Abstract
High-throughput screening is the dominant method to identify lead compounds in drug discovery. As such, the makeup of screening libraries will largely dictate the biological targets that can be modulated and the therapeutics that can be developed. Unfortunately, most compound screening collections consist principally of planar molecules with little structural or stereochemical complexity, compounds that do not offer the arrangement of chemical functionality necessary for modulation of many drug targets. Here we describe a novel, general, and facile strategy for the creation of diverse compounds with high structural and stereochemical complexity using readily available natural products as synthetic starting points. We show, through evaluation of chemical properties including fraction of sp3 carbons, ClogP, and the number of stereogenic centers, that these compounds are significantly more complex and diverse than those in standard screening collections, and guidelines are given for the application of this strategy to any suitable natural product.
Collections of small molecules are routinely used in high-throughput screens to find new drug leads. In fact, from 1999 to 2008, 45 of the 50 first-in-class small molecule new molecular entity FDA approvals originated from a screen.1 The composition of compound screening collections therefore has a significant impact on the types of drugs that come to market and the efficiency by which next-generation therapeutics are developed.
Many high-throughput screening (HTS) success stories involve biological targets that can be modulated by low molecular weight, relatively planar organic compounds with high sp2 character and low, if any, stereochemical complexity. For example, kinases are outstanding drug targets whose enzymatic activity is typically inhibited at the ATP binding site by organic compounds with no stereogenic centers and high aromatic content.2–3 However, for more complex biological targets, tremendous challenges in lead identification still exist. For example, disruptors of protein-protein interactions4 and inhibitors of transcription factors5 are rarely small, planar compounds with little sterochemical complexity. Thus, most HTS campaigns versus these targets using compounds present in standard screening collections will fail. In addition, compounds active in certain therapeutic areas (e.g., antibacterials) tend to be larger and more complex than the average screening compound.6 For the reasons described above and many others, there is a pressing need for the creation of compounds that are structurally complex and diverse.7
Recognizing this need, creative strategies to rapidly generate collections of complex molecules have appeared. One approach is diversity-oriented synthesis (DOS), where simple starting materials are coupled to make diverse structures that are more natural product-like in terms of size, percentage of sp3 carbons, and number of stereogenic centers.8–12 Other methods include the synthesis of natural product-inspired scaffolds that can be efficiently and differentially decorated,13–14 skeletal diversifications,15–17 use of natural product-derived building blocks for combinatorial synthesis,18 biology-oriented synthesis,19 and the synthesis of chiral and conformationally constrained oligomers.20
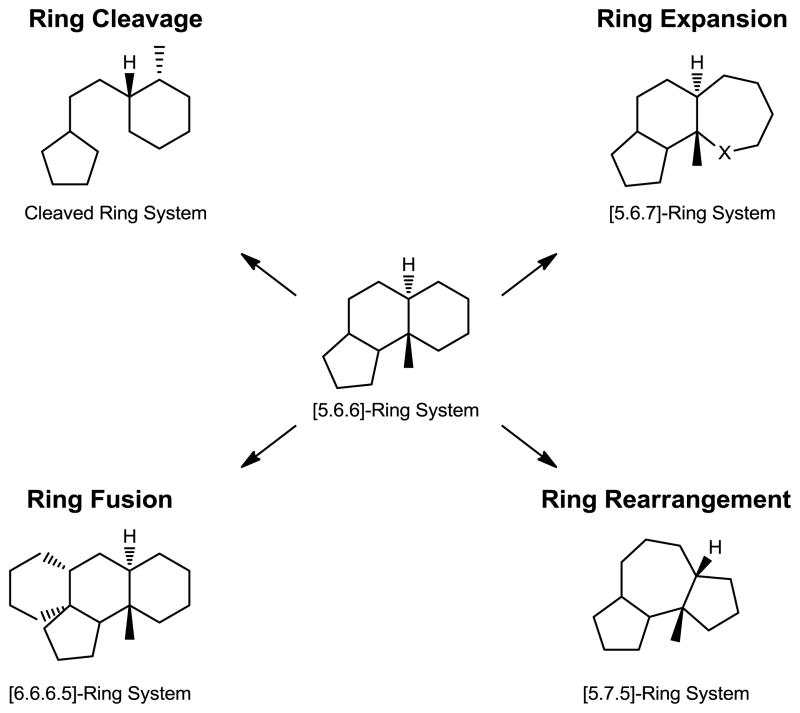
Here we report a new approach for the rapid creation of complex and diverse small molecules. In this process structurally complex natural products are converted, in an average of three chemical steps, to markedly different core scaffolds that are distinct from each other and from the parent natural product. Using chemoselective reactions, the core ring structures of readily available natural products are systematically altered via ring system distortion reactions, i.e., ring cleavage, ring expansion, ring fusion, ring rearrangements, or combinations thereof (Figure 1). Importantly, this method stands in contrast to traditional optimization campaigns whose goals are to enhance the inherent biological activity or improve drug-like properties of a natural product (e.g., erythromycin to azithromycin, penicillin to amoxicillin, etc). To demonstrate this ring distortion strategy, we have selected three readily available and well-studied natural products from different structural classes: gibberellic acid (diterpene), adrenosterone (steroid), and quinine (alkaloid) (Figures 2, 3, and 4, respectively). However, dozens of readily available natural products could be converted into diverse and complex molecules using this strategy. This method takes inspiration from the manner in which nature creates certain complex natural products, using a common intermediate to generate scores of compounds that are very different from one another.21
Figure 1.
Ring distortion reactions can be used to readily convert natural products to complex and diverse scaffolds.
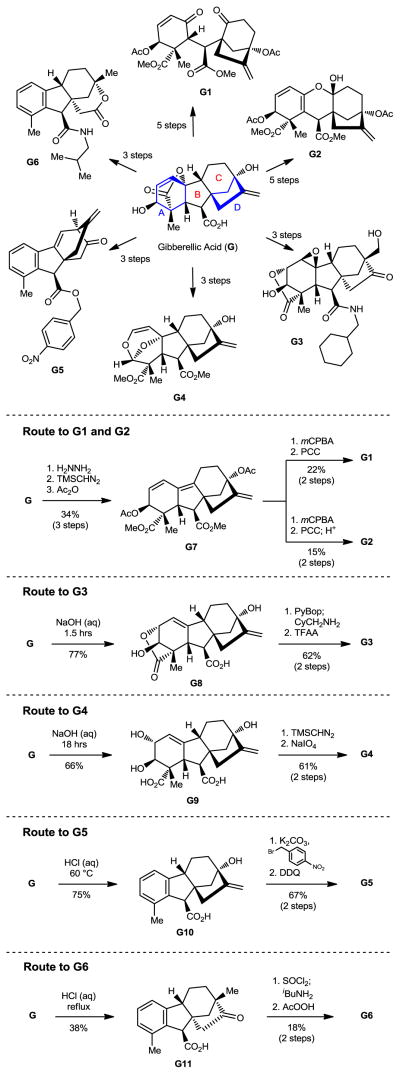
Figure 2.
Application of ring distortion reactions in the synthesis of complex and diverse small molecules from gibberellic acid (G).
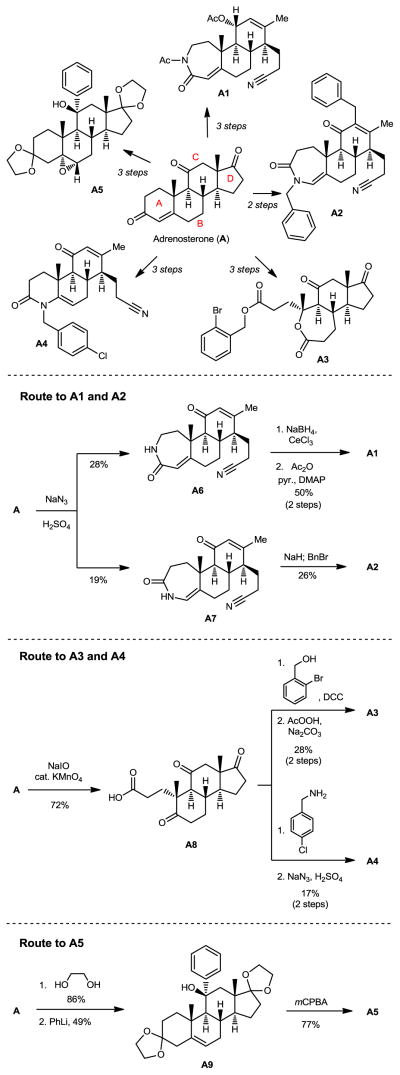
Figure 3.
Application of ring distortion reactions in the synthesis of complex and diverse small molecules from adrenosterone (A).
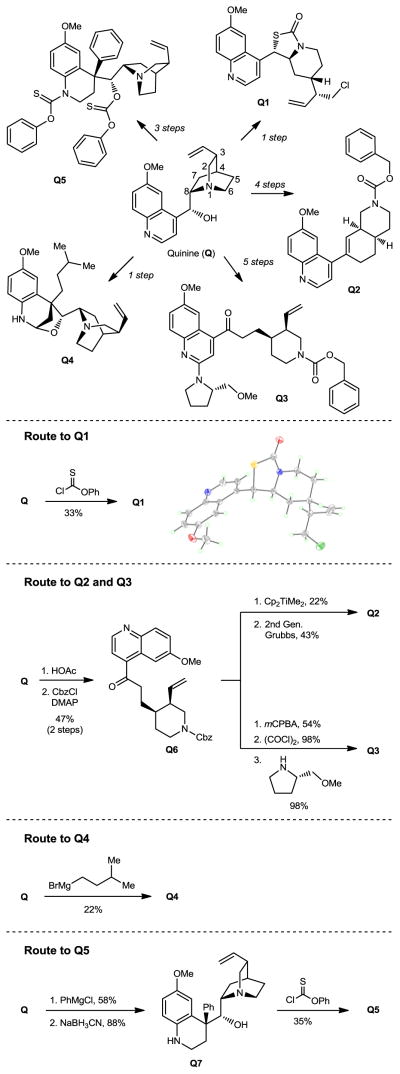
Figure 4.
Application of ring distortion reactions in the synthesis of complex and diverse small molecules from quinine (Q).
RESULTS AND DISCUSSION
Diversifying gibberellic acid
Gibberellic acid (G, Figure 2) is a plant hormone isolated from Gibberella fujikuroi22 and produced industrially23 on the ton scale. Gibberellic acid contains a tetracyclic diterpene core with a fused lactone, two allylic alcohols, an exocyclic olefin, and a carboxylic acid, enabling selective and independent functionalization of each ring of the core structure via a variety of ring system distortion reactions. We have exploited these structural features in concert with known degradation reactions of G24–27 in the construction of complex and diverse scaffolds in three to five steps from gibberellic acid (Figure 2, G1–G6).
Hydrazine-promoted elimination of the lactone on G,27 followed by methylation and acetylation, affords triene G7. Treatment with mCPBA yields an intermediate epoxide with complete selectivity for the tetrasubstituted olefin which, when subjected to oxidative cleavage conditions using PCC, produces diketone G1. Following exposure of G1 to silica or acid, the A-ring ketone tautomerizes and collapses onto the C-ring ketone to form ketal G2; this can be achieved directly from the epoxide precursor using PCC and an acidic workup.
Exposure of gibberellic acid to basic conditions leads to lactone rearrangement and the generation of alkene G8.26 Amidation of G8, followed by treatment with trifluoroperacetic acid provides G3 via epoxidation of both alkenes and Wagner-Meerwein rearrangement to afford the primary alcohol. Prolonged exposure of G to base leads to the cleavage of the lactone ring to provide diol G9.28 Methylation of the carboxylic acids, followed by oxidative cleavage of the diol with sodium periodate and intramolecular [4+2] cycloaddition provides acetal G4.
Treatment of gibberellic acid with dilute hydrochloric acid results in the elimination of the lactone and decarboxylation to aromatize the A-ring, enabling the isolation of allo-gibberic acid (G10).24 Esterification followed by oxidative rearrangement with DDQ29 gives [2.2.2]-bicycle G5. Exposure of gibberellic acid to refluxing hydrochloric acid results in aromatization and a Wagner-Meerwein rearrangement, forming gibberic acid (G11).24 Amidation through an intermediate acyl chloride followed by Baeyer-Villiger oxidation produces lactone G6.
Diversifying adrenosterone
Adrenosterone (A, Figure 3) is a steroid hormone that is produced in the adrenal cortex of mammals.30 Adrenosterone’s structurally complex steroidal framework contains five contiguous stereogenic centers; in addition, each of the four individual carbocyclic rings of adrenosterone is functionalized with an enone or ketone. Though embedded in the A-ring, the enone is also connected to the B-ring as an exocyclic double bond, while the C- and D-rings are each functionalized with a ketone. These key functional groups provide synthetic handles for known chemical reactivity, and can be strategically manipulated to synthesize novel, diverse, and complex chemical scaffolds in three or fewer steps (Figure 3, A1–A5).
During these synthetic investigations of adrenosterone a novel, substrate-dependent Schmidt reaction was discovered that effected both ring expansion and ring cleavage in a single synthetic transformation. Subjecting adrenosterone to Schmidt conditions for one hour gives two constitutional isomers (A6 and A7 in Figure 3) resulting from a tandem D-ring cleavage and A-ring expansion. Final dehydration of the subsequent primary amide in concentrated sulfuric acid results in the observed cyano groups in A6 and A7.
Enamide A6 and lactam A7 were each elaborated to novel complex molecular scaffolds. Lactam A6 undergoes a stereoselective Luche reduction of the C-ring enone, which following treatment with acetic anhydride in pyridine with catalytic DMAP affords A1. Treatment of enamide A7 with sodium hydride followed by benzyl bromide results in enone A2.
Althought not known specifically for adrenosterone, oxidative cleavage of certain steroidal A-ring enones can be effected using NaIO4 and catalytic KMnO4.31 The oxidative cleavage of adrenosterone’s A-ring with NaIO4 and KMnO4 gives acid A8, which upon treatment with 2-bromobenzyl alcohol and DCC provides the corresponding ester. A selective ring expansion at the B-ring ketone using a Baeyer-Villiger reaction with peracetic acid provides lactone A3. Acid A8 condenses with 4-chlorobenzylamine upon heating in ethanol to provide the corresponding A-ring substituted enamide. A final ring cleavage reaction of the D-ring using our Schmidt protocol yields A4.
Adrenosterone undergoes a double ring fusion reaction (at the A- and D-rings) upon treatment with ethylene glycol and catalytic p-TsOH.32 The resulting ketone reacts with phenyllithium to give A9.33–34 Final treatment of A9 with mCPBA results in epoxide ring fusion at the B-ring to yield A5.
Diversifying quinine
Quinine (Q, Figure 4), an alkaloid isolated from the bark of the genus Cinchona, is available in high purity and low cost due to its use as an anti-malarial therapeutic, food additive, and catalyst scaffold.35 Unlike other natural products employed herein, quinine is composed of two discrete ring systems; however, the stereochemical complexity and diverse functionality (a tertiary amine, a secondary alcohol, an olefin, and a quinoline) of quinine make it amenable to selective ring system distortion to create diverse molecular scaffolds (Figure 4, Q1–Q5).
In the course of these investigations we discovered an unprecedented, tandem ring cleavage/ring fusion of Q effected by treatment with thionochloroformate. Ring cleavage of the quinuclidine by O-phenyl thionochloroformate occurs selectively at N1-C2. In addition to the expected ring cleavage and chloride addition, this reaction also leads to diastereoselective rearrangement of the free alcohol and thiocarbamate to form S-thiocarbamate Q1 as a single diastereomer, as confirmed by x-ray crystallography.
In contrast to the selectivity observed with O-phenyl thionochloroformate, acid-catalyzed Hofmann-type elimination of quinine is known to occur exclusively at N1-C8,36 and addition of benzyl chloroformate to the crude degradation product results in ketone Q6, which was elaborated to form two unique structures (Q2–Q3). Petasis methylenation of ketone Q6 followed by 1,2-ring fusion via ring-closing metathesis using second generation Grubbs catalyst forms [4.4.0]-bicycle Q2. Quinoline N-oxidation of Q6 using mCPBA, chlorination with oxalyl chloride, and nucleophilic displacement of the chloride by (S)-2-(methoxymethyl)pyrrolidine provides amine Q3.
Further ring system distortions of the quinoline ring were accomplished through the addition of Grignard reagents.37 Exposure of quinine to isoamylmagnesium bromide in toluene results in nucleophilic addition to the quinoline ring followed by hemiaminal ether formation to provide Q4 as a single diastereomer. Alternatively, reduction of the hemiaminal ether formed through the addition of phenylmagnesium chloride to quinine with sodium cyanoborohydride provides tetrahydroquinoline Q7. Treatment of Q7 with O-phenyl thionochloroformate results in bis-acylation to form Q5 as the major product with no observed chlorine incorporation.
Compound Analysis
In contrast to most standard small molecule library constructions in which simple starting materials are built up into more complex products, an important consequence of starting with natural products is that all intermediates are structurally complex and worthy of inclusion in the final library in their own right. For example, in the course of synthesizing the compounds depicted in Figures 2–4, there were 19, 18, and 12 complex structures produced, respectively (all structures are shown in Supplementary Figure 1). Thus, detailed in Figures 2–4 is the synthesis of 49 structurally and stereochemically complex small molecules from three readily available natural products. Most of these compounds are created in good yield, and importantly, all compounds are created on multi-milligram scale allowing for full structural characterization (see Supplementary Information) and multiple biological screens. In addition, each of these compounds possesses sites for diversification, allowing for the facile and rapid creation of dozens of complex compounds (as described further below).
Advances in chemoinformatics have enabled evaluation of massive chemical and biological data sets, allowing for a rough determination of the structural features of small molecules that correlate with biological activity. It is apparent that many compounds in screening collections have non-trivial liabilities, including non-specific reactivity and a propensity to aggregate, leading to false positives and complicating the development of a hit into a drug.38 When specific disease areas are examined, the problem is more acute. For example, analysis of compounds that kill Gram-negative bacterial pathogens shows an average ClogP of −0.1,39 a realm occupied by vanishingly few compounds in commercial screening collections.
In an attempt to quantify the structural complexity and diversity of the novel compounds created through this paradigm, structural features known to track with biological activity were analyzed. A recent study has examined eight structural parameters (molecular weight, ClogP, polar surface area, rotatable bonds, hydrogen bond donors/acceptors, complexity, and fraction sp3 carbons (Fsp3)) of compounds synthesized by medicinal chemists over the last 50 years, and then compared them to marketed drugs.40 One of the most important conclusions from this analysis is that medicinal chemists are creating compounds with lower-than-ideal Fsp3, and with ClogP values that are higher than ideal. We have calculated Fsp3 and ClogP of the compounds described herein and have compared them to compounds in large screening collections. For this analysis we have used a 150,000-member compound collection from the ChemBridge MicroFormat Library – a standard commercial screening collection and one used by others in comparison analyses.41
Fsp3 is the number of sp3-hybridized carbon atoms in a compound divided by the sum of sp3- and sp2-hybridized carbon atoms.42 Studies have shown the benefits of higher Fsp3, including lower melting points and enhanced aqueous solubility,43 and have revealed that “discovery” compounds have lower Fsp3 than actual drugs (0.36 vs 0.47).43 The analysis of medicinal compounds synthesized over the last 50 years has shown that average Fsp3 is declining,40,44 a result attributed in part to the increasing ease of sp2-sp2 coupling reactions.43 ClogP is often used as a rough measure of lipophilicity; among other things, compounds with higher ClogP values tend to have non-ideal solubility, promiscuity, and off-target toxicity.45 Analysis has shown that the average ClogP for all medicinal compounds synthesized since 1985 has gone up significantly, and is higher than the average for marketed drugs.40,45 Indeed, a survey of 18 pharmaceutical companies from 2000–2010 shows the majority are still synthesizing compounds with a mean ClogP over 4.44 As shown by the histograms in Figure 5a, the new compounds described herein have an average Fsp3 of 0.59, which is considerably higher than those in the commercial collection (avg = 0.23). In a similar vein, the average ClogP for these compounds is 1.1 log units lower than those in the commercial screening set (2.90 vs. 3.99; Figure 5b), corresponding to a 12-fold reduction in hydrophobicity.
Figure 5.
Compounds created through the complexity-to-diversity (CtD) method have markedly different properties from those in commercial screening collections, and are structurally diverse from each other and from the partent natural products. For this analysis, a 150,000 compound collection from ChemBridge Corporation was utilized and compared to the 49 CtD compounds (synthesized in Figures 2–4) for a) fraction of sp3-hybridized carbons (Fsp3), b) calculated logP (ClogP), and c) number of stereogenic centers per compound. d) Tanimoto similarity coefficients for CtD compounds relative to the three natural products and to each other, where 1.0 represents perfect similarity. For full Tanimoto matrix analysis on the 49 compound set, please see Supplementary Figure 3.
The presence of stereogenic centers in a compound can also be used as a surrogate for molecular complexity. Compounds with stereogenic centers may interact more specifically with their chiral receptors, and compounds with low or no stereogenic centers are more prone to attrition during the various stages of drug discovery.43 Commercial screening collections are dominated by achiral compounds; for example, of the 150,000 compound ChemBridge collection, 82% have no stereogenic centers and 14% have a single stereocenter, leaving only 4% of these compounds with multiple stereogenic centers (Figure 5c). Obviously, the synthesis of complex and diverse compounds using natural products as input materials offers a tremendous advantage in this regard. Of the 49 compounds disclosed herein, all have two or more stereogenic centers, with the median number being five (Figure 5c).
While visual inspection of the structures in Figures 2, 3, and 4 readily reveals considerable structural diversity, we have applied a similarity metric to make more quantitative comparisons. For this, Tanimoto coefficients46 were generated in Discovery Studio (Accelrys) using ECFP_6 molecular fingerprints.47 Final structures in Figures 2–4 were used as the reference input for every other compound in the set, and a similarity score was obtained for each pair on a scale from 0 to 1, with 1 representing perfect similarity. As shown by the data in Figure 5d, the final compounds depicted in Figures 2–4 are very different both from one another and from the parent natural products. The G, A, and Q compound sets are expected to be quite different from one another; however, even within the sets the compounds show low Tanimoto coefficients (average for G set = 0.15, average for A set = 0.15, average for Q set = 0.22), indicating considerable structural diversity. For calibration purposes, this analysis was also performed on structures representing simple modifications to the parent compounds. As expected, these minor structural changes afford higher similarity scores (average of 0.67, see Supplementary Figure 2), consistent with work of others using Tanimoto coefficients.48 For the similarity matrix of the full 49 compound set, please see Supplementary Figure 3.
To demonstrate that traditional derivatization strategies can be applied to even these highly complex compounds containing an array of chemical moieties, 12 of the 49 compounds were selected for further functionalization via the synthesis of small libraries. As shown in Supplementary Figure 4, small collections of imides, N-benzylated amides, aryl amides, amides, lactones, secondary and tertiary alcohols, epoxides, triazoles, ureas, and sulfonamides were readily created from these twelve small molecules, and in this manner an additional 120 highly complex compounds were synthesized.
Guidelines
This new complexity-to-diversity approach has been demonstrated with three complex natural products; however, the same logic and methods can be applied to a multitude of other natural products. In general, compounds most amenable to diversification through this method will be available in suitable quantities (either from commercial sources or through isolation) and possess orthogonal functional groups allowing for ring system distortion and diversification through chemoselective reactions. As exemplified with gibberellic acid, adrenosterone, and quinine, certain common ring distortion strategies facilitate the rapid diversification (in ≤5 synthetic steps) of complex natural products:
Ring cleavage reactions. Ring cleavage reactions enable dramatic structural changes in one chemical step, and is the most utilized tactic in our complexity-to-diversity approach. A benefit of ring cleavage reactions is that they typically provide new functional groups that can be further diversified. Examples include the base-promoted hydrolysis of the lactone on isogibberellic acid (G9), oxidative cleavage on adrenosterone (A8), and N-C cleavage on quinine (Q1 & Q6).
Ring expansion reactions. Ring expansion reactions are useful in forming novel ring skeletons or as a prelude to ring cleavage reactions. There are several options for chemical reactions that induce ring expansion, with the Baeyer-Villiger and Schmidt reactions being powerful methods to target ketone and enone functionalities for ring expansion. Ring expansion, as a ring system distortion tactic, has been successfully applied in the synthesis of target structures G6, A3, A6, and A7.
Ring fusion reactions. Ring fusion reactions can provide further diversification by connecting disparate structural elements in the pre-existing ring system or simple addition of a new, constrained ring to the ring system. Various modes of ring fusion were used to demonstrate this tactic on each natural product. For example, the ring-closing metathesis product Q2 (1,2-ring fusion), formation of the [4+2] cycloaddition product G4, and formation of bis-ketal A9 (1,1-ring fusion) result from ring fusion reactions. Ring substitution reactions can also be an example of ring fusion. In the example of ring substitution, the composition of the ring changes without altering the ring size, as exemplified by the creation of the A-ring enamide in A4.
Ring rearrangement reactions. Ring rearrangement reactions that dramatically reorganize the core structure will be dictated by the natural product and are thus applicable on a case-by-case basis. This tactic is illustrated with gibberellic acid in the Wagner-Meerwein rearrangements of G3 and G11, or the DDQ oxidation of G10. These transformations are facilitated by the propensity of the tertiary alcohol in gibberellic acid’s C-ring to form a ketone upon carbon migration, altering the molecular topology.
Historically, natural products or their close analogues have been considered as end points in the drug discovery process. Indeed, this thinking has been quite fruitful; for example, 41% of anticancer drugs and 65% of antibacterial drugs are natural products or very close derivatives.49 The features that make natural products different from most synthetic compounds (e.g., high Fsp3, low ClogP, presence of stereogenic centers) give these compounds a propensity to bind to their macromolecular target with high affinity and specificity, while retaining the solubility and cell permeability needed for a therapeutic agent. The approach described herein uses natural products not as the end point, but as the starting point for the discovery process, starting with compounds inherently biased for biological success and systematically transforming them into diverse compounds of equal complexity. Beyond the three demonstrations herein, Supplementary Figure 5 presents ten other complex and readily available natural products that would be highly suited for manipulation through complexity-to-diversity, and this strategy is generalizable to scores of additional natural products.
Certain chemical properties, such as molecular complexity and multiple stereogenic centers, are extremely difficult to build in when producing large collections of compounds for high-throughput screening. The systematic application of ring distortion reactions on appropriate natural product starting materials offers a convenient approach to rapidly generate large numbers of complex and diverse small molecules. These compounds possess a high degree of molecular complexity, as shown by examination of Fsp3 and number of stereogenic centers, and are structurally diverse, as indicated by Tanimoto similarity analysis. Depending on the exact application, specific structural features (e.g., MW, ClogP, H-bond donors/acceptors, etc) can be programmed in by careful selection of diversification reactions and building blocks. This method to construct complex and diverse small molecules can rapidly provide compounds with properties suitable for a wide variety of biological and medicinal applications.
Methods
Full experimental details and characterization data for all new compounds are included in the Supplementary Information.
Supplementary Material
Acknowledgments
We are grateful to the Office of Naval Research (N00014-09-1-0249) and the University of Illinois for support of this work. R.W.H. III is an American Cancer Society postdoctoral fellow. K.C.M. is a National Science Foundation predoctoral fellow and a Robert C. and Carolyn J. Springborn graduate fellow. M.F.R. is a member of the NIH Chemistry-Biology Interface Training Grant (NRSA 1-T-32-GM070421). We would like to thank Dr. Danielle Gray for X-ray analysis of Q1, Dr. Matthew Brichacek for technical assistance with selected 2-D NMR experiments in addition to many helpful discussions and Dr. Ryan J. Rafferty for technical assistance. We would also like to thank Maaz Ahmed and Amer Al-khouja for the synthesis of various intermediates during these investigations.
Footnotes
Author contributions P.J.H. and R.W.H. III conceived the study. R.W.H. III designed and executed the synthesis of the adrenosterone compound set and constructed libraries based on compounds A1, A2, A3, A5, A9, A12, and A15. K.C.M. designed and executed the synthesis of the gibberellic acid compound set and constructed libraries based on G10, G11, and G19. R.W.H. designed and executed the synthesis of the quinine compound set and constructed the library based on Q1. T.A.F. performed computational analyses of all compound sets in this manuscript. M.F.R. constructed libraries of compounds A3, A12, G10, and G16. P.J.H. supervised this research and wrote this manuscript with the assistance of R.W.H. III, K.C.M., R.W.H. and T.A.F.
Competing financial interests The authors declare no competing financial interests.
References
- 1.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 2.Flaherty KT, Yasothan U, Kirkpatrick P. Vemurafenib. Nat Rev Drug Discov. 2011;10:811–812. doi: 10.1038/nrd3579. [DOI] [PubMed] [Google Scholar]
- 3.Tsai J, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domling A. Small molecular weight protein-protein interaction antagonists: an insurmountable challenge? Curr Opin Chem Biol. 2008;12:281–291. doi: 10.1016/j.cbpa.2008.04.603. [DOI] [PubMed] [Google Scholar]
- 5.Grivas PD, Kiaris H, Papavassiliou AG. Tackling transcription factors: challenges in antitumor therapy. Trends Mol Med. 2011;17:537–538. doi: 10.1016/j.molmed.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 7.Kodadek T. The rise, fall and reinvention of combinatorial chemistry. Chem Commun (Camb) 2011;47:9757–9763. doi: 10.1039/c1cc12102b. [DOI] [PubMed] [Google Scholar]
- 8.Schreiber SL. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science. 2000;287:1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 9.Galloway WR, Isidro-Llobet A, Spring DR. Diversity-oriented synthesis as a tool for the discovery of novel biologically active small molecules. Nat Commun. 2010;1:80. doi: 10.1038/ncomms1081. [DOI] [PubMed] [Google Scholar]
- 10.Spaller MR, Burger MT, Fardis M, Bartlett PA. Synthetic strategies in combinatorial chemistry. Curr Opin Chem Biol. 1997;1:47–53. doi: 10.1016/s1367-5931(97)80107-x. [DOI] [PubMed] [Google Scholar]
- 11.Clemons PA, et al. Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles. Proc Natl Acad Sci U S A. 2010;107:18787–18792. doi: 10.1073/pnas.1012741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui J, et al. Creation and manipulation of common functional groups en route to a skeletally diverse chemical library. Proc Natl Acad Sci U S A. 2011;108:6763–6768. doi: 10.1073/pnas.1015253108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelish HE, Westwood NJ, Feng Y, Kirchhausen T, Shair MD. Use of biomimetic diversity-oriented synthesis to discover galanthamine-like molecules with biological properties beyond those of the natural product. J Am Chem Soc. 2001;123:6740–6741. doi: 10.1021/ja016093h. [DOI] [PubMed] [Google Scholar]
- 14.Goess BC, Hannoush RN, Chan LK, Kirchhausen T, Shair MD. Synthesis of a 10,000-membered library of molecules resembling carpanone and discovery of vesicular traffic inhibitors. J Am Chem Soc. 2006;128:5391–5403. doi: 10.1021/ja056338g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar N, Kiuchi M, Tallarico JA, Schreiber SL. Small-molecule diversity using a skeletal transformation strategy. Org Lett. 2005;7:2535–2538. doi: 10.1021/ol0504345. [DOI] [PubMed] [Google Scholar]
- 16.Balthaser BR, Maloney MC, Beeler AB, Porco JA, Snyder JK. Remodelling of the natural product fumagillol employing a reaction discovery approach. Nature Chem. 2011;3:969–973. doi: 10.1038/nchem.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopp F, Stratton CF, Akella LB, Tan DS. A diversity-oriented synthesis approach to macrocycles via oxidative ring expansion. Nat Chem Biol. 2012;8:358–365. doi: 10.1038/nchembio.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niggemann J, Michaelis K, Frank R, Zander N, Hofle G. Natural product-derived building blocks for combinatorial synthesis. Part 1. Fragmentation of natural products from myxobacteria. J Chem Soc Perkin Trans. 2002;1:2490–2503. [Google Scholar]
- 19.Lachance H, Wetzel S, Kumar K, Waldmann H. Charting, navigating, and populating natural product chemical space for drug discovery. J Med Chem. 2012;55:5989–6001. doi: 10.1021/jm300288g. [DOI] [PubMed] [Google Scholar]
- 20.Aquino C, et al. A biomimetic polyketide-inspired approach to small-molecule ligand discovery. Nat Chem. 2012;4:99–104. doi: 10.1038/nchem.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connor SE, Maresh JJ. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep. 2006;23:532–547. doi: 10.1039/b512615k. [DOI] [PubMed] [Google Scholar]
- 22.Curtis PJ, Cross BE. Gibberellic acid. A new metabolite from the culture of filtrates of gibberella fujikuroi. Chem Ind. 1954:1066. [Google Scholar]
- 23.Rodrigues C, Vandenberghe LP, de Oliveira J, Soccol CR. New perspectives of gibberellic acid production: a review. Crit Rev Biotechnol. 2012;32:263–273. doi: 10.3109/07388551.2011.615297. [DOI] [PubMed] [Google Scholar]
- 24.Cross BE. Gibberellic acid. Part I. J Chem Soc. 1954:4670–4676. [Google Scholar]
- 25.Mulholland TPC. Gibberellic Acid. Part 9. The structure of allogibberic acid. J Chem Soc. 1958:2693–2701. [Google Scholar]
- 26.Henderson JH, Graham HD. A possible mechanism for biological and chemical activity of gibberellic acid. Nature. 1962;193:1055–1056. doi: 10.1038/1931055a0. [DOI] [PubMed] [Google Scholar]
- 27.Grove JF, Mulholland TPC. Gibberellic Acid. Part 12. The stereochemistry of allogibberic acid. J Chem Soc. 1960:3007–3022. [Google Scholar]
- 28.Cross BE, Grove JF, Morrison A. Gibberellic Acid. 18. Some rearrangements of ring A. J Chem Soc. 1961:2498–2515. [Google Scholar]
- 29.Cross BE, Markwell RE. Rearrangements of the gibbane skeleton during reactions with 2,3-di-chloro-5,6-dicyanobenzoquinone. J Chem Soc Perk T. 1973;1:1476–1487. [Google Scholar]
- 30.Idler DR, Schmidt PJ, Bitners I. Isolation and identification of adrenosterone in salmon (Oncorhynchus nerka) plasma. Can J Biochem Physiol. 1961;39:1653–1654. [Google Scholar]
- 31.Borthakur M, Boruah RC. A microwave promoted and Lewis acid catalysed solventless approach to 4-azasteroids. Steroids. 2008;73:637–641. doi: 10.1016/j.steroids.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein S, Littell R, Williams JH. Steroidal Cyclic Ketals. IV. The Conversion of 11-Keto- to 11α-Hydroxysteroids. The Preparation of 11-Epi-hydrocortisone, and Δ4-Androstene-11α-ol-3,17-dione. J Am Chem Soc. 1953;75:1481–1482. [Google Scholar]
- 33.Lecomte V, Stephan E, LeBideau F, Jaouen G. Improved addition of organolithium reagents to hindered and/or enolisable ketones. Tetrahedron. 2003;59:2169–2176. [Google Scholar]
- 34.Stephan E, Brossat M, Lecomte V, Bouit PA. Synthesis of the 11 beta-hydroxymethyl-androst-4-en-3,17-dione. Tetrahedron. 2006;62:3052–3055. [Google Scholar]
- 35.Song CE. Cinchona Alkaloids in Synthesis and Catalysis. Wiley; 2009. pp. 1–10. [Google Scholar]
- 36.Smith AC, Williams RM. Rabe rest in peace: confirmation of the rabe-kindler conversion of D-quinotoxine into quinine: experimental affirmation of the Woodward-Doering formal total synthesis of quinine. Angew Chem Int Ed Engl. 2008;47:1736–1740. doi: 10.1002/anie.200705421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hintermann L, Masuo R, Suzuki K. Solvent-controlled leaving-group selectivity in aromatic nucleophilic substitution. Org Lett. 2008;10:4859–4862. doi: 10.1021/ol801962a. [DOI] [PubMed] [Google Scholar]
- 38.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 39.O’Shea R, Moser HE. Physicochemical properties of antibacterial compounds: implications for drug discovery. J Med Chem. 2008;51:2871–2878. doi: 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
- 40.Walters WP, Green J, Weiss JR, Murcko MA. What do medicinal chemists actually make? A 50-year retrospective. J Med Chem. 2011;54:6405–6416. doi: 10.1021/jm200504p. [DOI] [PubMed] [Google Scholar]
- 41.Feher M, Schmidt JM. Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. J Chem Inf Comput Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 42.Yan A, Gasteiger J. Prediction of aqueous solubility of organic compounds by topological descriptors. QSAR Comb Sci. 2003;22:821–829. [Google Scholar]
- 43.Lovering F, Bikker J, Humblet C. Escape from flatland: increasing saturation as an approach to improving clinical success. J Med Chem. 2009;52:6752–6756. doi: 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]
- 44.Leeson PD, St-Gallay SA. The influence of the ‘organizational factor’ on compound quality in drug discovery. Nat Rev Drug Discov. 2011;10:749–765. doi: 10.1038/nrd3552. [DOI] [PubMed] [Google Scholar]
- 45.Leeson PD, Springthorpe B. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat Rev Drug Discov. 2007;6:881–890. doi: 10.1038/nrd2445. [DOI] [PubMed] [Google Scholar]
- 46.Rogers DJ, Tanimoto TT. A Computer Program for Classifying Plants. Science. 1960;132:1115–1118. doi: 10.1126/science.132.3434.1115. [DOI] [PubMed] [Google Scholar]
- 47.Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 48.Huggins DJ, Venkitaraman AR, Spring DR. Rational methods for the selection of diverse screening compounds. ACS Chem Biol. 2011;6:208–217. doi: 10.1021/cb100420r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.