule medicinal chemistry have been effectively translated into the design of peptide-derived ligands that target PPI. Several methods have been offered to conformationally define short peptides to bioactive conformations thereby improving their potency and stability9,10,11,12. As a result, constraining the lead peptide to the bound conformation is usually considered as the first step during the optimization process13. Given that there are few examples of rigidified i-PPI the rules governing the design of rigid ligands have yet to be fully revealed.
Constraining the backbone of the lead peptide to the bound conformation is one of the well-defined design criteria. This has been illustrated by three distinct examples where constraining the backbone to the bound conformation improved the binding affinity10,14,15. It is argued that prepaying the entropic penalty by imposing structural constraints to the backbone is a viable strategy to optimize i-PPI peptides. Our laboratory is interested developing i-PPI against cancer related targets16,17,18,19. One such PPI is through the TKB domain of the E3 ubiquitin ligase, Cbl, which plays an important role in the regulation of protein-tyrosine kinase levels within the cell20,21,22,23.
We recently reported that a pentapeptide, pYTPEP, binds Cbl(TKB) with low-μM affinities24. Here, we report the crystal structure of Cbl(TKB) in complex with the pentapeptide, pYTPEP (PDB ID: 4GPL). Analysis of the complex led to the conclusion that the PEP region adopts the poly-L-proline type II (PPII) conformation. Unlike the common secondary structures, α-helices and β-sheets, which are stabilized by hydrogen bonding, the PPII structure is stabilized by acyclic conformational control elements25,26,27,28,29.
Structure activity relationship (SAR) studies with pYTPXP peptides showed that both the backbone conformation (pYTPPP more active than pYTPAP) and the glutamic acid side chain functionality (pYTPEP more active than pYTPAP) contribute to Cbl(TKB) interaction. Based on these observations and analyses of the crystal structure (PDB ID: 4GPL) we synthesized the unnatural amino acid called proline template glutamic acid (ptE). The ptE analog is built on a 3-azabicyclo[3.1.0]hexane core wherein the Cα-Cβ bond is constrained to the bound conformation by the pyrrolidine ring while the Cβ-Cγ and glutamic acid are constrained to the bound conformation by a cyclopropane ring30,31. The peptide pYTP(ptE)P would therefore encapsulate the structural constraints required for not only the backbone of the PEP region (PPII conformation) but also the glutamic acid side chain functionality to adopt the bound conformation. We expected the pYTP(ptE)P to adopt the PPII conformation in solution and estimated that this preorganization of the PEP region will result in nearly an order of magnitude increase in Cbl(TKB) affinity. The pentapeptide, pYTP(ptE)P was indeed found to adopt the PPII conformation in solution as determined by NMR. Contrary to our estimation, pYTP(ptE)P showed over an order of magnitude loss of activity. Together the data suggests that constraining the backbone alone (PPP vs PAP) improves the binding affinity, however, constraining both the backbone and side chain (PptEP vs PPP or PEP) has a negative effect. Based on our results, we propose that over-rigidification of i-PPI against Cbl(TKB) could have a detrimental effect during the inhibitor design process.
Results
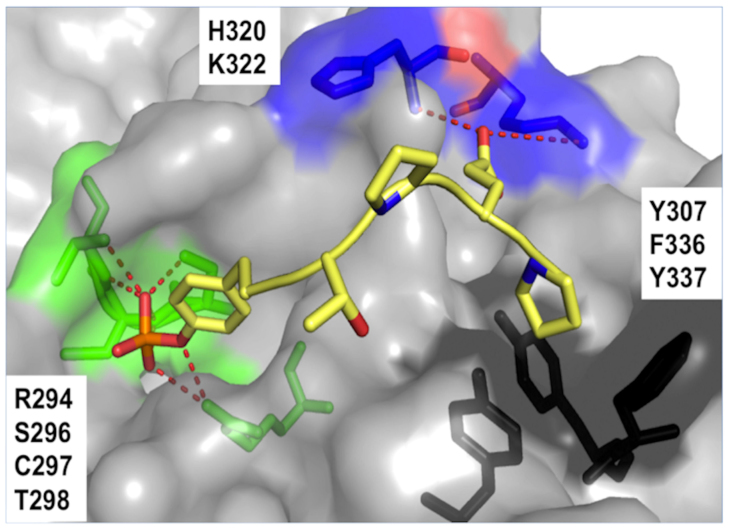
To determine the binding site and binding mode of the pentapeptide, the co-crystal structure of the Cbl(TKB)-pYTPEP complex was solved. We found that the pentapeptide occupied the exact same binding site as the 12-mer peptide (Fig. 1). The N-terminal pTyr presents numerous H-bonds with the protein surface and the p + 4 Pro occupies a shallow, hydrophobic groove. Additionally, the p + 3 Glu favorably interacts with Cbl(TKB) via H-bonds.
Figure 1. Structure of the complex between Cbl(TKB) (gray surface) and the pentapeptide (yellow sticks, PDB ID 4GPL).
The interacting residues at the Cbl(TKB):peptide interface are labeled and are represented as green sticks for the pTyr site, blue sticks for the p+3 Glu site, and black sticks for the p+4 Pro site. H-bonds are shown as red dashes.
Visual examination of the pentapeptide complex did not reveal any obvious cause for our report on the ~4-fold improvement in binding affinity compared to the dodecapeptide. Molecular Dynamics (MD) simulations, however, attributed the improvement to the more costly desolvation penalty for the dodecamer than that of the pentapeptide (Table S1).
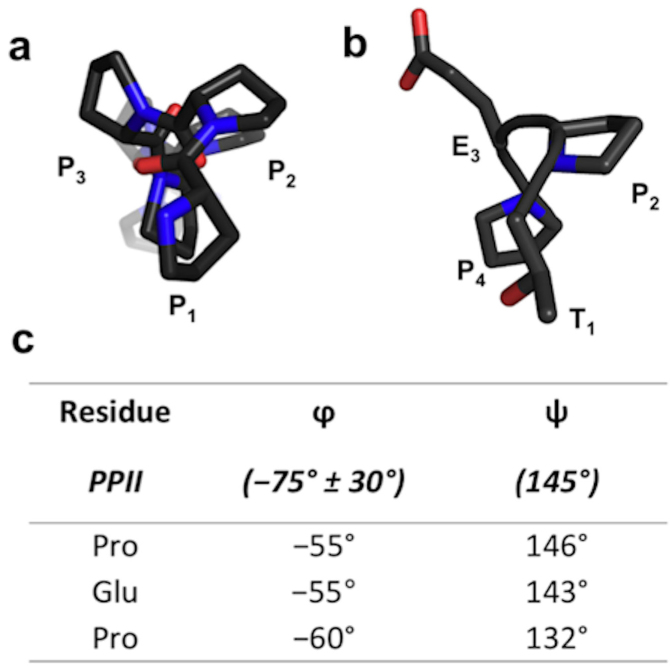
Inspection of the structure also revealed that the backbone geometry of the pentapeptide is nearly indistinguishable from the docecapeptide (Fig. S1). This result experimentally confirms that the pentapeptide adopts the same binding mode as the dodecapeptide. Indeed, this particular conformation is commonly known as the poly-L-proline type II (PPII) helix. The lesser known of the secondary structures found in proteins, PPII helices adopt extended, left-handed helical structures. The PPII helix is characteristic of Pro oligomers and, unlike the more common secondary structures, is stabilized by acyclic conformational control elements rather than intramolecular H-bonds. As a result, proline dimers and oligomers will spontaneously adopt such a conformation. More precisely, PPII is defined by ideal backbone dihedral angles: Φ = −75° and ψ = −145°. In an ideal PPII helix, there is a 3-fold symmetry parallel to the helical axis and every 4th residue is at the same point (Fig. 2a). The pentapeptide, when viewed parallel to the helical axis exhibits the 3-fold symmetry characteristic of the PPII helix (Fig. 2b). The dihedral angles of the bound pentapeptide, indeed, conformed to those of an ideal PPII helix (Fig. 2c). These observations, in conjunction with previous reports, indicate that the bioactive conformation of Cbl(TKB)-binding peptides is the PPII helix.
Figure 2. The bioactive conformation of the pentapeptide in complex with Cbl(TKB).
(a) Pro oligomers present a 3-fold symmetry about the helical axis in an ideal PPII helix. (b) The bound conformation of the pentapeptide presents a characteristic 3-fold symmetry about the helical axis. (c) Dihedral angles extracted from the bound conformation of the pentapeptide adopt those of an ideal PPII helix.
SAR studies with a panel of pentapeptides that bind Cbl(TKB)24 in conjunction with the structural data derived from longer versions of these peptides32,33,34 suggest that deviation from the PPII conformation at the p+3, p+4 and p+5 results in a significant loss of binding affinity (~2–3 orders of magnitude). These observations led us to conclude that the peptide backbone geometry may be a contributing factor in determining Cbl(TKB) binding. As a result, we hypothesized that imposing PPII structural constraints onto the pentapeptide would stabilize the bound conformation and improve binding affinity.
To test this hypothesis, we generated two additional peptides (Table 1), which report on the contribution of backbone constraints to Cbl(TKB) binding. Here, we have used our previously reported fluorescence polarization (FP) assay to quantify Cbl(TKB)-peptide binding35. FP remains a valuable, simple, and cost-effective approach that has a large dynamic range for inhibitor affinities under the same reaction conditions. Using a fluorescently labeled probe and Cbl(TKB), binding relationships for the peptides were generated by FP measurements. Peptide 1 binds Cbl(TKB) with an inhibition constant, Ki = 1.3 μM (Table 1), which is consistent with our previously reported ITC data24. Peptide 2 replaces the p+3 Glu with Ala, which maintains flexibility at the C-terminal PXP region but eliminates the Glu side chain interaction. This modification resulted in a Ki = 18.1 μM, greater than an order of magnitude loss in affinity relative to 1. It is known that proline oligomers including dimers adopt the PPII conformation in solution. Peptide 3 replaces the p+3 Glu with Pro yielding a PPP motif at the C-terminus which we expect to induce PPII conformation in solution. The presence of NOE signals between the α-proton of the ith proline and δ-proton of the (i+1)th proline indicates PPII conformation26. Evidence of peptide 3 to adopt the PPII conformation in solution was confirmed through NOE NMR experiments (Fig. S2). This modification resulted in a Ki = 5.0 μM, which is a 3-fold improvement in affinity relative to peptide 2. Although both peptides 2 and 3 resulted in a loss of affinity for Cbl(TKB), the trend in binding energies indicates that inducing PPII conformation in peptide 3 recaptures more than half the binding affinity lost by peptide 2. This is consistent with the reported studies that show constraining the backbone conformation of the lead peptide to the bound conformation results in increased binding affinity. Clearly the remaining loss was due to a lack of Glu side chain interaction with Cbl(TKB). This is not surprising given that residues on Cbl(TKB) with the potential to make hydrogen bonds with the Glu side chain were observed in the crystal structure.
Table 1. Peptide numbering, sequence, inhibition constants, and binding energy as determined by fluorescence polarization (FP). The variable residues at the p+3 position are highlighted in bold. IC50 values were obtained from competitive binding experiments, converted to Ki according to the Coleska-Wang equation42, and ΔG calculated from the relationship, ΔG = RT ln Ki. Each value represents an average of 4 separate experiments.
| # | Peptide Sequence | Ki (μM) | ΔG (kcal·mol−1) |
|---|---|---|---|
| 1 | pYTPEP | 1.3 ± 0.6 | −7.95 ± 0.21 |
| 2 | pYTPAP | 18.1 ± 5.7 | −6.53 ± 0.22 |
| 3 | pYTPPP | 5.0 ± 2.2 | −7.32 ± 0.30 |
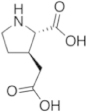
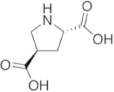
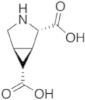
To maintain both backbone constraint and sidechain interaction, a focused panel of unnatural amino acids was examined (Table 2). The amino acid analogues are known as proline templated glutamic acid (ptE) residues. Each contains a pyrrolidine ring with an appended Glu sidechain. The ptEs are so named according to the position of the acidic moiety. A computational study involving energy minimization was used to prioritize our chemical synthesis. To that end, we generated tripeptide (PXP) repeats with each ptE at the middle position. The energy of the tripeptides was minimized and overlaid onto the bound conformation of 1 (Fig. S3). The dihedral angles of the sidechain were extracted and compared to those of the bound peptide, 1 (Table 2). Examining the sidechain dihedral angles (χ1 and χ2) revealed that ptE built on the bicyclic core had the same sidechain dihedral angles as that of the peptide in the bound conformation. We, therefore, concluded that the pYTP(ptE)P peptide will not only preorganize the backbone to the bound conformation but the side chain rigidity will prime the carboxylic acid into an optimal interacting geometry.
Table 2. Sidechain dihedral angle calculation based on energy minimization. Modeled tripeptides containing each proline templated glutamamic acid variant at the center position were energy minimized and the sidechain dihedral angles (χ1 and χ2) were extracted and compared to the sidechain dihedral angles of Glu within the bound conformation of the pentapeptide, 1.
 |
 |
 |
|---|---|---|
| 3-ptE | 4-ptE | ptE |
| Χ1 | Χ2 | |
| PDB ID: 4GPL | (−55°) | (−150°) |
| 3-ptE | −128° | −58° |
| 4-ptE | 10° | −151° |
| ptE | −62° | −167° |
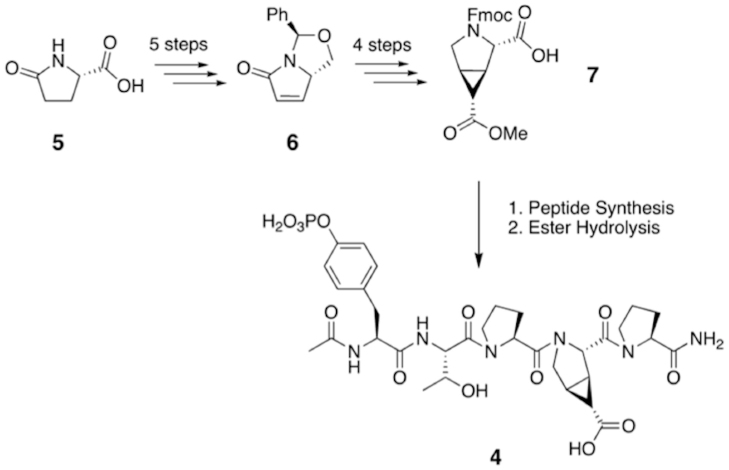
The selected ptE was synthesized in 9 steps from commercially available pyroglutamic acid by adapting known methodologies31,36. The synthetic route to generate ptE is summarized in Figure 3. The final product, 7, is a fully protected compound suitable for peptide synthesis. The modified amino acid 7 was then incorporated into the active pYTPXP sequence at the p+3 position using solid phase Fmoc-peptide synthesis. The methyl ester on the sidechain was hydrolyzed under basic conditions, yielding the free acid and peptide 4. We expect 4 to induce both PPII structure and set the Glu sidechain to the bound conformation. Indeed peptide 4 adopted the PPII conformation as evidenced by the NMR NOE cross peaks (Fig. S2).
Figure 3. Synthetic route to generate 4.
L-pyroglutamic acid, 5, was used to generate the unsaturated bicyclic ring system 6 over a series of 5 steps. 6 then yielded a fully protected cyp-ptE, 7, suitable for peptide synthesis. 7 was incorporated into the pentapeptide sequence pYTPXP, where X denotes the variable amino acid position, using standard solid phase peptide synthesis methodology. Treatment of the purified peptide with NaOH converted the methyl ester to the desired acid, yielding 4.
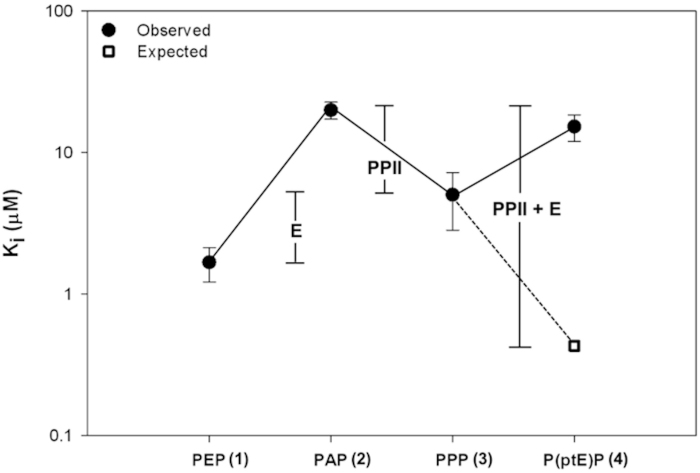
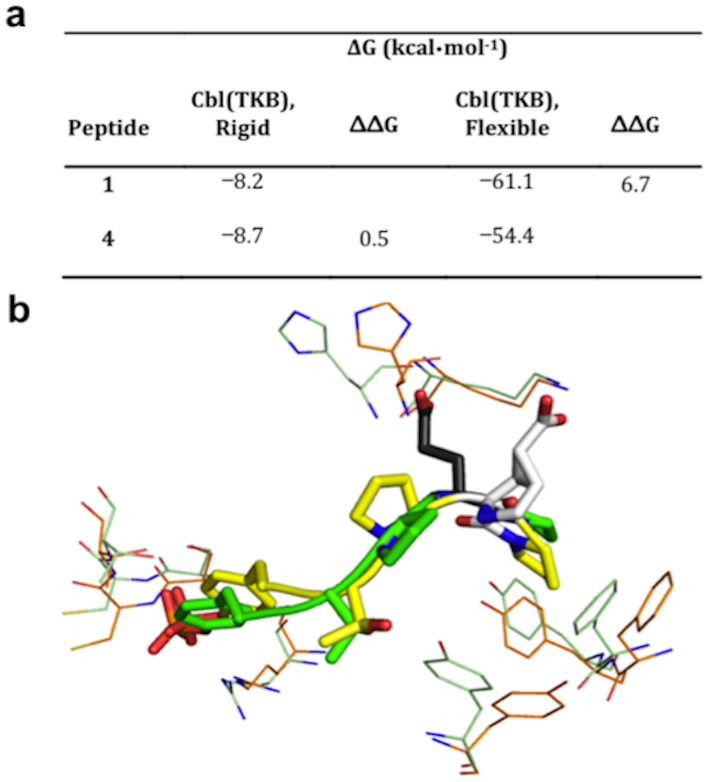
Based on the SAR data we estimated that peptide 4 will have ~ 3-fold improvement in the inhibition constant, Ki (Fig. 4). This estimation was based on the fact that 3 partially recaptured activity lost in 2. Therefore addition of the Glu sidechain functionality to 3 by incorporating ptE would further improve the binding affinity. However the result showed the opposite effect, as 4 was resulted in a Ki = 15.2 μM, which represents ~12-fold decrease in affinity. Together these data suggest that constraining the backbone of the peptide to the bound conformation is beneficial however constraining both the backbone and sidechain to the bound conformation is detrimental.
Figure 4. Inhibition constants, Ki, of pentapeptides in complex with Cbl(TKB).
The x-axis is labeled according to the C-terminal sequence within the peptide pYTPXP. The white box represents the estimated Ki value of 0.43 μM for the P(ptE)P peptide.
To explore a possible cause of the observed loss of affinity, we used a retrospective, computational study with MD and docking simulations. We assessed binding of peptides (1 and 4) to both a rigid receptor and a flexible receptor. When Cbl(TKB) rigidity was maintained, we observed that peptide 4 had a favorable binding energy (ΔΔG) compared to 1 (Fig. 5a). This improvement was, indeed, the expected scenario, however, experimental evidence did not reflect this to be the case. Instead, when Cbl(TKB) was flexible, we observed a favorable ΔΔG of binding for 1 compared to 4, which is consistent with the experimental findings. The simulations with flexible Cbl(TKB) provide a possible molecular basis for the loss of affinity for peptide 4 observed experimentally. Inspection of the computed binding mode of 4 indicates that the ptE residue orients away from productive Cbl(TKB) interaction (Fig. 5b) and is solvent exposed, which is a potential mechanism for the observed loss of activity. Given these pieces of computational information, it seems that the rigid peptide, 4 is unable to occupy the binding site for productive interaction due to the overly rigid structure. Therefore, flexibility in the peptide sidechain is possibly a requirement for the binding interaction. Together these data suggest that Cbl(TKB) binding to the peptide requires flexibility, which is consistent with an induced-fit model.
Figure 5. Computational studies of peptides 1 and 4 binding to Cbl(TKB) using MD and docking simulations.
(a) Comparison of 1 and 4 on of the change in binding energy when receptor is rigid or flexible. (b) Bound conformation as determined by MD. Green and orange lines indicate rigid and flexible Cbl(TKB), respectively. Green and yellow sticks represent 1 and 4, respectively. The variable amino acid at the p+3 position is shown as black sticks for peptide 1 and white sticks for peptide 4.
Discussion
We report the crystal structure of Cbl(TKB) bound to a pentapeptide pYTPEP that binds with low-μM affinity. Analysis of the available structures suggests that the PEP region adopts the PPII conformation in the bound state. Consistent with the other systems, constraining the backbone of the PEP to the PPII conformation resulted in an increase in binding affinity. However the loss of glutamic acid side chain resulted in reduced activity. To address this we synthesized an unnatural amino acid ptE which, when incorporated into the peptide, resulted in constraining both the backbone and the sidechain to the bound conformation.
Several methods have been reported to conformationally define peptides in an effort to improve binding affinity. One such strategy involves aliphatic sidechain “stapling” to force the selected peptide sequence to adopt an α-helical conformation9. In this case, the peptide backbone is rigid while the sidechains remain flexible. Similarly, another reported strategy imposes sidechain constraints while maintaining backbone flexibility. This method has made use of extended β-amino acids to force selected residues to adopt the bound geometry observed in crystal structures15. In both scenarios, extrinsic control elements are employed to bias the geometry of the peptide. The proline-templated amino acid (PTAA) strategy combines both backbone and side chain constraints using intrinsic conformational control elements. The intrinsic conformational control elements have been analogously applied in the hydrogen-bond surrogate (HBS) approach, wherein strategically placed covalent bonds replace hydrogen bonds to induce α-helical conformation11. Given this background, we estimated that the PTAA modification would result in a peptide mimic with increased binding affinity.
Contrary to our expectation, experimental results show that the incorporation of ptE resulted in a loss of binding affinity. A computational study to delineate this difference suggests that peptide binding to Cbl(TKB) supports an induced-fit model. There are two proposed mechanisms to describe protein-ligand binding reactions: conformational selection and folding-after-binding37. The conformational selection mechanism requires that the ligand adopt the bound conformation a priori. The folding-after-binding mechanism requires that the ligand must be unfolded and, subsequently, adopt the bound conformation upon complexation with the protein partner. In this context, the peptide must remain flexible for the side chain to properly orient while transitioning to form the Cbl(TKB)-peptide complex. As a result, our data supports the fold-after-binding mechanism for Cbl(TKB)-peptide binding. Taken together, the induced-fit and fold-after-binding models suggest that constraining both the peptide backbone and the sidechain are detrimental to productive Cbl(TKB) binding interactions. That is, over-rigidification of peptide ligands targeting Cbl(TKB) may not be optimal during inhibitor design. This information can guide future medicinal chemistry efforts to improve the binding affinity to generate potent Cbl(TKB) inhibitors as chemical probes or potential therapeutics.
Methods
Protein purification
Cbl(47–351) in a pET28a plasmid was transformed into Rosetta2(DE3) cells (Novagen), grown in LB broth to an OD600 of 0.8, and induced by addition of 0.5 mM IPTG followed by incubation at 18°C overnight for protein expression. The harvested cell pellet was suspended in buffer (20 mM HEPES, 500 mM NaCl, 2.5 mM CaCl2, 25 mM imidazole, pH 7.7, with a protease inhibitor cocktail), and lysed by 3 passes in an Emulsiflex C5 (Avestin, Inc). The lysate was clarified by centrifugation at 25,000 × g for 30 minutes at 4°C.
Cbl(TKB) was initially purified by nickel chromatography (His-Trap FF column, GE Healthcare) on an AKTAfplc system using the initial buffer as the wash buffer, followed by a 20 column volume gradient to 600 mM imidazole to elute His-Cbl. The eluted protein was quantitated (Bradford assay; Bio-Rad Laboratories, Inc), then treated with thrombin to remove the 6x-His tags. This Cbl pool was treated with benzamidine sepharose resin to remove the thrombin and nickel resin to collect His tags and uncleaved His-Cbl, then filtered to remove resin beads.
The supernatant containing free Cbl(TKB) was concentrated to 10 mg·mL−1 using spin concentrators (9000 MWCO; Pierce Biotechnology), and loaded in 40 mg aliquots onto a Superdex75 16/60 size exclusion column (GE Healthcare). The purified Cbl was eluted in 1 mL fractions in 20 mM HEPES, 500 mM NaCl, 5 mM β-mercaptoethanol, at pH 8.0. Before crystallographic use, the clean protein fractions, as determined from a Coomassie gel, were individually assayed by dynamic light scattering before combining the monomodal, monodisperse fractions into a protein pool.
Crystallization
Purified Cbl(TKB) was concentrated to 10 mg·mL−1 and pYTPEP peptide was added at a 1:2 molar ratio (Cbl:pYTPEP). The mixture was allowed to react overnight in an ice bucket. Crystals grew in a 1 μL:1 μL ratio hanging drop with a reservoir solution containing 0.25 M potassium sodium tartrate tetrahydrate and 20% w/v PEG 3350. The crystal grew in three days at room temperature.
X-ray diffraction data collection
The crystal was then transferred to a solution of 5 μL reservoir and 5 μL 50% glycerol. The crystal was mounted on a MiTeGen micromesh and plunged into a N2 gas stream at 100 K. Data were collected using a Rigaku FR-E superbright Cu Kα rotating-anode generator operating at 45 kV and 45 mA, fitted with a quarter-χ goniometer and VariMaxHR optics. Diffraction images were collected on an R-AXIS IV++ image-plate detector.
A full native data set to 3.0 Å was collected with 1° oscillations with 64 images at a distance of 200 mm with 30 min exposure times. The data were processed using CrystalClear (d*trek)3 (Fig. S4 and Table S2a).
Structure solution and model refinement
Chain A from PDBID 2CBL32 was used for a molecular replacement search model. Molecular replacement was performed at 3 Å resolution with MOLREP in the CCP4i suite38,39, followed by rigid body refinement with REFMAC540 and resulted in a R value of 30.80% and Rfree of 24.09%. Restrained refinement with REFMAC5 resulted in R = 22.15% and Rfree = 28.30%. The resulting model and electron density map were examined with COOT41. The pYTPEP peptide was modeled into Fo-Fc electron density. Then with cycles of REFMAC5 restrained refinement and examination of 2Fo-Fc and Fo-Fc electron density maps the protein model was corrected, Pro47 was added and 36 water molecules were added. Final refinement resulted in R = 20.03% and Rfree = 25.87% (Table S2). Atomic coordinates and structure factors of the Cbl(TKB)-pentapeptide complex have been deposited in the Protein Data Bank with the accession number 4GPL.
Proline-templated glutamic acid synthesis
Full synthetic details and NMR spectra are available in the supplementary information (Fig. S5, Fig S6, and Fig. S7).
Peptide synthesis and purification
The peptide was synthesized using standard Fmoc-chemistry on Rink Amide NovaGel™ resin (0.25 mmol) (EMD) using N-α-Fmoc-protected amino acids (Aapptec) or unnatural N-α-Fmoc protected amino acids (3B Scientific Corporation or Fisher Scientific) and TBTU-HOBt coupling chemistry on a Focus XC synthesizer (Aapptec). Fmoc-acid (5 eq) and TBTU/HOBt (4eq) (Chem-Impex international, INC) were dissolved in 2–3 mL of NMP. DIEA (Sigma) (15 eq) was added to the mixture and incubated for 5 min. This mixture was then added to Fmoc-deprotected peptide resin and allowed to couple for 1 h. Each coupling step was monitored using the Kaiser test (Sigma). To avoid derivatives with deletion, after the coupling step the N-terminal extremities were capped with a 5% acetic anhydride (Sigma), 5% DIEA, 5% HOBt, and 85% NMP. After each coupling and deprotection step, the resin was thoroughly washed with DMF, MeOH and DCM. At the end of the synthesis, the N-terminus of the desired peptide was acetylated as described above. The peptides were then cleaved from the resin using trifluroacetic acid (TFA) (Sigma)/TIS (Sigma)/water (95:2.5:2.5) over a 3 h period. The crude peptides were precipitated in cold ether and air-dried overnight.
Purification was performed on a preparative Agilent LC system (Agilent Technologies) using an Agilent C18 reverse-phase Zobrax 300SB-C18 column (21.2 × 150 mm, 5 micron). Buffer A was water with 0.05% TFA and buffer B was acetonitrile with 0.05% TFA. Gradient was buffer B from 5 to 40% in 20 min then 40 to 100% in 5 min at 20 mL·min−1 flow rate. The peptide fractions were lyophilized on a Sharp freeze −110 (Aapptec). The purity of the peptides was determined by HPLC analysis with a Agilent C18 reverse phase column (4.6 × 50 mm, 3.5 micron) with similar buffers but a gradient from 5 to 50% B in 20 min and a gradient from 50 to 100% B in 5 min with a 1 mL·min−1 flow rate. Electrospray ionization mass spectrometry was carried out on an Agilent HPLC-MS system. The resulting peptide was treated with 4 eq of NaOH for 2 hr. The reaction was quenched with 3 eq HCl and lyophilized. The peptide product was then diluted with 250 μL D2O. Amino acid analysis was used to determine the concentration and purity of each peptide used in this study.
Determination of Ki and ΔG by competitive fluorescence polarization
The competition experiments were performed as previously described24,35. Briefly, titrated amounts of unlabeled peptides were added to the 384-well plates. Then, the mixture of Cbl(TKB) protein and fluorescently-labeled pentapeptide probe at 10 μM and 100 nM, respectively, were added to each well. The plates were allowed to equilibrate at 25°C for 10 min before reading. The IC50 were calculated using the four-parameter logistic equation and, subsequently, used as input values to calculate the Ki (dissociation constant of inhibitor) from the Coleska-Wang equation42. ΔG was calculated according to the equation ΔG = RT lnKi, where R is the universal gas constant in kcal·mol−1 and T is the temperature in Kelvin. All of the data represent the average of 4 independent experiments.
Molecular dynamics and docking simulations
Starting from the crystal structure of Cbl(TKB) in complex with a nonapeptide, SDGpYTPEPA (PDB ID: 2CBL32), we constructed three different peptide-Cbl complex systems. The pentapeptide, pYTPEP, was prepared simply by removing the extra residues SDG and A at the ends of the original nonapeptide. The residues TLN were built and added to the N-terminus of the nonapeptide in VegaZZ43. Both peptides were modified to have an acyl-protected C-terminus and amidated N-terminus. The PDB structure of the pentapeptide, pYTP(ptE)P, was generated using ChemDraw 3D and aligned to pYTPEP in the complex.
We performed Molecular dynamics (MD) simulations on the three complexes, three free peptides, and one apo Cbl domain. The AMBER 99SB force field was used, along with the standard simulation packages, AMBER11 and NAMD 2.844,45,46. For the phosphotyrosine and calcium ion (Ca2+), we used the parametrization of Homeyer et al.47 and Bradbrook et al.48, respectively. The antechamber module and GAFF49 were used to obtain force field parameters for the modified residue, ptE. After the energy minimization, the systems were solvated using a box of TIP3P water50 of at least 12 Å around the complex by tleap module in AMBER11. The systems were composed of about 51,000 atoms. To neutralize the system charge, chloride counterions (Cl−) were inserted. The energy minimizations of 1000 steps and 5000 steps were performed for water and the system, respectively. After 20 ps of solvent equilibrium, the system was gradually heated from 50 K to 300 K. All production runs were performed for 20 ns at 300 K. Postanalysis trajectories from 2–20 ns were considered. The temperature was maintained at 300 K by Langevin dynamics. The particle mesh Ewald (PME) was used for the long-range electrostatic interactions, while the Van der Waals interactions were smoothly cutoff between 10 and 12 Å. A time step of 2.0 fs was used.
Molecular docking of the peptides was performed using AutoDock Vina as previously described24,51. Briefly, the published crystal structure (PDB ID: 1FBV33) with the ligand removed was used as the rigid macromolecule. The Lamarckian genetic algorithm was used to search for minimum energy ligand conformations and orientations. A point grid of 28 × 20 × 26 Å and centered relative to the center of the macromolecule was used for the docking simulation. The size and location of the point grid were selected to include the pYTPEP docking site and to permit free rotation of the docked peptides. A total of 9 unique docking results were generated and the lowest energy conformation that placed the pTyr in the reported site was used for analysis. Ligand structures were designed in ChemDraw 3D and charges were placed on the functional groups that are ionized at pH 7. Torsions were assigned to the ligands by AutoDock Tools. All 3D molecular structures were generated using PyMOL52.
Author Contributions
AN conceived the project. EAK performed the FP titrations, NMR experiments, and docking simulations. QC synthesized the ptE. SK synthesized the peptides. CK and GEB purified, crystallized and solved the structure of Cbl(TKB)-peptide complex. MK and CAC performed the MD simulations. EAK and AN performed data analysis, prepared the figures, and wrote the manuscript. The final manuscript was edited and revised through contributions by all authors.
Supplementary Material
The paradox of conformational constraint in the design of Cbl(TKB)-binding peptides
Acknowledgments
This publication was made possible by grants from the National Center for Research Resources (5P20RR016469) and the National Institute for General Medical Science (NIGMS) (8P20GM103427), a component of the National Institutes of Health (NIH) and its contents are the sole responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. This work was also funded by the Nebraska Research Initiative and the Eppley Cancer Center Support Grant P30CA036727. We would like to thank Jeff Lovelace for technical assistance.
References
- Oltersdorf T. et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435, 677–681 (2005). [DOI] [PubMed] [Google Scholar]
- Vassilev L. T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004). [DOI] [PubMed] [Google Scholar]
- Pflugrath J. W. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr 55, 1718–1725 (1999). [DOI] [PubMed] [Google Scholar]
- Arkin M. R. & Wells J. A. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov 3, 301–317 (2004). [DOI] [PubMed] [Google Scholar]
- Joseph P. R. et al. Structural characterization of BRCT-tetrapeptide binding interactions. Biochem Biophys Res Commun 393, 207–210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussie P. H. et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274, 948–953 (1996). [DOI] [PubMed] [Google Scholar]
- Mullard A. Protein-protein interaction inhibitors get into the groove. Nat Rev Drug Discov 11, 173–175 (2012). [DOI] [PubMed] [Google Scholar]
- Shakespeare W. C. SH2 domain inhibition: a problem solved? Curr Opin Chem Biol 5, 409–415 (2001). [DOI] [PubMed] [Google Scholar]
- Baek S. et al. Structure of the stapled p53 peptide bound to Mdm2. J Am Chem Soc 134, 103–106 (2011). [DOI] [PubMed] [Google Scholar]
- Udugamasooriya G., Saro D. & Spaller M. R. Bridged peptide macrocycles as ligands for PDZ domain proteins. Org Lett 7, 1203–1206 (2005). [DOI] [PubMed] [Google Scholar]
- Patgiri A., Jochim A. L. & Arora P. S. A hydrogen bond surrogate approach for stabilization of short peptide sequences in alpha-helical conformation. Acc Chem Res 41, 1289–1300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Liao W. & Arora P. S. Enhanced metabolic stability and protein-binding properties of artificial alpha helices derived from a hydrogen-bond surrogate: application to Bcl-xL. Angew Chem Int Ed Engl 44, 6525–6529 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner J., Qu H. & Hruby V. J. Peptidomimetics, a synthetic tool of drug discovery. Curr Opin Chem Biol 12, 292–296 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchey L. K., Porter J. R., Ghosh I. & Arora P. S. High specificity in protein recognition by hydrogen-bond-surrogate alpha-helices: selective inhibition of the p53/MDM2 complex. Chembiochem 11, 2104–2107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorbe J. E. et al. Thermodynamic and structural effects of conformational constraints in protein-ligand interactions. Entropic paradoxy associated with ligand preorganization. J Am Chem Soc 131, 16758–16770 (2009). [DOI] [PubMed] [Google Scholar]
- Lokesh G. L., Muralidhara B. K., Negi S. S. & Natarajan A. Thermodynamics of phosphopeptide tethering to BRCT: the structural minima for inhibitor design. J Am Chem Soc 129, 10658–10659 (2007). [DOI] [PubMed] [Google Scholar]
- Yuan Z., Kumar E. A., Kizhake S. & Natarajan A. Structure-activity relationship studies to probe the phosphoprotein binding site on the carboxy terminal domains of the breast cancer susceptibility gene 1. J Med Chem 54, 4264–4268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z. et al. Exploiting the P-1 pocket of BRCT domains toward a structure guided inhibitor design. ACS Med Chem Lett 2, 764–767 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonov A. et al. Dual-fluorophore quantitative high-throughput screen for inhibitors of BRCT-phosphoprotein interaction. Anal Biochem 375, 60–70 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschard P. & Park M. Escape from Cbl-mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell 3, 519–523 (2003). [DOI] [PubMed] [Google Scholar]
- Ogawa S. et al. Deregulated intracellular signaling by mutated c-CBL in myeloid neoplasms. Clin Cancer Res 16, 3825–3831 (2010). [DOI] [PubMed] [Google Scholar]
- Sanada M. et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature 460, 904–908 (2009). [DOI] [PubMed] [Google Scholar]
- Mohapatra B. et al. Protein tyrosine kinase regulation by ubiquitination: Critical roles of Cbl-family ubiquitin ligases. Biochim Biophys Acta 1833, 122–139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar E. A. et al. Peptide truncation leads to a twist and an unusual increase in affinity for casitas B-lineage lymphoma tyrosine kinase binding domain. J Med Chem 55, 3583–3587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan P. M. & McGavin S. Structure of poly-L-proline. Nature 176, 470–478 (1955). [Google Scholar]
- MacArthur M. W. & Thornton J. M. Influence of proline residues on protein conformation. J Mol Bio 218, 397–412 (1991). [DOI] [PubMed] [Google Scholar]
- Stapley B. J. & Creamer T. P. A survey of left-handed polyproline II helices. Protein Sci 8, 587–595 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R. & Madalengoitia J. S. Conformational stability of proline oligomers. Tetrahedron Lett 37, 6235 (1996). [Google Scholar]
- Zhang R., Brownewell F. E. & Madalengoitia J. S. A(1,3) like strain as a key conformational control element in the design of poly-L-proline type II peptide mimics. J Am Chem Soc 120, 3894 (1998). [Google Scholar]
- Zhang R. et al. Poly-L-proline type II peptide mimics as probes of the active site occupancy requirements of cGMP-dependent protein kinase. J Pept Res 66, 151–159 (2005). [DOI] [PubMed] [Google Scholar]
- Mamai A., Zhang R., Natarajan A. & Madalengoitia J. S. Poly-L-proline type II peptide mimics based on the 3-azabicyclo[3.1.0]hexane system. J Org Chem 66, 455–460 (2001). [DOI] [PubMed] [Google Scholar]
- Meng W., Sawasdikosol S., Burakoff S. J. & Eck M. J. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature 398, 84–90 (1999). [DOI] [PubMed] [Google Scholar]
- Zheng N., Wang P., Jeffrey P. D. & Pavletich N. P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 (2000). [DOI] [PubMed] [Google Scholar]
- Ng C. et al. Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates. Embo J 27, 804–816 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar E. A., Charvet C. D., Lokesh G. L. & Natarajan A. High-throughput fluorescence polarization assay to identify inhibitors of Cbl(TKB)-protein tyrosine kinase interactions. Anal Biochem 411, 254–260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamai A., Hughes N. E., Wurthmann A. & Madalengoitia J. S. Synthesis of conformationally constrained arginine and ornithine analogues based on the 3-substituted pyrrolidine framework. J Org Chem 66, 6483–6486 (2001). [DOI] [PubMed] [Google Scholar]
- Kiefhaber T., Bachmann A. & Jensen K. S. Dynamics and mechanisms of coupled protein folding and binding reactions. Curr Opin Struct Biol 22, 21–29 (2012). [DOI] [PubMed] [Google Scholar]
- Vagin A. & Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr 56, 1622–1624 (2000). [DOI] [PubMed] [Google Scholar]
- Winn M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolovska-Coleska Z. et al. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem 332, 261–273 (2004). [DOI] [PubMed] [Google Scholar]
- Pedretti A., Villa L. & Vistoli G. VEGA - An open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. Journal of Computer-Aided Molecular Design 18, 10.1023/B: JCAM.0000035186.90683.f2 (2004). [DOI] [PubMed] [Google Scholar]
- Hornak V. et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D. A. et al. The Amber biomolecular simulation programs. J Comput Chem 26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. C. et al. Scalable molecular dynamics with NAMD. J Comput Chem 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homeyer N., Horn A. H., Lanig H. & Sticht H. AMBER force-field parameters for phosphorylated amino acids in different protonation states: phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J Mol Model 12, 281–289 (2006). [DOI] [PubMed] [Google Scholar]
- Bradbrook G. M. et al. X-ray and molecular dynamics studies of concanavalin-A glucoside and mannoside complexes - Relating structure to thermodynamics of binding. Journal of the Chemical Society - Faraday Transactions 94, 1603–1611 (1998). [Google Scholar]
- Wang J., Wolf R. M., Caldwell J. W., Kollman P. A. & Case D. A. Development and testing of a general amber force field. J Comput Chem 25, 1157–1174 (2004). [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W. & Klein M. L. Comparison of simple potential functions for simulating liquid water. Journal of Chemical Physics 79, 926–935, 10.1063/1.445869 (1983). [Google Scholar]
- Trott O. & Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano W. L. PyMOL User's Guide. (DeLano Scientific LLC, 2004). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The paradox of conformational constraint in the design of Cbl(TKB)-binding peptides