Abstract
The encapsulation and delivery of short interfering RNA (siRNA) has been realized using lipid nanoparticles1,2, cationic complexes3,4, inorganic nanoparticles5–8, RNA nanoparticles9,10 and dendrimers11. Still, the instability of RNA and the relatively ineffectual encapsulation process of siRNA remain critical issues towards the clinical translation of RNA as a therapeutic1,12,13. Here we report the synthesis of a delivery vehicle that combines carrier and cargo: RNA interference (RNAi) polymers that self-assemble into nanoscale pleated sheets of hairpin RNA, which in turn form sponge-like microspheres. The RNAi-microsponges consist entirely of cleavable RNA strands, and are processed by the cell’s RNA machinery to convert the stable hairpin RNA to siRNA only after cellular uptake, thus inherently providing protection for siRNA during delivery and transport to the cytoplasm. More than half a million copies of siRNA can be delivered to a cell with the uptake of a single RNAi-microsponge. The approach could lead to novel therapeutic routes for siRNA delivery.
RNA interference (RNAi) is a powerful tool for suppressing gene expression, and much research has been directed at efforts to develop an efficient delivery method for short interfering RNA (siRNA; refs 1,2). Conventional complexation or encapsulation of siRNA with polymers or lipids can often require multi-step synthesis of carriers5,13,14 or relatively ineffectual encapsulation processes; furthermore, such approaches often involve introducing a significant amount of an additional component, which can lead to greater potential for immunogenic response or toxicity. Furthermore, the amount of siRNA per carrier is limited owing to the rigidity of double-stranded siRNA, low surface charge of individual siRNA, and low loading efficiency5,9, making RNAi encapsulation particularly challenging. Furthermore, RNAi requires specialized synthesis and is often available only in small quantities at high cost, making it a very costly cargo that is delivered with fairly low efficiency carriers. A different and potentially valuable approach would be to use the DNA/RNA machinery15–18 provided by nature to generate RNAi in polymeric form, and in a manner that actually assembles into its own compact delivery cargo system. In this way, the RNAi is generated in a stable form with multiple copy numbers at low cost, and distributed in a form that can readily be adapted for systemic or targeted delivery.
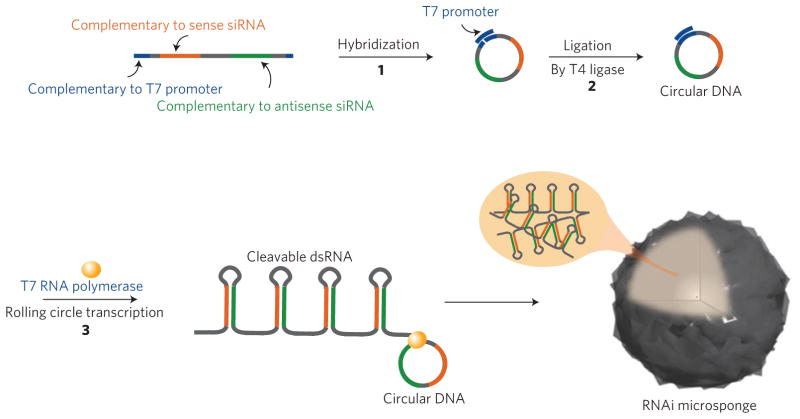
By taking advantage of new RNA synthetic methods16–19 for the generation of nanostructures via rational design20–24, we use an enzymatic RNA polymerization to form condensed RNA structures that contain predetermined sequences for RNAi by rolling circle transcription25 (RCT). Seyhan et al. has previously used the RCT route for generating multimeric transcripts26; however, the resulting RNA does not self-assemble into nanostructures, but forms solubilized lower molecular weight strands. By rational design, we use RNA polymerase to generate elongated pure RNA strands as polymers that can self-assemble into organized nano- to microstructures, which is key for efficient delivery and high cargo capacity, offering the combined benefit of low off-target effects and low toxicity4. Using a new approach, we use the T7 promoter as a primer so that extremely high molecular weight RNA strands can be produced. As shown in Fig. 1, long linear single-stranded DNA encoding complementary sequences of the antisense and sense sequences of anti-luciferase siRNA are first prepared. Because both ends of the linear DNA are also partially complementary to the T7 promoter sequence, the long strand is hybridized with a short DNA strand containing the T7 promoter sequence to form circular DNA [1] (see Supplementary Table S1). The nick in the circular DNA is chemically closed with a T4 DNA ligase [2]. The closed circular DNA is then used to produce RNA transcripts via RCT, encoding both antisense and sense sequences of anti-luciferase siRNA, yielding hairpin RNA structures [3] (see Supplementary Fig. S1). The hairpin RNA structures can actively silence genes when converted to siRNA by Dicer. From in vitro RCT of the circular DNA, we can obtain multiple tandem copies of the sequence in coils of single-stranded and double-stranded RNA transcripts. Although the products might be compared to DNA toroidal condensates27,28, in this case, there is no charged condensing element that assists in the formation of the structure.
Figure 1. Schematic showing the process of rolling circle transcription (RCT) for the self-assembled RNAi-microsponge.
To perform RCT (ref. 35), circular DNA needs to be synthesized first (see Supplementary Information). Linear single-stranded DNA that includes antisense and sense sequences of anti-luciferase siRNA is hybridized with an equal molar amount of short DNA strands containing the T7 promoter sequence. The nick in the circular DNA was chemically closed by T4 DNA ligase. By RCT of the closed circular DNA, multiple tandem repeats of hairpin RNA structures from both antisense and sense sequences are generated to be able to form spherical sponge-like structures.
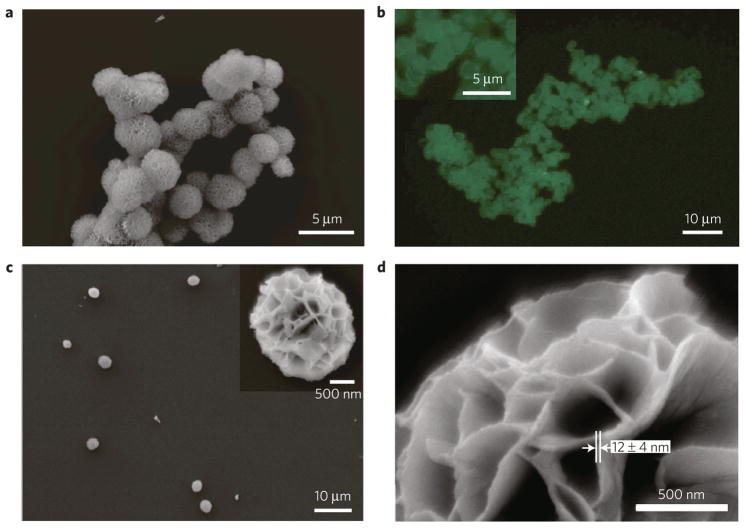
The RNA transcripts form porous sponge-like superstructures with a nanoscopic structure readily visible in the scanning electron microscope (SEM) image (Fig. 2a). Because of the structural similarity, we refer to the resulting RNA product as an RNAi-microsponge. Unlike conventional nucleic acid systems29, our RNAi-microsponge exhibits a densely packed molecular scale structure without the use of an additional agent. We confirmed that the RNAi-microsponges are composed of RNA by staining with SYBR II and labelling with Cyanine 5-dUTP, and observing the resulting bright green and red fluorescence, respectively (Fig. 2b and Supplementary Fig. S2). Also, we provide further evidence with an RNase digestion experiment at various concentrations of RNase. The results clearly show the rate-dependent degradation of the RNA-microsponge at high concentrations of RNase (Supplementary Fig. S3). Monodisperse RNA-microsponges were prepared with short sonication (Fig. 2c and Supplementary Information). The particles exhibit a uniform size of 2 μm, and consistent nanoscale pleated or fan-like spherical morphology. On the basis of the molecular weight and concentration, each RNAi-microsponge contains approximately a half million tandem copies of RNA strands that are cleavable with Dicer (see Supplementary Information). A higher magnification SEM image of the RNA particles reveals that the sponge-like structure is constructed from RNA sheets which are approximately 12 ± 4 nm thick (Fig. 2d).
Figure 2. Characterization of the RNAi-microsponge.
a, SEM image of RNAi-microsponges. b, Fluorescence microscope image of RNAi-microsponges after staining with SYBR II, an RNA-specific dye. c, d, SEM images of RNAi-microsponges after sonication. Low-magnification image of an RNAi-microsponges (c). High-magnification image of an RNAi-microsponge (d).
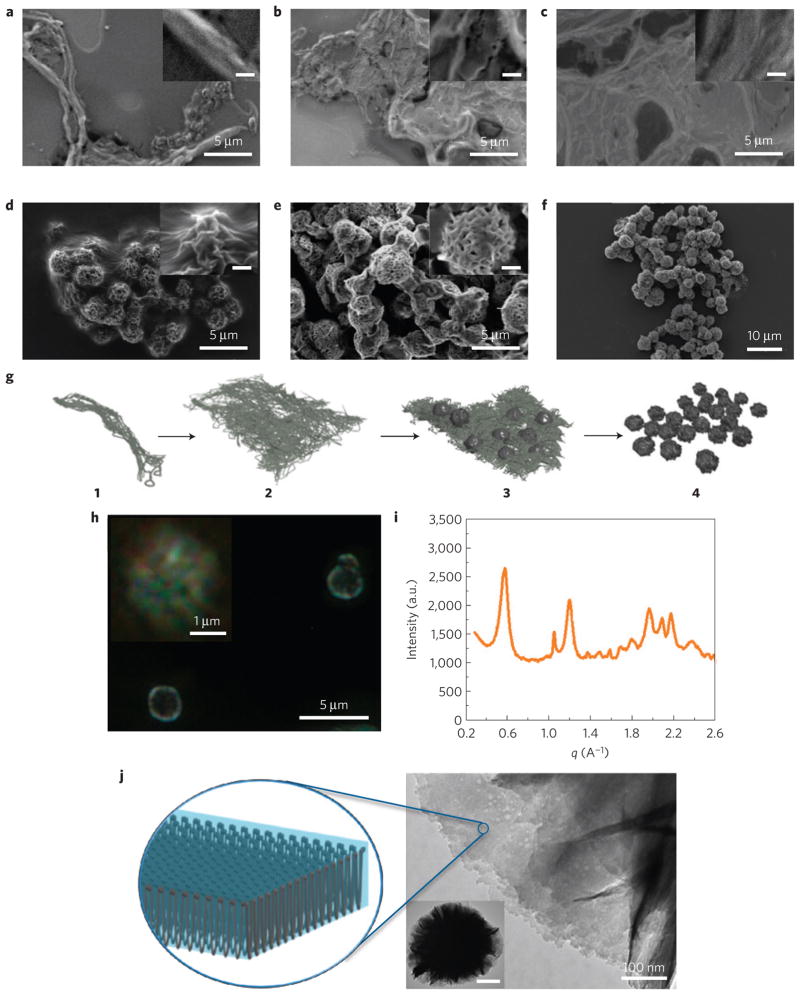
To examine the formation of the sponge-like spherical structures from their RNA strand building blocks, time-dependent experiments were performed during the RCT polymerization. The morphologies of the RNA superstructures were revealed by SEM after 1 h, 4 h, 8 h, 12 h, 16 h and 20 h RCT reaction time. As shown in Fig. 3a, the RCT products first form a fibre-like structure in the early stages of the polymerization. After further reaction time, a sheet-like structure is formed (Fig. 3b). At the 8 h time-point, the sheet-like structure became thicker and began to exhibit a densely packed internal structure (Fig. 3c). Wrinkled and semi-spherical structures begin to appear on the sheet structures in the 12 h reaction sample (Fig. 3d and Supplementary Fig. S4). After 16 h, the morphology of the RNA polymer product transforms into interconnected globular superstructures in which the sheets are re-organized into a complex buckled and folded internal structure (Fig. 3e). These spherical structures start to separate into individual particles, and after 20 h, the final spherical sponge-like structures were observed (Figs 3f and 2a). Based on the SEM images from time-dependent experiments, a schematic cartoon of the process of formation of the sponge-like superstructure is suggested in Fig. 3g. The final structure is reminiscent of the lamellar spherulite structures that are formed by highly crystalline polymers when nucleated in the bulk state or solution30. In the case of traditional synthetic polymers such as polyethylene or polyethylene oxide, the thickness of the lamellar sheets corresponds to the dimensions of chain-folded polymer molecules. It is possible that as the RNA polymer is continuously generated during the RCT reaction, and reaches very high molecular weight at high localized concentrations, a similar ordering and assembly process occurs here31. Thus far, such a self-assembled crystalline superstructure has not been observed for RNA polymers. The crystalline structure of RNAi-microsponges was confirmed by polarizing optical microscopy (POM); under crossed polarizers, birefringence of the individual particles is observed (Fig. 3h). In comparison to the SEM image (inset of Fig. 2c), it seems that the RNA sheet has a crystal-like ordered structure (inset of Fig. 3h). X-ray diffraction further confirmed the crystalline structure of the RNAi-microsponge (Fig. 3i). The crystallite thickness is estimated to be ~7.4 nm, as determined from the Scherrer equation (Supplementary Table S2 and Supplementary Information). This finding is consistent with the thickness from SEM images, although the resolution of SEM is not as sensitive at the nanoscale. Furthermore, transmission electron microscope (TEM) images (Fig. 3j and Supplementary Fig. S5) showing densely assembled RNA sheet structures in the RNAi-microsponge support the proposed structure, as shown in schematic in Fig. 3j. Similar to liquid crystal phases from duplex DNA (ref. 32), the high molecular weight of RNA polymers with periodic RNA duplexes leads to the formation of crystal-like ordered structures. The melting experiment using POM with a heating stage shows that the RNAi-microsponge is pretty stable up to 150 °C, which is much higher than the melting temperature of any double helix DNA or RNA molecules, suggesting that the formation of the RNAi-microsponge is dominantly based on the ordered crystalline structure of RNA polymers (Supplementary Fig. S6). The assembly of the RNA polymer was also observed when polymerized at different concentrations of the rolling circle DNA polymerizing or initiating units (Supplementary Fig. S7). At lower concentrations, individual branched dendritic polycrystals were formed in solution, but they did not assemble into microparticles until a critical concentration of DNA was achieved. The concentration dependence, the appearance of more traditional crystalline structures at low concentration, as well as the observed crystallite thickness of 7.4 nm for the sponge layer structures, which corresponds to the length of the rigid 21 base pair (bp) RNA repeat sequence, were all consistent with phenomena observed for the formation of spherulitic superstructures of chain-folded lamellar sheets30.
Figure 3. Formation of sponge-like spherical structures purely with RNA strands.
a–e, SEM images of RNA products of time-dependent RCT at 37 °C for 1 h (a), 4 h (b), 8 h (c), 12 h (d) and 16 h (e). Inset: scale bar, 500 nm. f, Image of mature RNAi-microsponges after 20 h RCT. g, Schematic of the formation of RNAi-microsponges. The spherical sponge-like structure is formed through a series of preliminary structures. A tandem copy of RNA strands from the RCT reaction are entangled and twisted into a fibre-like structure 1. As the RNA strands grow, they begin to organize into lamellar sheets that gradually become thicker 2; as the internal structure of the sheets begins to get very dense, some of the RNA sheets begin to grow in the z direction, possibly owing to the limited packing area for the RNA polymer as it is produced by the reaction. This process could generate a wrinkled semi-spherical structure on the sheet 3. Finally, the entire structure begins to pinch off to form individual particles consisting of gathered RNA sheets 4. h, Polarized optical microscopy of RNAi-microsponges. i, X-ray diffraction pattern of RNAi-microsponges. j, TEM images of RNAi-microsponges and schematic representation of the proposed crystal-like ordered structure of an RNA sheet in the microsponge. Inset: scale bar, 500 nm.
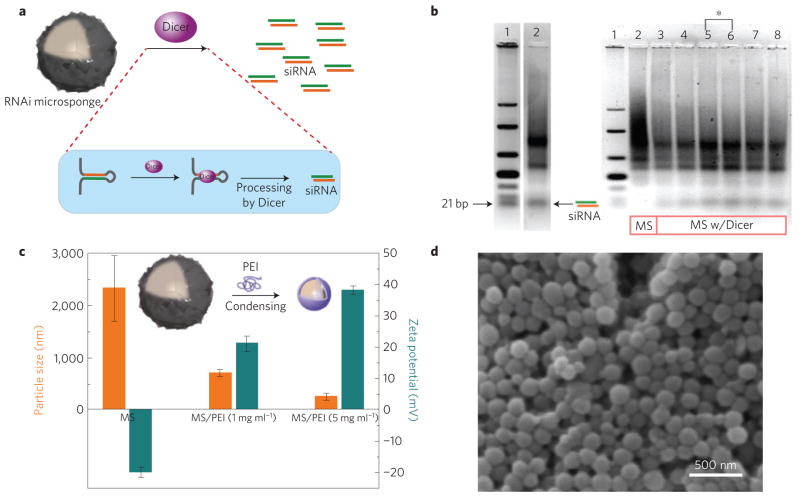
The RNAi-microsponges have a highly localized concentration of RNA strands, as they essentially consist of near 100% potential RNAi. For this reason, these systems should be an effective means to deliver and generate siRNA through intracellular processing mechanisms. The RNA structures were designed to be cleaved by the enzyme Dicer by cutting double-stranded RNA into approximately 21 nucleotide RNA duplexes in the cytoplasm33, where it can be converted to siRNA by the RNA-induced silencing complex (RISC) for gene silencing (Fig. 4a). To confirm Dicer cleavage of RNAi-microsponges, they were incubated with recombinant Dicer and the products were analysed by gel electrophoresis (Fig. 4b). In the presence of recombinant Dicer, RNAi-microsponges yielded 21-bp products (Fig. 4b, left), whereas there are no RNA strands as small as the 21-bp siRNA without Dicer treatment (lane 2 of Fig. 4b, right). Owing to the amount of cleavable RNA strands and the size of the RNAi-microsponge, recombinant Dicer required at least a 36 h reaction time to generate the maximum amount of siRNA (lanes 3–8 of Fig. 4b, right). 9.5% (w/w) of RNAi-microsponges were converted to siRNA, indicating that 21% of the cleavable double-stranded RNA was actually diced to siRNA (Supplementary Table S3 and Supplementary Information). Dicer also produced the two or three repeat RNA units (Fig. 4b). With these results, we estimate that each individual RNAi-microsponges can yield ~102,000 siRNA copies (see Supplementary Information).
Figure 4. Generating siRNA from RNAi-microsponges by the RNAi pathway, and condensing RNAi-microsponges for transfection.
a, Schematic of the generation of siRNA from RNAi-microsponges by Dicer in the RNAi pathway. b, Gel-electrophoresis result after Dicer reaction. On the left, lanes 1 and 2 indicate double-stranded RNA ladder and RNAi-microsponges (MS) after treatment with Dicer (1 unit) for 36 h, respectively. On the right, lanes 1 and 2 indicate double-stranded RNA ladder and RNAi-microsponges without Dicer treatment. Lanes 3–8 correspond to 12 h, 24 h, 36 h and 48 h reaction with 1 unit of Dicer, and 36 h reaction with 1.25 and 1.5 units of Dicer, respectively. Increasing the amount of Dicer did not help to generate more siRNA (lane 7 and 8 of b). The amount of generated siRNA from RNAi-microsponges was quantified relative to double-stranded RNA standards. 21% of the cleavable double-stranded RNA was actually diced to siRNA because Dicer also produced the two or three repeat RNA units. The results suggest the possibility that in a more close-packed self-assembled structure, some portion of the RNA is not as readily accessed by Dicer. c, Particle size and zeta potential before and after condensing RNAi-microsponges with PEI. d, SEM image of further condensed RNAi-microsponges with PEI. The size of the RNAi-microsponges was significantly reduced by linear PEI because the RNAi-microsponges with high charge density would be more readily complexed with oppositely charged polycations. The porous structure of the RNAi-microsponges disappeared following condensation.
To enhance the cellular uptake of the RNA particle, the synthetic polycation, polyethylenimine (PEI) was used to condense the RNAi-microsponge and generate a net positively charged outer layer34. Owing to the high negative charge density of the RNAi-microsponge, cationic PEI was readily adsorbed onto the particles by electrostatic interaction. The change of particle surface charge (zeta potential) from −20 mV (RNAi-microsponge) to +38 mV (RNAi-microsponge/PEI) indicates the successful assembly of RNAi-microsponges with PEI (Fig. 4c). The size of the particles decreased significantly to 200 nm from the original average size of approximately 2 μm (Fig. 4c). The shrinking was also confirmed by SEM, showing approximately 200 nm monodisperse particles (Fig. 4d and Supplementary Fig. S8). It is worth noting that a single PEI-layered RNAi-microsponge still contains the same number of cleavable RNA strands, thus yielding an extremely high siRNA density. To the best of our knowledge, this represents the highest number of siRNA molecular copies encapsulated in a nanoparticle; typically the loading of siRNA can be challenging for standard polymeric carriers.
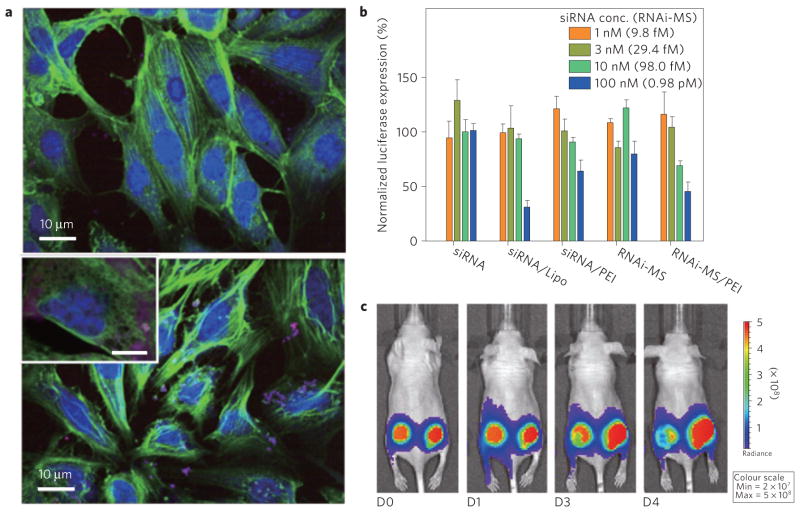
To confirm the cellular transfection of the RNA particles, red fluorescence labelled RNAi-microsponge/PEI was incubated with T22 cells. RNAi-microsponge/PEI particles exhibited significant cellular uptake by the cancer cell line, compared with the uncondensed RNAi-microsponges (Fig. 5a). Because the RNAi-microsponge was designed to generate siRNA for silencing of firefly luciferase expression, the drug efficacy was determined by measuring the fluorescence intensity of cell lysate after transfection (Fig. 5b and Supplementary Fig. S9). As expected, naked siRNA did not show any significant gene silencing up to 100 nM siRNA, whereas RNAi-microsponges showed slightly reduced gene expression at 980.0 fM. PEI-layered RNAi-microsponges efficiently inhibited the firefly luciferase expression down to 42.4% at a concentration of 980 fM. The RNAi-MS/PEI delivery system shows better silencing efficiency than siRNA/PEI. The level of gene knockdown was also evaluated with in vivo optical images of firefly luciferase expression in tumours after intratumoral injection of RNAi-microsponge/PEI (Fig. 5c and Supplementary Fig. S10). As can be seen in Fig. 5c, after four days the level of firefly luciferase expression in the tumour was significantly reduced for the PEI-layered RNAi-microsponges; however, there is no significant decrease in firefly luciferase expression with a control RNA-microsponge/PEI that does not knock down luciferase (Supplementary Fig. S11). Note that extremely low numbers (2.1 fmol) of RNAi-microsponge/PEI particles were used to achieve significant gene silencing efficiency—roughly three orders of magnitude less carrier was required to achieve the same degree of gene silencing as a conventional particle-based vehicle6. Compared with other strategies, siRNA delivery using our RNAi-microsponges provides synergistic effects for loading efficiency, drug efficacy, and low cytotoxicity (Fig. 5b, c and Supplementary Fig. S12).
Figure 5. Transfection and gene silencing effect.
a, Intracellular uptake of red fluorescent dye-labelled RNAi-microsponges without PEI (top) and RNAi-microsponge/PEI (bottom). To confirm the cellular transfection of RNA particles, both types of particles, labelled for red fluorescence, were incubated with T22 cells. Fluorescence labelled RNAi-microsponges without a PEI outer layer showed relatively less cellular uptake by the cancer cell line (T22 cells) suggesting that the larger size and strong net negative surface charge of RNAi-microsponges probably prevents cellular internalization. Inset: scale bar, 5 μm. b, Suppression of luciferase expression by siRNA, Lipofectamine complexed with siRNA (siRNA/Lipo), siRNA complex with PEI (siRNA/PEI), RNAi-microsponge (RNAi-MS), and RNAi-microsponge condensed by PEI (RNAi-MS/PEI). The same amount of siRNA is theoretically produced from RNAi-microsponges at the concentration in parentheses. c, In vivo knockdown of firefly luciferase by RNAi-MS/PEI. Optical images of tumours after intratumoral injection of RNAi-MS/PEI into the left tumour of a mouse and PEI solution only as a control into the right tumour of the same mouse.
We demonstrated a new class of siRNA carrier, the RNAi-microsponge, which introduces a new self-assembled structure that provides a route for the effective delivery of siRNA. The RNAi-microsponge presents a means of rapidly generating large amounts of siRNA in a form that assembles directly into a drug carrier that can be used for direct transfection simply by coating with a positively charged polyion. Given the high cost of therapeutic siRNA and the need for high levels of efficiency, this approach could lead to much more directly accessible routes to therapies involving siRNA. The siRNA, which is highly prone to degradation during delivery, is protected within the microsponge in the crystalline form of polymeric RNAi. We can significantly reduce the difficulties of achieving high loading efficiency for siRNA using this approach. The microsponges are able to deliver the same transfection efficiency with a three orders of magnitude lower concentration of siRNA particles than typical commercially available nanoparticle-based delivery8. Furthermore, the ease of modification of the RNA polymer composition enables the introduction of multiple RNA species for combination therapies. The RNAi-microsponge presents a novel materials system in general owing to its unique morphology and nanoscale structure within the polymer particle, and provides a promising self-assembling material that spontaneously generates a dense siRNA carrier for broad clinical applications of RNAi delivery using the intrinsic biology of the cell.
Methods
In vitro RCT by T7 RNA polymerase to create RNA-microsponges
Ligased circular DNA templates (0.3 μM) were incubated with T7 RNA polymerase (5 units μl−1) at 37 °C for 20 h in the reaction buffer (8 mM Tris-HCl, 0.4 mM spermidine, 1.2 mM MgCl2, and 2 mM dithiothreitol) including 2 mM ribonucleotide tri-phosphate in the final concentration. To fluorescently label the RNA particle, Cyanine 5-dUTP (0.5 mM) was added. The resultant solution was pipetted several times and then sonicated for 5 min to break any possible connection of the particles. The solution was centrifuged at 6,000 r.p.m. for 6 min to remove the supernatant. Then, RNase-free water was added to wash the particles. The solution was sonicated again for 1 min, then centrifuged. This washing step was repeated three more times to remove the reagents of RCT. Measurement of the RNA-microsponge concentration was conducted by measuring fluorescence using Quant-iT RNA BR assay kits (Invitrogen). 10 μl of RNA-microsponge solution or standard solution was incubated with 190 μl of working solution for 10 min at room temperature. The fluorescence was measured at 630/660 nm using a Fluorolog-3 spectrofluorometer (Horiba Jobin Yvon).
Treatment of RNAi-microsponges with recombinant Dicer
RNAi-microsponges were digested with from 1 unit to 1.5 unit recombinant Dicer (Genlantis, San Diego, CA) in 12 μl of reaction solution (1 mM adenosine tri-phosphate, 5 mM MgCl2, 40% (v/v) Dicer reaction buffer). The samples treated for different reaction time from 12 h to 48 h were collected and were then inhibited by adding Dicer stop solution (Genlantis, San Diego, California).
Degradation experiments of RNAi-microsponges
RNA-microsponges were incubated for 24 h in 10% of serum at 37 °C. Degradation experiments with various concentrations of RNase were also performed for 24 h at 37 °C (NEB, Ipswich, Massachusetts).
Characterization of RNAi-microsponges
JEOL JSM-6060 and JSM-6070 scanning electron microscopes were used to obtain high-resolution digital images of the RNA-microsponges. The sample was coated with Au/Pd. A JEOL 2000FX transmission electron microscope was used to obtain the internal structure of the RNA particle. A Zeiss AxioSkop 2 MAT fluorescent microscope was used to image green fluorescently stained RNA-microsponges by SYBR II. To characterize the crystalline structure of RNA-microsponges, laboratory X-ray powder diffraction (XRD) patterns were recorded using a PANalytical X’Pert Pro diffractometer, fitted with a solid-state X’Celerator detector. The diffractometer uses Cu Kα radiation (λ(Kα1) = 1.5406 Å, λ(Kα2) = 1.5433 Å, weighted average λ = 1.5418 Å) and operates in a Bragg geometry. The data were collected from 5° to 40° at a scan rate of 0.1° min−1.
Assembly of the PEI layer on RNAi-microsponges
For assembly of the outer layer, RNA-microsponges were mixed with PEI solution, used at a final concentration of up to 5.0 mg ml−1. Free PEI was easily removed by centrifugation at 13,700 r.p.m. for 30 min. This step was repeated two more times. The PEI-layered RNA particles were resuspended in PBS solution (pH 7.4) or MilliQ water.
In vitro siRNA knockdown experiments
T22 cells were maintained in growth media comprised of Minimum Essential Media-Alpha Modification (MEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. Three days before knockdown experiments, cells were seeded in 6-well plates at 30,000 cells per well. Two days before transfection, each well was co-transfected with 3.5 g each of pRL-CMV and gWiz-Luc using Fugene-HD according to the manufacturers instructions. One day before transfection, cells were trypsinized and re-seeded in 96-well plates at an initial seeding density of 2,000 cells per well. Cells were allowed to attach and proliferate for 24 h. All knockdown experiments were performed in triplicate. 50 μl of fluorescently labelled RNAi-MS and RNAi-MS/PEI were added to 250 μl phenol-free Opti-MEM at the final concentration of up to 21.2 fM. Lipofectamine/siRNA complexes were formed at a 4:1 ratio (v/w). The growth media were removed and Opti-MEM was added to cells, followed by RNAi-microsponges or complexes in PBS, for a total volume of 150 μl per well, with no less than 100 μl Opti-MEM per well. Cells were incubated with siRNA constructs for 4 h, after which the media were removed and replaced with 10% serum-containing growth medium. A luciferase assay was performed using the Dual-Glo Luciferase Assay Kit (Promega, Madison, Wisconsin) and measured on a Perkin Elmer Plate 1420 Multilabel Counter plate reader. Green flourescent protein expression was measured after quenching the luciferase signal with the Stop-and-Glo reagent from Promega.
In vivo siRNA knockdown experiments
T22-Luc is a genetically defined mouse ovarian cancer cell line (p53−/−, Akt, myc) that stably expresses luciferase after infection with pMSCV-puro-firefly luciferase viral supernatant and selecting the cells in a medium containing 2.0 μg ml−1 of puromycin for one week. T22-Luc tumours were induced on both hind flanks of female nude mice (five weeks old) with a single injection of 2–5 million cells in 0.1 ml media. After the tumours grew to ~100 mm (ref. 3) in volume, intratumoral injections of RNAi-microsponges were given in volumes of 50 μl. To determine the degree of luciferase knockdown, D-Luciferin (Xenogen) was given intraveneously (tail vein injection, 25 mg kg−1) and bioluminescence images were collected on a Xenogen IVIS Spectrum Imaging System (Xenogen, Alameda, California) 10 min after injection. Living Image software Version 3.0 (Xenogen) was used to acquire and quantitate the bioluminescence imaging data sets.
Supplementary Material
Acknowledgments
Supported by the National Institutes of Health (NIH) NIBIB Grant R01-EB008082, an American Recovery and Reinvestment (ARRA) grant, the National Science Foundation Grant, Division of Materials Research Polymers Program #0705234 and a Nanotechnology grant from the Koch Institute for Integrative Cancer Research. We also thank the Institute for Soldier Nanotechnologies (ISN) and Center for Materials Science Research (CMSE) for use of facilities.
Footnotes
Author contributions
J.B.L. and P.T.H. designed the experiments. J.B.L., J.H., D.K.B. and Z.P. carried out experiments. J.B.L., J.H., D.K.B., Z.P. and P.T.H. contributed to analysis of the data. J.B.L. and P.T.H. wrote the manuscript.
The authors declare no competing financial interests.
Supplementary information accompanies this paper on www.nature.com/naturematerials.
Reprints and permissions information is available online at www.nature.com/reprints.
References
- 1.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nature Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 2.Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science. 2008;319:627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mok H, Lee SH, Park JW, Park TG. Multimeric small interfering ribonucleic acid for highly efficient sequence-specific gene silencing. Nature Mater. 2010;9:272–278. doi: 10.1038/nmat2626. [DOI] [PubMed] [Google Scholar]
- 4.Liu L, et al. Self-assembled cationic peptide nanoparticles as an efficient antimicrobial agent. Nature Nanotechnol. 2009;4:457–463. doi: 10.1038/nnano.2009.153. [DOI] [PubMed] [Google Scholar]
- 5.Lee JS, et al. Gold, poly(β-amino ester) nanoparticles for small interfering RNA delivery. Nano Lett. 2009;9:2402–2406. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elbakry A, et al. Layer-by-layer assembled gold nanoparticles for siRNA delivery. Nano Lett. 2009;9:2059–2064. doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- 7.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J Am Chem Soc. 2009;131:2072–2073. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, et al. Multifunctional magnetoplasmonic nanoparticle assemblies for cancer therapy and diagnostics (theranostics) Macromol Rapid Commun. 2010;31:228–236. doi: 10.1002/marc.200900793. [DOI] [PubMed] [Google Scholar]
- 9.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nature Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum Gene Ther. 2005;16:1097–109. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taratula O, et al. Surface-engineered targeted PPI dendrimer for efficient intracellular and intratumoral siRNA delivery. J Control Release. 2009;140:284–293. doi: 10.1016/j.jconrel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 13.Davis ME, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akinc A, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nature Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seeman NC. Nanomaterials based on DNA. Annu Rev Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chworos A, et al. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 17.Guo P. The emerging field of RNA nanotechnology. Nature Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo P. RNA nanotechnology: Engineering, assembly and applications in detection, gene delivery and therapy. J Nanosci Nanotechnol. 2005;5:1964–1982. doi: 10.1166/jnn.2005.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grabow WW, et al. Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano Lett. 2011;11:878–887. doi: 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price AD, Zelikin AN, Wark KL, Caruso F. A biomolecular ‘ship-in-a-bottle’: Continuous RNA synthesis within hollow polymer hydrogel assemblies. Adv Mater. 2010;22:720–723. doi: 10.1002/adma.200903411. [DOI] [PubMed] [Google Scholar]
- 21.Afonin KA, et al. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nature Nanotechnol. 2010;5:676–682. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khaled A, et al. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo P, et al. Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Mol Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 24.Shu D, et al. Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett. 2004;4:1717–1723. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daubendiek SL, Ryan K, Kool ET. Rolling-circle RNA-synthesis—circular oligonucleotides as efficient substrates for T7 RNA-polymerase. J Am Chem Soc. 1995;117:7818–7819. doi: 10.1021/ja00134a032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seyhan AA, Vlassov AV, Johnston BH. RNA interference from multimeric shRNAs generated by rolling circle transcription. Oligonucleotides. 2006;16:353–363. doi: 10.1089/oli.2006.16.353. [DOI] [PubMed] [Google Scholar]
- 27.Richards KE, Williams RC, Calendar R. Mode of DNA packing within bacteriophage heads. J Mol Biol. 1973;78:255–259. doi: 10.1016/0022-2836(73)90114-9. [DOI] [PubMed] [Google Scholar]
- 28.Hendrix RW. Bacteriophage DNA packaging: RNA gears in a DNA transport machine. Cell. 1998;94:147–150. doi: 10.1016/s0092-8674(00)81413-0. [DOI] [PubMed] [Google Scholar]
- 29.Trubetskoy VS, Loomis A, Hagstrom JE, Budker VG, Wolff JA. Layer-by-layer deposition of oppositely charged polyelectrolytes on the surface of condensed DNA particles. Nucleic Acids Res. 1999;27:3090–3095. doi: 10.1093/nar/27.15.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banks W, Thomson G, Sharples A. Formation of spherulites in polyethylene. Nature. 1962;194:542–544. [Google Scholar]
- 31.Tang ZY, Zhang ZL, Wang Y, Glotzer SC, Kotov NA. Self-assembly of CdTe nanocrystals into free-floating sheets. Science. 2006;314:274–278. doi: 10.1126/science.1128045. [DOI] [PubMed] [Google Scholar]
- 32.Nakata M, et al. End-to-end stacking and liquid crystal condensation of 6 to 20 base pair DNA duplexes. Science. 2007;318:1276–1279. doi: 10.1126/science.1143826. [DOI] [PubMed] [Google Scholar]
- 33.Tijsterman M, Ketting RF, Plasterk RH. The genetics of RNA silencing. Annu Rev Genet. 2002;36:489–519. doi: 10.1146/annurev.genet.36.043002.091619. [DOI] [PubMed] [Google Scholar]
- 34.Decher G. Fuzzy nanoassemblies: Toward layered polymeric multicomposites. Science. 1997;277:1232–1237. [Google Scholar]
- 35.Diegelman AM, Kool ET. Generation of circular RNAs and trans-cleaving catalytic RNAs by rolling transcription of circular DNA oligonucleotides encoding hairpin ribozymes. Nucleic Acids Res. 1998;26:3235–3241. doi: 10.1093/nar/26.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.