Abstract
Unnatural amino acids can be genetically incorporated into proteins in live cells by using an orthogonal tRNA/aminoacyl-tRNA synthetase pair. Here we describe a method to efficiently express the orthogonal tRNA and synthetase in Saccharomyces cerevisiae, which enables unnatural amino acids to be genetically incorporated into target proteins in yeast with high efficiency. We also describe the use of a yeast strain deficient in the nonsense-mediated mRNA decay, which further increases the unnatural amino acid incorporation efficiency when a stop codon is used to encode the unnatural amino acid. These strategies will facilitate the investigation of proteins and their related biological processes in yeast by exploiting the novel properties afforded by unnatural amino acids.
Keywords: Unnatural amino acid, Yeast, Orthogonal tRNA, Orthogonal synthetase, Amber suppression, Polymerase III promoter, Nonsense-mediated mRNA decay, Green fluorescent protein
1. Introduction
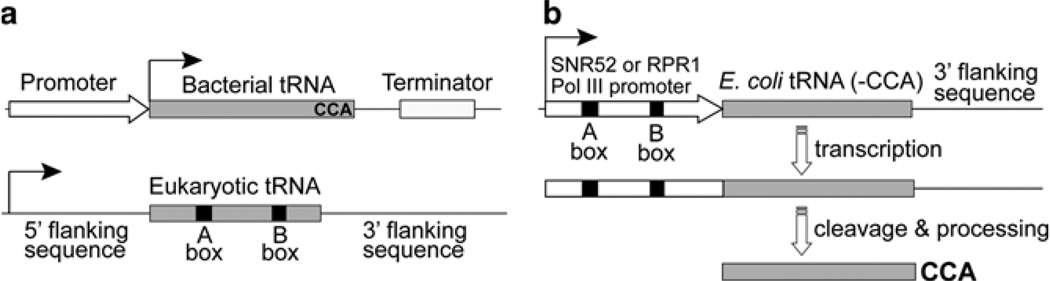
To genetically incorporate an unnatural amino acid into proteins in yeast, an orthogonal tRNA/aminoacyl-tRNA synthetase pair needs to be expressed (1, 2). This tRNA/synthetase pair does not cross-talk with endogenous tRNA/synthetase pairs and is engineered to be specific for the desired unnatural amino acid. One challenge is to efficiently express the orthogonal tRNA in yeast (1, 3). Most orthogonal tRNAs used in yeast are derived from bacteria. However, bacteria and yeast differ significantly in tRNA transcription and processing (4) (Fig. 1a). Bacterial tRNAs are transcribed by the sole RNA polymerase (Pol) through promoters upstream of the tRNA gene. The transcription of yeast (and other eukaryotic) tRNAs by Pol III depends principally on promoter elements within the tRNA known as the A- and B-box (4). The A- and B-box identity elements are conserved among eukaryotic tRNAs, but are lacking in many bacterial tRNAs. In addition, bacterial tRNA genes encode full tRNA sequences, whereas eukaryotic tRNAs have the 3′-CCA trinucleotide enzymatically added after transcription (4). Therefore, transplanting bacterial tRNA directly into the tRNA gene cassette in yeast does not generate functional tRNA.
Fig. 1.
A general method for efficient expression of orthogonal bacterial tRNAs in yeast. (a) Gene elements and organization for tRNA transcription in bacteria (top) and in eukaryotic cells (bottom) are different. (b) Expression of prokaryotic tRNAs in yeast by using an external SNR52 or RPR1 Pol III promoter. These promoters contain the eukaryotic consensus A- and B-box sequences and thus can drive the RNA transcription by Pol III. After the primary RNA is transcribed, the promoter is cleaved to generate the mature tRNA.
We developed a general method to express bacterial tRNAs in yeast (3), which involves placing an external Pol III promoter containing the consensus eukaryotic A- and B-box sequences upstream of the target bacterial tRNA gene (Fig. 1b). The 3′-CCA trinucleotide of the tRNA is excluded, and the tRNA(−CCA) gene is followed by a 3′-flanking sequence of an endogenous yeast tRNA. A primary RNA consisting of the promoter and the tRNA is transcribed, and the promoter is cleaved posttranscriptionally to yield the mature tRNA. Two yeast Pol III promoters, the RPR1 promoter and the SNR52 promoter, have been shown to efficiently drive the expression of Escherichia coli tRNAs in yeast. The expressed E. coli tRNA is six- to ninefold more active in translation than the same tRNA transcribed by using the SUP4 5′-flanking sequence. Alternative methods using a strong Pol II promoter with tandem tRNA repeats (5) or the yeast tRNAArg fused upstream of the target tRNA (6, 7) have also been developed. We will focus on the SNR52/RPR1 promoter method here, as this method works with different E. coli tRNAs and has been reproducibly used in different laboratories (8, 9). A similar approach involving the use of type-3 Pol III promoters also works efficiently in mammalian cells (10).
The amber stop codon, UAG, is often introduced into the gene of interest to specify the site at which the unnatural amino acid is to be incorporated. The Nonsense-mediated mRNA decay (NMD) pathway mediates the rapid degradation of mRNAs that contain premature stop codons in yeast (11), whereas no such pathway exists in E. coli. When stop codons are used to encode unnatural amino acids, NMD could result in a shorter lifetime for the target mRNA and thus a lower protein yield in yeast. We generated an NMD-deficient yeast strain (LWUPF1Δ) by knocking out the upf1 gene from the yeast genome, and found that this strain indeed increases the unnatural amino acid incorporation efficiency in comparison to the wild-type (wt) yeast (3).
To demonstrate this method, we describe here the procedures to incorporate a fluorescent unnatural amino acid 2-amino-3-(5-(dimethylamino) napththalene-1-sulfonamido) propanoic acid (abbreviated as DanAla) into the green fluorescent protein (GFP) at position 39. As shown in Fig. 2, the orthogonal E. coli leucyl amber suppressor tRNA () will be expressed using the SNR52 promoter, and the orthogonal DanAla-specific synthetase (DanAlaRS) (12) will be expressed using the GPD promoter. The target GFP gene with a UAG codon at site 39 will be expressed using the ADH1 promoter in another plasmid. A His6 tag is appended to the C-terminus of GFP for Western detection and affinity purification. The two plasmids will be co-transformed into the wt or LWUPF1Δ yeast strain. The incorporation of DanAla into GFP will be verified with the generation of GFP fluorescence and quantified using flow cytometry or Western blot. DanAla-containing GFP proteins will be extracted from cells and purified using Ni–NTA chromatography.
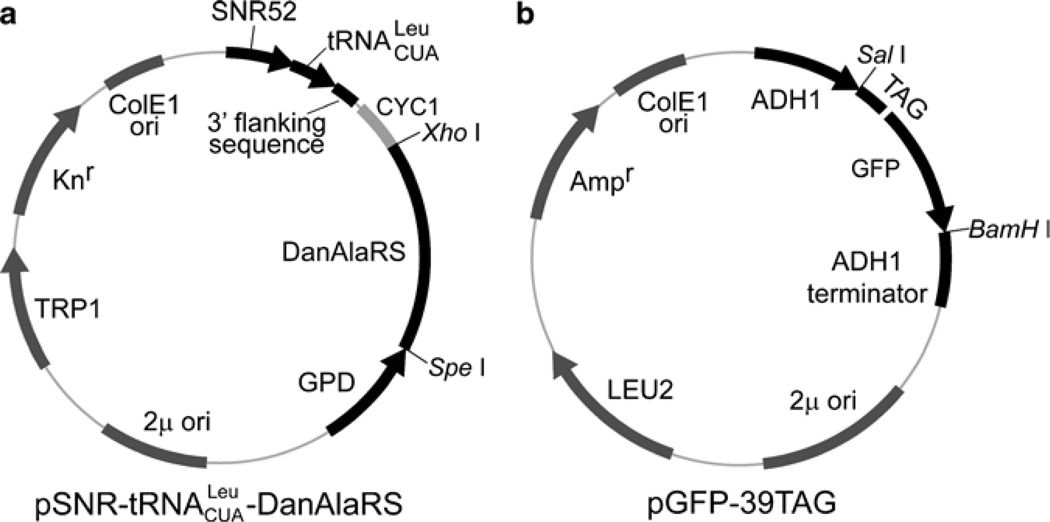
Fig. 2.
Schematic illustration of the expression plasmids. (a) Expression plasmid for the orthogonal tRNA/synthetase. The orthogonal tRNA (without 3′-CCA trinucleotide) is expressed by the SNR52 promoter; the 3′-flanking sequence of SUP4 is appended at the 3′ end. The orthogonal synthetase gene is expressed by the GPD promoter; mutant synthetase genes specific for different unnatural amino acids can be cloned to replace the DanAlaRS gene by using the unique SpeI and XhoI sites. (b) Expression plasmid for the target gene with the UAG codon introduced at the site of interest. Other genes can be cloned into this vector by using the SalI and BamHI sites.
2. Materials
Prepare all solutions with ultrapure water and use analytical grade reagents unless indicated otherwise.
2.1. Stock Solutions
40% d-glucose (1 L): dissolve 400 g of d-glucose in 750 mL water. Adjust the final volume to 1 L. Autoclave at 121°C for 15 min.
50% PEG-3350 (100 mL): add 50 g PEG-3350 in 90 mL water, warm the solution to 50°C to help PEG-3350 to dissolve. Make up the final volume to 100 mL. Autoclave at 121°C for 15 min.
1 M Tris–HCl, pH 8.0 (1 L): dissolve 121.1 g Tris in 800 mL water; adjust pH to 8.0 with HCl. Make up the final volume to 1 L. Autoclave at 121°C for 15 min.
0.5 M EDTA, pH 8.0 (1 L): dissolve 186.1 g Na2EDTA 2H2O in 800 mL water, adjust pH to 8.0 with NaOH. Make up the final volume to 1 L. Autoclave at 121°C for 15 min.
10× TE (1 L): mix 880 mL water, 100 mL 1 M Tris–HCl, pH 8.0, 20 mL 0.5 M EDTA, pH 8.0 completely. Autoclave at 121°C for 15 min.
10× LiAc (1 M, 100 mL): dissolve 10.2 g lithium acetate dihydrate in 90 mL water; adjust pH to 7.5 with dilute acetic acid. Bring volume up to 100 mL. Autoclave at 121°C for 30 min.
500× DanAla (500 mM, 1 mL): weigh 168.5 mg of DanAla, add 750 µL of water and vortex to mix. Add concentrated HCl dropwise until the solution become clear. Add water to bring to 1 mL (see Note 1).
25× EDTA-free protease inhibitor: dissolve one tablet of EDTA-free protease inhibitor cocktail tablet (Roche) in 1 mL of 1× PBS.
2.2. Media
YPD Medium (1 L): 20 g/L Bacto-peptone, 10 g/L yeast extract, 20 g/L d-glucose, pH 6.5. Weigh 20 g bacto-peptone and 10 g yeast extract, transfer to a 1-L graduated cylinder or a glass beaker, and add water to 950 mL. Mix completely and adjust pH to 6.5. Autoclave at 121°C for 15 min. Let the medium cool below 50°C and add 50 mL of sterile 40% d-glucose stock solution.
SD/−Leu/−Trp Medium (1 L): dissolve 6.7 g yeast nitrogen base without amino acids and 0.64 g-Leu/−Trp dropout supplement (see Note 2) in 950 mL water; mix to dissolve. Adjust pH to 5.8 if necessary. Autoclave at 121°C for 15 min. Let the medium cool below 50°C and add 50 mL of sterile 40% d-glucose stock solution.
SD/−Leu/−Trp/+DanAla Medium (1 L): dissolve 6.7 g yeast nitrogen base without amino acids and 0.64 g-Leu/−Trp drop-out supplement (see Note 2) in 948 mL water; mix to dissolve. Adjust pH to 5.8 if necessary. Autoclave at 121°C for 15 min. Let the medium cool below 50°C; add 50 mL of sterile 40% d-glucose stock solution and 2 mL 500× DanAla stock solution.
2.3. Agar Plates
YPD Agar medium (1 L): 20 g/L Bacto-peptone, 10 g/L yeast extract, 20 g/L d-glucose, 20 g/L agar, pH 6.5. Weigh 20 g of bacto-peptone, 10 g of yeast extract and 20 g of agar, transfer to a 1-L graduated cylinder or a glass beaker, and add water to 950 mL. Mix completely and adjust pH to 6.5 (see Note 3). Autoclave at 121°C for 15 min. Let the medium cool to ~55°C, and then add 50 mL of sterile 40 % d-glucose stock solution. Mix completely and pour plates. Allow the medium to solidify at room temperature (see Note 4).
SD/−Leu/−Trp agar Medium (1 L): dissolve 6.7 g of yeast nitrogen base without amino acids, 0.64 g of Leu/−Trp dropout supplement and 20 g agar in 950 mL of water. Mix completely and adjust pH to 5.8 if necessary. Autoclave at 121°C for 15 min. Let the medium cool to ~55°C, and then add 50 mL of sterile 40% d-glucose stock solution. Mix completely and pour plates. Allow the medium to solidify at room temperature.
SD/−Leu/−Trp/+DanAla Agar medium (1 L): dissolve 6.7 g of yeast nitrogen base without amino acids, 0.64 g of Leu/−Trp dropout supplement and 20 g of agar in 948 mL of water. Mix completely and adjust pH to 5.8 if necessary. Autoclave at 121°C for 15 min. Let the medium cool to ~55°C, and then add 50 mL of sterile 40% d-glucose stock solution and 2 mL of 500× DanAla stock solution. Mix completely and pour plates. Allow the medium to solidify at room temperature.
2.4. Transformation Reagents
1× TE buffer (100 mL): add 10 mL 10× TE stock solution into 90 mL sterilized water and mix completely.
Carrier DNA (100 mL): dissolve 200 mg of salmon sperm DNA in 100 mL of 1× TE buffer by stirring at 4°C for 1–2 h. Aliquot to 1 mL. Denature it in boiling water bath for 10 min and chill on ice before use (see Note 5).
TE/Lithium acetate solution (30 mL): add 3 mL of 10× TE, 3 mL of 10× LiAc into 24 mL of sterilized water, mix completely (see Note 6).
PEG/LiAc solution (10 mL): add 1 mL of 10× TE, 1 mL 10× LiAc into 8 mL of 50% PEG-3350 stock solution. Mix completely (see Note 6).
2.5. Other Solutions
1× PBS (pH 7.4): dilute 10× PBS (Roche) to 1× with sterilized water.
Washing buffer: 1× PBS, 0.15 mM NaCl, 20 mM imidazole.
Elution buffer: 1× PBS, 0.15 mM NaCl, 250 mM imidazole.
3. Methods
3.1. Plasmid Construction
3.1.1. Construct the Expression Plasmid for the Orthogonal tRNA/Synthetase (Fig. 2a)
Prepare the gene cassette for expressing the orthogonal . The final cassette consists of the following: PstI – SNR52 promoter – – 3′-flanking sequence – SalI. This cassette can be made using overlapping PCR or ordered directly from companies providing gene synthesis services. See Note 7 for sequence details.
Digest the above gene cassette with PstI and SalI, ligate it with the precut plasmid p-TyrRS (3) to make pSNR- -TyrRS (see Note 8).
PCR amplify the DanAlaRS gene using primer FW29 5′-AGC TCG AGT TAG CCA ACG ACC AGA TTG AG-3′ and FW30 5′-AGA CTA GTA TGC AAG AGC AAT ACC GCC CG-3′. Digest the PCR product with SpeI and XhoI, and ligated into the precut pSNR- -TyrRS to make pSNR- -DanAlaRS (see Note 8).
3.1.2. Construct the Expression Plasmid for the Target Gene, pGFP-39TAG (Fig. 2b) and pGFP
A plasmid containing the 2µ ori, LEU2, Ampr, the ColE1 ori, and MCS is used as the backbone to construct the pGFP-39TAG plasmid (3).
Use site-directed mutagenesis to introduce Tyr39TAG mutation into the EGFP gene. Amplify the mutant GFP-39TAG gene with primer JT171 5′-TAG TCG GAT CCT CAG TGA TGG TGA TGG TGA TGC TTG TAC AGC TCG TCC ATG CC-3′ and primer JT172 5′-TAG TCG TCG ACA TGG ATT ACA AAG ATG ATG ATG ATA AAG TGA GCA AGG GCG AGG AG-3′to add a His6 tag at the C-terminus.
Flank the PCR product with the ADH1 promoter and ADH1 terminator, and clone the whole gene cassette into the backbone plasmid using the HindIII and EcoRI sites to make pGFP-39TAG (see Note 9).
Use similar procedures to make the pGFP plasmid, which expresses the wt EGFP gene without the 39TAG mutation.
3.2. Prepare Yeast Competent Cells
The following procedures are applicable to both wt yeast and the NMD-deficient LWUPF1Δ strain. To determine if the LWUPF1Δ strain would be helpful for your target protein expression, see Note 10.
Streak the frozen glycerol stock of the yeast strain onto an YPD agar plate (see Note 11).
Incubate the plate at 30°C until yeast colonies are >2 mm in diameter.
Scrape the entire colony into 1-mL YPD medium in a Falcon tube, vortex to disperse it (see Note 12).
Transfer the medium into a glass tube and add 4-mL fresh YPD medium.
Incubate in a shaker at 30°C, 230–270 rpm overnight.
Measure OD600 of the overnight culture, and inoculate into 50-mL fresh YPD medium to OD600 = 0.15 in a 500-mL flask.
Incubate in a shaker at 30°C, 230–270 rpm till OD600 = 0.3–0.5. This will take 3–5 h for a strain with a doubling time of 2 h.
Harvest cells in a 50-mL conical tube at 1,500 × g for 5 min at room temperature.
Resuspend the pellet in 10 mL sterile 1× TE.
Pellet the cells at 1,500 × g for 5 min at room temperature.
Resuspend the pellet gently in 10-mL sterile TE/lithium acetate.
Pellet the cells at 1,500 × g for 5 min at room temperature.
Resuspend the pellet gently in 0.5 mL sterile TE/lithium acetate.
3.3. Transformation with Lithium Acetate
Incubate the competent cells at 30°C for 30 min (see Note 13).
Boil 10 µg carrier DNA for 10 min, and quickly chill in ice water.
Mix the carrier DNA with 2 µg pSNR- -DanAlaRS and 2 µg pGFP-39TAG in a sterile 1.5-mL Eppendorf tube (see Note 14). Set up a control tube with carrier DNA only.
Add 150-µL yeast competent cells to each tube (see Note 15).
Incubate at 30°C for 30 min.
Add 840 µL PEG/LiAc solution (see Note 16).
Incubate at 30°C for 30 min.
Heat-shock in a water bath at 42°C for 15 min.
Pellet cells at 20,000 × g for 5–10 s at room temperature, and remove the PEG/LiAc solution sterilely as quickly as you can.
Immediately add 0.2 mL 1× TE to resuspend the pellet by pipetting it up and down gently.
Spread on SD/−Leu/−Trp agar plate to select the transformants (see Note 17).
Incubate at 30°C until colonies are >1 mm in diameter.
3.4. Re-streak Transformed Colonies
Pick ten single colonies and re-streak on SD/−Leu/−Trp agar plate.
Incubate at 30°C until colonies are >2 mm in diameter.
3.5. Verify DanAla Incorporation (see Note 18)
Re-streak single clones on SD/−Leu/−Trp and SD/−Leu/−Trp/+DanAla agar plates.
Incubate at 30°C until colonies are >1 mm in diameter.
Excite the plates with 480-nm light.
Observe the plates through a viewing filter glass that passes light >500 nm.
Green colonies will be seen on SD/−Leu/−Trp/+DanAla plate only. Take a photo with a digital camera if needed.
3.6. Quantify DanAla Incorporation Efficiency (see Note 18)
Pick a single colony from the SD/−Leu/−Trp agar plate, suspend in 1 mL SD/−Leu/−Trp medium, and vortex to disperse.
Transfer the medium into a glass tube and add 4-mL fresh SD/−Leu/−Trp medium.
Incubate in a shaker at 30°C, 230–270 rpm overnight.
Measure OD600 of the culture, and inoculate into 10 mL fresh SD/−Leu/−Trp/+DanAla medium to OD600 = 0.2 in a 50-mL flask; inoculate into 10 mL fresh SD/−Leu/−Trp without DanAla medium to OD600 = 0.2 in a 50-mL flask as the negative control.
Incubate in an orbital shaker at 30°C, 230–270 rpm for 6 h.
Pellet cells at 1,500 × g for 5 min at room temperature.
Wash cells once with 1× PBS and resuspend in 1× PBS.
Perform steps 1–7 for yeast cells transformed with the pSNR- -DanAlaRS and pGFP in SD/−Leu/−Trp medium as a positive control (see Note 18).
Perform steps 1–7 for yeast cells without any plasmids in appropriate medium as a negative control.
Perform steps 1–7 for yeast cells transformed with pGFP-39TAG only in SD/−Leu medium as another negative control.
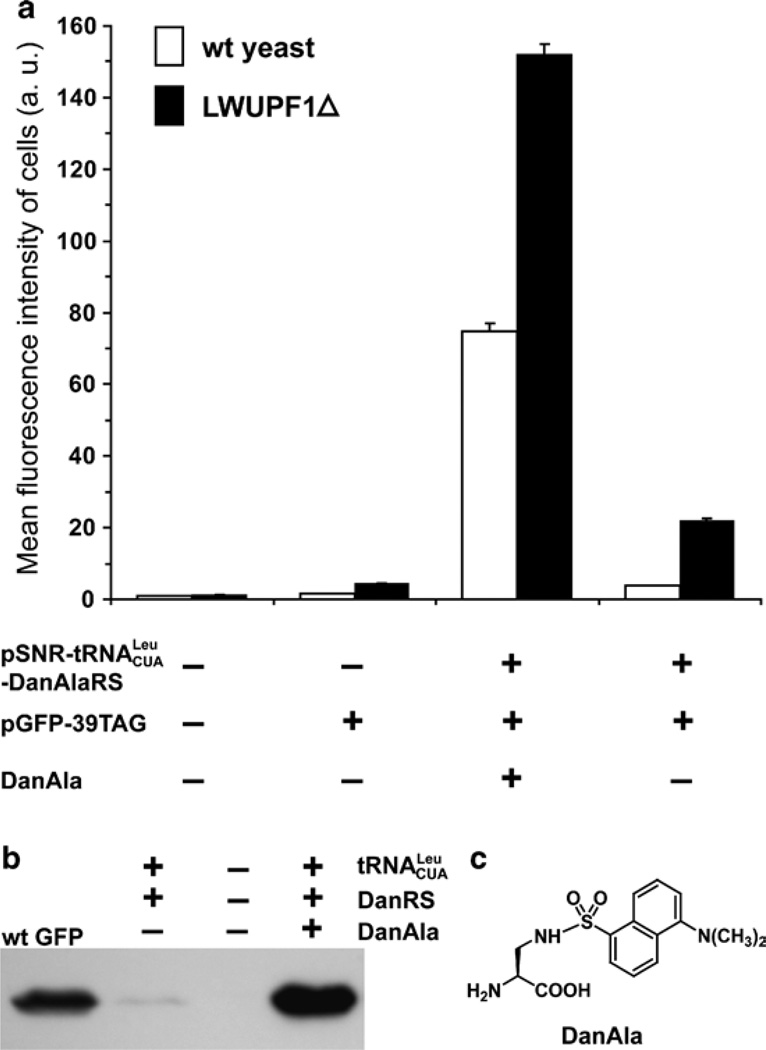
Analyze the fluorescence intensity of all cell samples with a flow cytometer (such as the FACScan from Becton and Dickinson). Use 488 nm as the excitation light and 530 nm (band-width = 30 nm) as the emission filter. Use the negative and positive control samples to adjust the detection sensitivity of the flow cytometer so that the fluorescence intensity readings of all samples are in a good dynamic range. DanAla is not excited at 488 nm. Determine the mean fluorescence intensity of each sample (Fig. 3a). Repeat steps 1–11 ≥3 times independently.
Calculate the DanAla incorporation efficiency using the mean fluorescence intensity of cells (see Note 18).
Fig. 3.
Analysis of DanAla incorporation into GFP in yeast. (a) Flow cytometric analysis of DanAla incorporation into GFP in wt yeast and the NMD-deficient LWUP1Δ strain. Error bars represent S.E.M., n = 3. (b) Western blot analysis of the DanAla-incorporated GFP expressed in the LWUPF1Δ strain. Cell lysates were separated by SDS-PAGE and probed with a monoclonal anti-His5 antibody (Invitrogen). A purified wt GFP with a C-terminal His6 tag was used as a positive control. (c) The structure of DanAla.
3.7. Western Blot Analysis (see Note 19)
Pick a single colony from SD/−Leu/−Trp agar plate, suspend in 1 mL SD/−Leu/−Trp medium, and vortex to disperse.
Transfer the medium into a glass tube and add 4 mL fresh SD/−Leu/−Trp medium.
Incubate in a shaker at 30°C, 230–270 rpm overnight.
Measure OD600 of the culture, and inoculate into 5-mL fresh SD/−Leu/−Trp/+DanAla medium to OD600 = 0.2 in a glass tube; also inoculate into 5-mL fresh SD/−Leu/−Trp medium without DanAla to OD600 = 0.2 in a glass tube as the negative control.
Incubate in an orbital shaker at 30°C, 230–270 rpm for 6 h.
Pellet cells at 1,500 × g for 5 min at room temperature.
Resuspend the pellet in 35 µL Y-PER (Pierce), and add 2 µL 25× EDTA stock solution.
Agitate at room temperature for 60 min.
Centrifuge at 20,000 × g for 10 min; keep the supernatant.
Load purified GFP-His6 protein as a positive control (see Note 20), 10 µL negative control, and 10 µL sample on 12% SDS-PAGE gel.
Transfer the proteins from gel to a PVDF membrane.
Detect with the Penta·His mouse monoclonal antibody (Invitrogen) followed by a goat anti mouse IgG-HRP antibody (see Note 21).
Develop the film with SuperSignal West Pico Chemiluminescent Substrate (Pierce) (Fig. 3b).
Strip the blot and re-probe with an antibody specific for a housekeeping protein for loading control.
3.8. Protein Expression and Purification
Pick a single colony from SD/−Leu/−Trp agar plate, suspend in 1-mL SD/−Leu/−Trp medium, and vortex to disperse.
Transfer the medium into a glass tube and add 4-mL fresh SD/−Leu/−Trp medium.
Incubate in a shaker at 30°C, 230–270 rpm overnight.
Measure OD600 of the culture, and inoculate into 250 mL fresh SD/−Leu/−Trp/+DanAla medium to OD600 = 0.1 in a 1-L flask.
Incubate in an orbital shaker at 30°C, 230–270 rpm for 48 h (see Note 22).
Pellet cells at 1,500 × g for 5 min at room temperature.
Resuspend the pellet in 10 mL Y-PER (Pierce), and add 1/2 tablet of EDTA-free protease inhibitor (Roche).
Agitate at room temperature for 20 min.
Sonicate for 2.5 min (80% power, 3 s on and 10 s off) by using a Sonic Dismembrator (Fisher Scientific).
Centrifuge at 20,000 × g for 10 min; keep the supernatant.
Resuspend the pellet in 10 mL Y-PER (Pierce), add 1/2 tablet of EDTA-free protease inhibitor (Roche).
Repeat steps 8–10, combine the supernatant.
Add 2 mL Ni–NTA slurry (Qiagen, pre-balanced with Y-PER), and incubate at 4°C for 1 h on a lab rocker.
Load the slurry in a column, and wash with 10-bed volumes of 1× PBS buffer, followed by 10-bed volumes of washing buffer.
Elute the DanAla-containing GFP with elution buffer, 1 mL, 3×.
Exchange the protein into 1× PBS buffer by using Centricon concentrators (Amicon).
Run SDS-PAGE to check the purity of the protein.
Determine the protein concentration by using the Bradford assay.
Acknowledgments
We thank Dr. Vicki Lundblad and members of the Lundblad lab for providing reagents and advice on yeast protocols. This work was supported by CIRM (RN1-00577-1) and NIH (1DP2OD004744).
Footnotes
Some unnatural amino acids may be difficult to dissolve; lower the stock concentration to 200 mM or 100 mM when necessary. Solubility is also dependent on the purity of the unnatural amino acid. If racemic mixture of unnatural amino acids is used, the concentration of the effective l-amino acid will be 50% of the calculated value. Use optical pure l-amino acids whenever possible.
Amino acid dropout supplements can be purchased from Clontech (Catalog number 630417). If purchasing from other suppliers, change the amount accordingly by following the product information.
The agar will not fully dissolve until it is autoclaved.
Prepare agar plates in advance. Unsleeve to dry the plates at room temperature for 1 day prior to plating.
Store the boiled carrier DNA at −20°C, which can be reboiled and reused for three times without loss of activity.
Prepare these solutions fresh prior to use.
The sequence for the SNR52 promoter is the following: tctttgaaaagataatgtatgattatgctttcactcatatttatacagaaacttgatgtttt ctttcgagtatatacaaggtgattacatgtacgtttgaagtacaactctagattttgtagtg ccctcttgggctagcggtaaaggtgcgcattttttcacaccctacaatgttctgttcaaaaga ttttggtcaaacgctgtagaagtgaaagttggtgcgcatgtttcggcgttcgaaact tctccgcagtgaaagataaatgatc. The sequence for the is the following: GCCCGGATGGTGGAATCGGTAGACACAAGGGATTCTAAATCCCTCGGCGTTCGCGCTGTGCGGGTTCAAGTCCCGCTCCGGGTA. Note the underlined anticodon CTA, which recognizes the UAG amber codon. The 3′-CCA trinucleotide of the tRNA is not included in the plasmid. The 3′-flanking sequence of the SUP4 is the following: TTTTTTTGTTTTTTATGTCT. We avoided introducing a restriction enzyme site between the SNR52 promoter and the tRNA because such mutations may impair the promoter strength and/or hamper the generation of the correct 5′ end of the tRNA. If a new tRNA does not show activity after being expressed using the SNR52 promoter, check if there is any mutation in the promoter and the tRNA.
Plasmids pSNR- -DanAlaRS and pSNR- -TyrRS harbor the orthogonal E. coli leucyl and tyrosyl amber suppressor tRNA, respectively, and are available from the Wang group (http://wang.salk.edu) upon request. When incorporating unnatural amino acids using orthogonal tRNA/synthetase pairs, the synthetase is evolved to be specific for different unnatural amino acids, but the orthogonal tRNA does not need to be changed and works with all these mutant synthetases. Therefore, mutant synthetases evolved from the E. coli TyrRS are all used with the E. coli tyrosyl amber suppressor tRNA (), and mutant synthetases evolved from the E. coli LeuRS are all used with the . To make an orthogonal tRNA/synthetase expression plasmid for the incorporation of a different unnatural amino acid, one just needs to replace the synthetase gene without recloning the tRNA gene. tRNA genes are generally difficult to be cloned, and the PstI and SalI sites are no longer unique in the final plasmid. The DanAlaRS and TyrRS gene can be swapped for other mutant synthetase genes using the SpeI (N-terminus) and XhoI (C-terminus) unique sites.
To express your gene of interest, any plasmid with LEU2 and 2µ ori can be used. A strong promoter such as the ADH1 promoter used here is preferred for high expression level. Through the above cloning procedure, we built in a unique SalI site at the N-terminus and a BamHI site at the C-terminus of the GFP gene in the pGFP-39TAG. This plasmid is also available through the Wang group (http://wang.salk.edu) as a convenient fluorescent positive control to monitor unnatural amino acid incorporation (see Subheadings 3.5 and 3.6) and to facilitate the cloning of your genes of interest.
The NMD-deficient LWUPF1Δ strain is available at the Wang group (http://wang.salk.edu) upon request. To determine whether the LWUPF1Δ strain will help the expression, check the location of the UAG codon in your gene of interest. NMD in yeast shows a polar effect of nonsense codon positions (13). The steady-state mRNA level is reduced by NMD more significantly when the nonsense codon is closer to the 5′ end than to the 3′ end of an mRNA. Consistently, the increase of unnatural amino acid incorporation efficiency in the LWUPF1Δ strain correlates with the position of the UAG codon: more than a twofold increase is measured when the amber codon is within the N-terminal two thirds of the gene, whereas no significant increase is detected when it is within the C-terminal fourth of the coding region.
For the LWUPF1Δ strain, add 0.5 mg/mL G418 into the YPD medium to keep the selective pressure. If your yeast strain has additional marker or plasmid, use the appropriate SD/drop out medium to keep selective pressure.
Different yeast strains grow at different rates. If colonies are small, or if you are inoculating a larger volume, use several colonies.
For the highest transformation efficiency, use competent cells within 1 h of their preparation.
The concentration of the plasmid is an important factor for the transformation efficiency. The smaller is the volume ratio of DNA mixture to competent cells, the higher will be the transformation efficiency. The total volume of the DNA mixture less than 10 µL is preferable.
The volume of the competent cells should ≥10× volume of the DNA mixture.
The PEG/LiAc solution should be freshly prepared before use.
In order to get single colonies, spread 190 µL suspension on one plate; then add 180 µL fresh 1× TE to the suspension left and spread on another plate.
The pGFP-39TAG can be used as the reporter plasmid to quickly verify the incorporation of an unnatural amino acid by the orthogonal tRNA/synthetase on agar plates, and to quantify the incorporation efficiency of the unnatural amino acid into GFP by using flow cytometry. Green fluorescence of GFP should be detected on cells when the unnatural amino acid is added to the agar plate. In the absence of the unnatural amino acid, no green fluorescence will be detected. To determine the incorporation efficiency, follow the procedures in Subheading 3.6. Measure the mean fluorescence intensity of cells transformed with pSNR- -DanAlaRS and pGFP-39TAG grown in 1 mM of DanAla (Int1), and those grown in the absence of DanAla (Int2). Also measure the mean intensity of cells transformed with pSNR- - LeuRS and pGFP-39TAG (Int3) and of cells transformed with pGFP-39TAG alone (Int4). Here Leu is incorporated by the orthogonal E. coli /LeuRS pair through amber suppression. The ratio defined by (Int1−Int2)/(Int3−Int4) will determine the relative incorporation efficiency of DanAla to Leu. To obtain the net incorporation efficiency of DanAla, measure the mean intensity (Int5) of cells transformed with pSNR- -DanAlaRS and pGFP (a plasmid identical to pGFP-39TAG except that the 39TAG is reverted to wt tyrosine codon TAC). The net DanAla incorporation efficiency is defined by (Int1−Int2)/(Int5−Int2). Note that this GFP reporter can be generally used to evaluate the incorporation of many unnatural amino acids, as the 39TAG site is permissive for GFP fluorescence. Also note that the incorporation efficiency of an unnatural amino acid can be protein-dependent and site-dependent.
Western blot analysis can also be used to determine the incorporation efficiency of the unnatural amino acid into the target protein. Use densitometry to measure the intensities of target protein bands and use the loading control for sample normalization. If the orthogonal tRNA/synthetase for your unnatural amino acid of interest has not been fully characterized before, it is also necessary to perform mass spectrometric analysis of the purified target protein with the unnatural amino acid incorporated (14). Tandem mass spectrometric analysis of protease digested peptides will determine the identity of the amino acid incorporated at the UAG site. A semiquantitative estimation of the incorporation fidelity can be obtained from the peptide intensities (5). Mass analysis of the intact protein will also reveal if common amino acids are incorporated at the UAG site and if there is any misincorporation at other sites in the target protein.
Alternatively, 10 µL of supernatant similarly prepared from cells transformed with pGFP can be used here.
If a primary antibody against the target protein is available, it can be used here for Western detection in replacement of the Penta·His antibody.
The incubation time for expression is protein dependent. We highly recommend a time course experiment to determine the optimal expression time for your target protein. Western blot analysis of cell lysates can be used to monitor target protein expression level conveniently.
References
- 1.Wang Q, Parrish AR, Wang L. Expanding the genetic code for biological studies. Chem Biol. 2009;16:323–336. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, Wang L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J Am Chem Soc. 2008;130:6066–6067. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- 4.Sprague KU. Transcription of eukaryotic tRNA genes. In: Soll D, RajBhandary UL, editors. tRNA: Structure, Biosynthesis, and Function. Washington, DC: ASM Press; 1995. pp. 31–50. [Google Scholar]
- 5.Chen S, Schultz PG, Brock A. An improved system for the generation and analysis of mutant proteins containing unnatural amino acids in Saccharomyces cerevisiae. J Mol Biol. 2007;371:112–122. doi: 10.1016/j.jmb.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 6.Otter CA, Straby KB. Transcription of eukaryotic genes with impaired internal promoters: the use of a yeast tRNA gene as promoter. J Biotechnol. 1991;21:289–293. doi: 10.1016/0168-1656(91)90049-2. [DOI] [PubMed] [Google Scholar]
- 7.Otter CA, Edqvist J, Straby KB. Characterization of transcription and processing from plasmids that use polIII and a yeast tRNA gene as promoter to transcribe promoter-deficient downstream DNA. Biochim Biophys Acta. 1992;1131:62–68. doi: 10.1016/0167-4781(92)90099-l. [DOI] [PubMed] [Google Scholar]
- 8.Majmudar CY, et al. Impact of non-natural amino acid mutagenesis on the in vivo function and binding modes of a transcriptional activator. J Am Chem Soc. 2009;131:14240–14242. doi: 10.1021/ja904378z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HS, et al. Genetic incorporation of a small, environmentally sensitive, fluorescent probe into proteins in Saccharomyces cerevisiae. J Am Chem Soc. 2009;131:12921–12923. doi: 10.1021/ja904896s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W, et al. Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat Neurosci. 2007;10:1063–1072. doi: 10.1038/nn1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- 12.Summerer D, et al. A genetically encoded fluorescent amino acid. Proc Natl Acad Sci USA. 2006;103:9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao D, Parker R. Computational modeling and experimental analysis of non-sense-mediated decay in yeast. Cell. 2003;113:533–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, et al. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]