Abstract
Biofilms can methylate mercury (Hg) at higher rates than unattached bacteria and are increasingly recognized as important Hg methylation sites in the environment. Our previous study showed that methylation rates in biofilm cultures were up to 1 order of magnitude greater than those in planktonic cultures of a sulfate-reducing bacterium. To probe whether the differential Hg methylation rates resulted from metabolic differences between these two cultures, Hg methylation assays following molybdate or chloroform inhibition (a specific inhibitor of the acetyl-CoA pathway) were conducted on biofilm and planktonic cultures of Desulfovibrio desulfuricans strains M8 and ND132. Molybdate was as effective in inhibiting Hg methylation as well as growth in both planktonic and biofilm cultures. The addition of chloroform only impacted Hg methylation in biofilm cultures, suggesting that different pathways are used for methylation in biofilm compared to planktonic cultures. To investigate this further, expression of the cooS gene, which encodes for carbon monoxide dehydrogenase, a key enzyme in the acetyl-CoA pathway, was compared in biofilm and planktonic cultures of ND132. Biofilm cultures showed up to 4 times higher expression of cooS than planktonic cultures. On the basis of these results, the acetyl-CoA pathway appears to play an important role in methylation in biofilm cultures of this organism, possibly by supplying the methyl group to Hg methylating enzymes; methylation in planktonic cultures appears to be independent of this pathway. This observation has important implications, particularly in developing reliable models to predict Hg methylation rates in different environments and perhaps eventually in being able to control this undesirable chemical transformation.
INTRODUCTION
Elevated concentrations of methylmercury (MeHg) in natural ecosystems, mostly produced in situ by specific microbial guilds, are of vital environmental concern because MeHg is a potent neurotoxin that can readily accumulate in aquatic and terrestrial food chains, posing a threat to wildlife and human health.1,2 Dissimilatory sulfate-reducing bacteria (SRB) and iron-reducing bacteria are now widely recognized as the primary methylators of mercury (Hg) in most aquatic systems;3–8 however, nearly all previous mechanistic methylation studies to date have been solely conducted with planktonic cultures of these methylating microbes, even though many acknowledge that the majority of bacteria in both natural and engineered environments (including those in soil) live as sessile cells, or biofilms.9,10 Indeed, attached microbial communities, such as periphyton in wetlands and microbial mats, are increasingly recognized as important sites for environmental Hg methylation.11–14
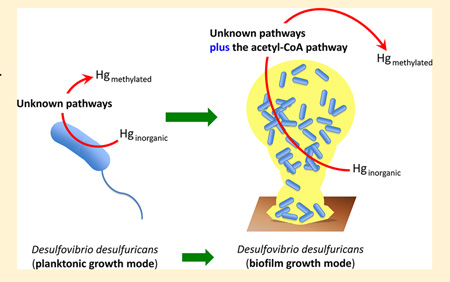
While much remains to be learned about the role of environmental biofilms in the Hg methylation process, at present biofilms are known to provide conditions conducive to the growth of anaerobic organisms (including methylating microbes) in systems that would be considered oxic at the bulk scale owing to reduced microzones that form during biofilm growth.15–18 The nutrient cycling and symbiotic relationships among organisms that occur in biofilms may also drive Hg methylation.19–21 For example, the coexistence of SRB with phototrophic sulfur-oxidizing bacteria may be of particular importance in MeHg formation because sulfur oxidizers consume sulfide,4,22–25 which may maintain favorable chemical speciation for Hg uptake by SRB and thus ultimately increase Hg methylation.26 The biofilm growth mode itself may be another determinant of a cell’s methylation rate, possibly because of different gene expression or metabolic activities that may occur in organisms growing as biofilms as opposed to unattached cultures. In fact, previous work with pure cultures of two strains of Desulfovibrio desulfuricans showed that Hg methylation rates were 1 order of magnitude higher when the cultures were grown as biofilms compared to when the same organisms were grown as planktonic cultures.27 However, the biochemical mechanism of in vivo Hg methylation remains elusive, particularly in biofilms.28–30
Earlier planktonic-culture studies on the mechanism of Hg methylation, conducted on one model strain of SRB, D. desulfuricans LS, indicated that MeHg synthesis was an enzymatically catalyzed process associated with the acetylcoenzyme A (acetyl-CoA) pathway.31–34 In this carbon metabolism pathway, carbon monoxide dehydrogenase, encoded by the cooS gene, is involved in the cleavage of acetyl-CoA and the further oxidation of the formed carbon monoxide to carbon dioxide.35 Other work showed that, although B12-containing methyl transferase plays a key role in MeHg formation in complete-oxidizing SRB, Hg methylation in incomplete oxidizers (such as D. desulfuricans) appeared to be independent of the acetyl-CoA pathway.36,37 The recent study by Parks et al. used rigorous genomic, genetic, and biochemical approaches to identify two genes, hgcA (which encodes a putative corrinoid protein) and hgcB (which encodes a 2[4Fe-4S] ferredoxin), necessary for Hg methylation.38 These genes were proposed as key components of the bacterial Hg methylation pathway, with the HgcA protein hypothesized to facilitate transfer of a methyl group to inorganic mercury(II) and the HgcB protein serving for HgcA turnover. The acetyl-CoA pathway is shown as a potential source of C1 units for the methylating enzymes in the proposed mechanistic model.38 While available evidence from these studies with planktonic cultures suggests that there may be multiple pathways for Hg methylation,36 pathways for methylation in biofilms have, to date, not been studied, and it is unknown whether there may be a metabolic basis for the high methylation rates observed in environmental biofilms of the two tested strains of D. desulfuricans.27
To investigate whether differential methylation rates in biofilm versus planktonic cultures of incomplete-oxidizing SRB resulted from metabolic differences between these two modes of growth, we extended our study by performing enzyme inhibition assays on planktonic and biofilm cultures of two strains of D. desulfuricans: the strain M8 is a brackish water strain of SRB used in our previous work,27 and the strain ND132 is a well-known Hg-methylating sulfate reducer whose genome has recently been sequenced and thus can be examined genetically.38–41 Both molybdate, a known inhibitor of sulfate reduction,3 and chloroform (CHCl3), a known inhibitor of corrinoid-containing enzymes, which play a critical part in the acetyl-CoA pathway,35 were included in this study to specifically probe the role of the acetyl-CoA pathway in Hg methylation. In addition, expression of cooS, which encodes for a key enzyme involved in the acetyl-CoA pathway, relative to the reference genes and cell count in biofilm and planktonic cultures of this organism was compared.
MATERIALS AND METHODS
Cultures and Growth Media
Both strains were cultured at room temperature (~22 °C) in a medium containing basal salts, vitamin mixture, selenite–tungstate, thiamine, and a nonchelated trace element mixture, buffered at pH 7.2 with 30 mM bicarbonate.42 Lactate (35 mM) and sulfate (28 mM) were chosen as the electron donor and acceptor, respectively. The salinity of the medium was adjusted to suit each strain: 10 and 1 g/L of NaCl for M8 and ND132, respectively. The medium was reduced with 0.25 mM titanium(III), which has been shown to not affect the growth of SRB.43,44 The medium was prepared anoxically under a gas stream passed through heated copper beads (for inhibition assays, CO2 and N2 in a ratio of 20:80 (%) were used, and for gene expression assays, H2, CO2, and N2 in a ratio of 5:15:80 (%) were used to match the headspace in the anoxic chamber). Planktonic cultures were maintained in serum bottles sealed with butyl rubber stoppers. Biofilm cultures were grown by inoculating sterile 50 mL Falcon tubes filled with 30 mL of a Hg-free medium and containing acid-washed, autoclaved glass microscopic slides. The Falcon tubes were kept in an anoxic chamber (Coy Laboratories) in an atmosphere consisting of H2, CO2, and N2 in a ratio of 5:15:80 (%). While the experimental biofilms were being established (over the course of 1 week for inhibition tests and 2 weeks for gene expression studies), 50% of the medium was replenished every 2 days to provide sufficient nutrients to maintain growth.
Prior to inoculation of experimental bottles, planktonic cultures were grown to exponential phase, pelleted, and resuspended in a fresh medium formulated as described in the previous paragraph but without sulfate in order to minimize sulfide formation prior to the start of the experiment. A total of 1 mL of inoculum was then transferred to 50 mL acid- washed serum bottles containing 30 mL of sulfate-containing assay media (again, formulated as described in the previous paragraph). Active biofilm-coated slides were transferred to acid-washed, sterile 50 mL Falcon tubes containing 30 mL of assay media after being rinsed three times in a fresh sulfate-free medium to eliminate unattached cells. A sulfate-free medium was used to eliminate sulfide transfer during inoculation because sulfide is known to inhibit methylation. During inhibition experiments, planktonic cultures were maintained on an orbital shaker at 160 rpm in the dark, while unstirred Falcon tubes containing biofilm-coated slides were covered with aluminum foil and kept in the anoxic chamber. For gene expression assays, all cultures were unstirred and uncovered and were kept at room temperature.
Enzyme Inhibition Tests
Stock chloroform solutions were first prepared in absolute (>99%) ethanol and then diluted twice (at least a 30-fold dilution each time) in a Hg-free medium. A final concentration of 50 µM chloroform was chosen.35,36 Similar to chloroform, molybdate was added from stock solutions and then diluted in the medium to the final concentration of 28 mM.36,45 Solutions of chloroform and molybdate were prepared and filter-sterilized immediately before being spiked into assay media.
Triplicate bottles/tubes of planktonic and biofilm cultures were prepared. An initial test was conducted using ND132 with cultures incubated for 7 h at room temperature, followed by 48 h Hg methylation assays (spiked with a 50 ng L−1 inorganic HgII standard). A second set of tests was conducted with both M8 and ND132 with all inhibitor-assayed cultures incubated for 7 h at room temperature, followed by 12 h Hg methylation assays (spiked with a 50 ng L−1 inorganic HgII standard). Each bottle was sampled for MeHg and cell density measurements at the beginning and end of the methylation assays. Killed cultures spiked with a Hg standard served as negative controls, while live cultures spiked only with Hg (i.e., no inhibitors added) served as positive controls and were included in each batch. Cultures that did not receive Hg and inhibitors served as blank assays to determine the background level of MeHg.
Gene Expression Tests
Assay media, as previously described, in half of the bottles/tubes were amended with 50 ng L−1 inorganic HgII and equilibrated overnight prior to inoculation with ND132. For each of the four conditions (live ± Hg and killed ± Hg), bottles and tubes were sacrificed in triplicate for each time point. Four replicate biofilm slides were scraped into one new tube with phosphate-buffered saline (PBS), thereby “pooling” the biofilm cells for a higher cell count (12 total biofilms per time point per condition). Inside the anoxic chamber, biofilm cells were collected by rinsing the slide with sterile PBS, vortexing for 30 s, scraping, rinsing, and vortexing again, as previously described by other researchers.9,46 The slide was removed once the PBS solution contained the biofilm cells. Samples from each of the new triplicate tubes were collected for MeHg, RNA, DNA, and cell counts. Samples were preserved and stored as follows: Hg samples were syringe-filtered, acidified (0.5% (v/v) HCl for total Hg; 0.2% (v/v) H2SO4 for MeHg), and stored at 8 °C; RNA was stabilized using 2:1 Qiagen RNAprotect Bacteria Reagent and refrigerated at 8 °C; DNA samples were unamended and frozen in −80 °C; samples preserved for cell counts received 5% (v/v) formaldehyde and were refrigerated at 8 °C.
RNA Extraction, Design of Degenerate Primer Sets, and Reverse Transcription (RT)-qPCR
RNA samples unamended with stabilizing reagent were immediately extracted for RNA content. mRNA was extracted (Qiagen RNeasy kit, Qiagen RNase-Free DNA Digestion kit), quantified (Nanodrop 2000), and amplified (Qiagen SYBR Green One-Step RT-qPCR kit, Applied Biosystems StepOnePlus Real-Time PCR System). The location of the cooS gene in ND132 was located through the EMBL database, and 130 base-pair (bp) amplicon primers were obtained using GeneBlast. Two sets of primers (150 and 190 bp) for 16S rRNA in ND132 were designed similarly (see Table 1) and are used to normalize cooS gene expression. It is noted that, although the use of reference genes is desirable, reference genes can vary significantly and can adapt to growth conditions.47,48 In such cases, geometric means of multiple references are used.49–51 Cycling conditions were as follows: 95 °C for 10 min followed by 45 cycles of 94 °C for 15 s, 60 °C for 1 min, and 72 °C for 30 s. Relative quantification of the qPCR results uses the 2−ΔΔCt method to compare differences in gene expression.52,53 Both amplification plots and melt curves were obtained during RT-qPCR. This expression was normalized to both cells (counted) and the geometric mean of expression of two primer sets from 16S rRNA.
Table 1.
Primers Used in RT-qPCR
| primer | forward sequence (5′ → 3′) | reverse sequence (5′ → 3′) |
|---|---|---|
| cooS | AGGGCGAGACCAAGGATTAC | GCGAAAAAGCACTCCATGAC |
| 16S rRNA primer set 1 | GGGGGAAACCCTGACGCAGC | TGCTGGCACGGAGTTAGCCG |
| 16S rRNA primer set 2 | CGACGCCGCGTGTAGGAAGA | ACGCACGCTTTACGCCCAGT |
MeHg and Cell Density Determination
MeHg analysis was performed via distillation/ethylation/gas chromatography–cold vapor atomic fluorescence spectrometry.54 Prior to distillation, interference resulting from sulfide was eliminated by acidifying samples with 9 N sulfuric acid and then purging with gold-coated sand-trap-filtered nitrogen (45 mL min−1 for approximately 25 min).27 The cell density from both planktonic and biofilm cultures was determined by direct counts using 4′,6-diamidino-2-phenylindole staining and epifluorescent microscopy in accordance with our previous protocols.27
RESULTS
Effects of Inhibitors in Both Planktonic and Biofilm Cultures
Our previous study comparing the Hg methylation rates on a per cell basis between planktonic and biofilm cultures of a methylating sulfate reducer, D. desulfuricans M8, indicated that the specific Hg methylation rate in biofilm cultures was approximately 1 order of magnitude higher than that in planktonic cultures.27 In the current study, the importance of the acetyl-CoA pathway in both planktonic and biofilm cultures of this strain, as well as another known Hg-methylating strain, D. desulfuricans ND132, was tested using specific inhibitors.
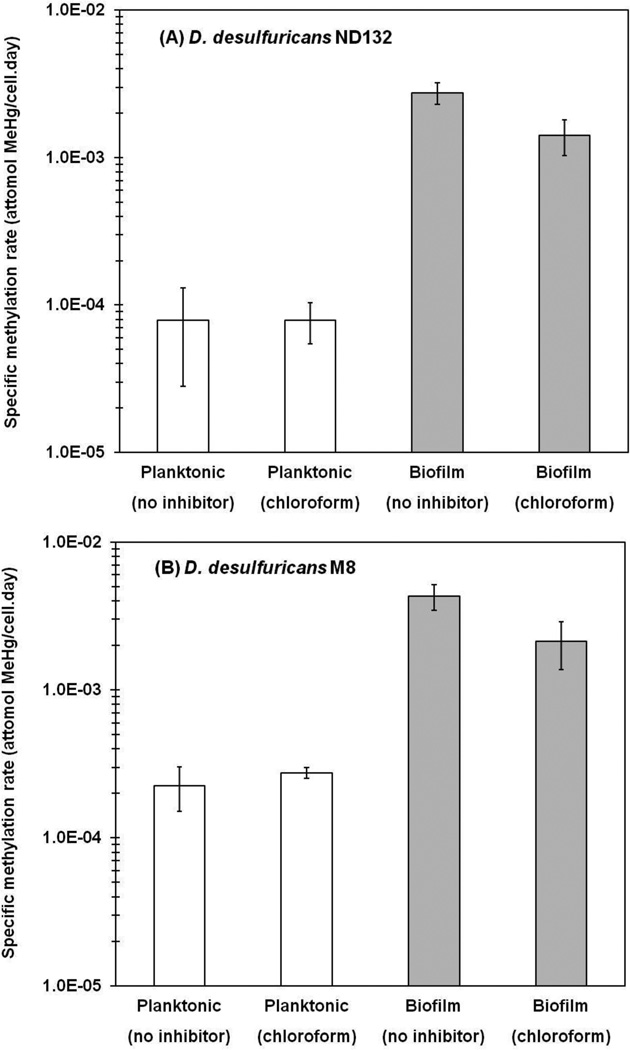
As expected, a similar and effective inhibition of growth and Hg methylation was observed in both planktonic and biofilm cultures of M8 and ND132 amended with molybdate, a known competitive inhibitor for sulfate in SRB.55 However, results show that chloroform had no significant influence on either growth or methylation in planktonic cultures. Growth rates were not significantly different in cultures with and without chloroform addition for ND132 (0.040 h−1) and M8 (0.051 h−1). Hg methylation was also not affected significantly (p > 0.05; t test) by the presence of chloroform (Figure 1).
Figure 1.
Observed methylation rates after a 12 h methylation test following a 7 h inhibition test in planktonic and biofilm cultures: (A) D. desulfuricans ND132; (B) D. desulfuricans M8. Error bars represent the standard deviations of triplicate assays.
Yet, unlike planktonic cultures, chloroform inhibition on Hg methylation was observed in biofilm cultures. Both ND132 and M8 produced lower MeHg concentrations in cultures incubated with chloroform, and the specific rates of Hg methylation in chloroform-amended cultures decreased by a factor of greater than 2 compared to cultures in the absence of inhibitors (p < 0.05; t test; Figure 1). Interestingly, chloroform had no influence on the growth of biofilm cultures, as seen in 0.025 h−1 (chloroform-free cultures) vs 0.025 h−1 (chloroform-amended cultures) of ND132 and 0.063 vs 0.069 h−1 of M8, respectively.
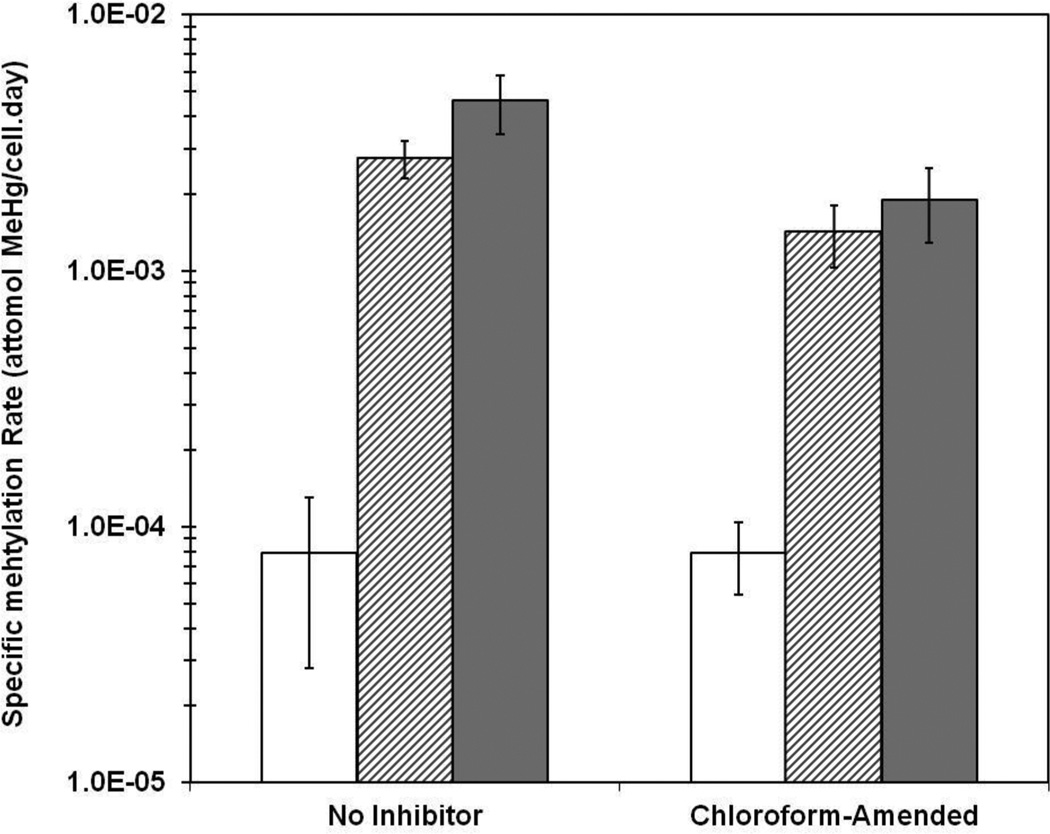
Figure 2 depicts a longer (2 day) inhibition experiment with ND132, in which an even greater difference in rates can be observed between biofilm and planktonic cultures. Because it is unclear whether cells that detached from biofilms during the experiments should be treated as biofilm or planktonic cells, specific Hg methylation rates illustrated in Figure 2 were calculated according to the case 1 method described in an earlier study, which assumed that the detached cells still methylate Hg at the same rate as the attached cells.27 It should be noted that case 1 calculations resulted in more conservative numbers compared to case 2 calculations, which account for methylation by the detached cells at the rate determined for the planktonic system so that only attached cells are regarded as true biofilm cells.
Figure 2.
Methylation rates after a 2-day methylation test following 7 h of exposure to inhibitors in planktonic and biofilm cultures of D. desulfuricans ND132. Blank columns represent rates of planktonic cultures. Light-filled columns are the rates of biofilm cultures according to case 1 calculations, assuming the detached cells still methylate Hg at the same rates as the biofilm cells. Dark-filled columns are the methylation rates of biofilm cells, accounting for methylation by detached cells at the rates determined for planktonic cultures (case 2). Error bars represent the standard deviations of triplicate assays.
Comparison of cooS Gene Expression
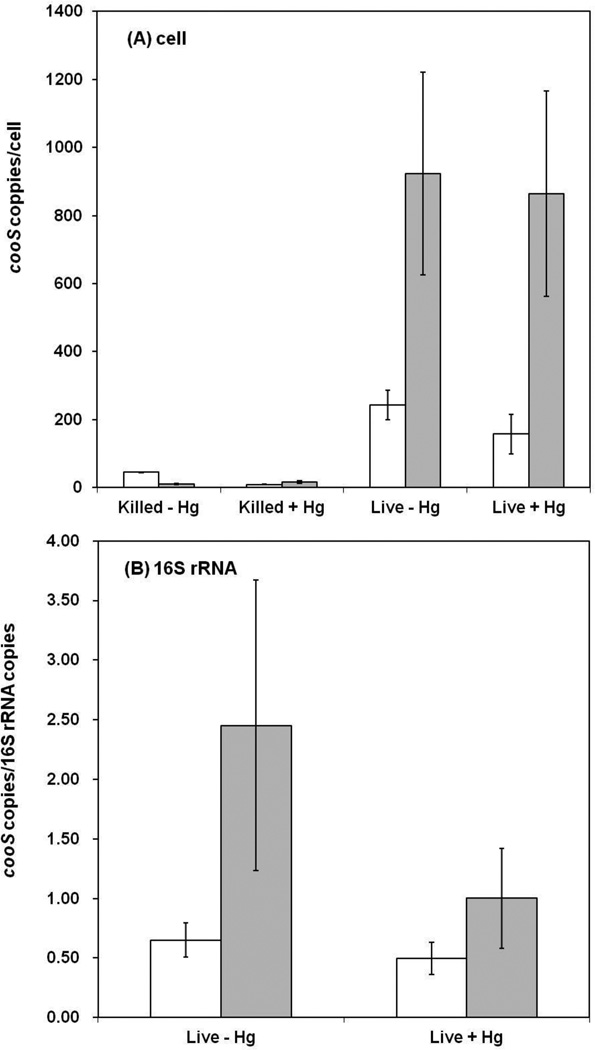
Given that the results from chloroform inhibition assays indicated a potential role of the acetyl-CoA pathway in biofilm cultures, the importance of Hg methylation by this pathway was further explored by comparing gene expression of cooS in biofilms and planktonic cultures of D. desulfuricans ND132. Expression of cooS normalized to the cell number (i.e., copies/cell) was approximately 4 times higher in biofilms compared to planktonic cultures (Figure 3A). The cell count error, as calculated by the square root of total cells by the number of total cells, was less than 6.5% for live biofilm and planktonic cultures, as well as for killed planktonic cultures. The error was as high as 13% for killed biofilm cultures, as a result of significantly fewer cells present to be counted. Primer dimerization did not appear to occur based on melting curve analysis. Similar results were observed when normalizing to the geometric mean of expression of two primer sets from 16S rRNA (Figure 3B).
Figure 3.
Comparison of cooS gene expression, based on the 2−ΔΔCt method for relative quantification, between planktonic and biofilm cultures of D. desulfuricans ND132 in the presence and absence of Hg. (A) Copies per cell are relative values based on changes in the threshold cycle. (B) Copies of cooS per copies of 16S rRNA are also based on relative values based on changes in the threshold cycle. Blank columns represent planktonic cultures. Dark columns represent biofilm cultures. Error bars represent the standard error of triplicate experimental bottles (each bottle was subjected to triplicate analysis).
DISCUSSION
While significant progress has been made toward identifying the biochemical mechanism that leads to MeHg production in bacterial cells, to date this mechanism has not been definitively identified.30,38,56 The recent study by Parks et al. identified genes necessary for Hg methylation in SRB, although the exact pathway(s) by which Hg is methylated in these organisms is not known for certain.38 Results of previous studies have suggested that the acetyl-CoA pathway is involved in this microbially mediated environmental process in methylating SRB, particularly when this pathway is used in methylating SRB for primary carbon metabolism (i.e., in complete-oxidizing SRB).31–34,36 Considering that other pathways of Hg methylation occurring independently of the acetyl-CoA pathway are still unclear,36,37,57 an understanding of whether metabolic differences between planktonic and biofilm cultures of Hg-methylating D. desulfuricans strains, in particular the role of the acetyl-CoA pathway in the production of MeHg in biofilm versus planktonic cultures, could help to elucidate the differential methylation rates observed in these two growth modes. Thus, enzyme inhibition and RT-qPCR assays specifically targeting the gene and enzyme required in the acetyl-CoA pathway were conducted in this study.
Data from chloroform inhibition tests with the strains M8 and ND132 revealed that Hg methylation was only inhibited in biofilm cultures. Consistent with work done by Ekstrom et al., methylation was not inhibited by chloroform in planktonic cultures of both of these incomplete oxidizers.36 In their work, Ekstrom et al. showed that, with 50 µM chloroform, Hg methylation was effectively inhibited (lower than the detection limit) in planktonically growing cultures of the complete oxidizer, Desulfococcus multivorans 1be1. In another set of their experiments, neither Hg methylation nor growth of an incomplete-oxidizing strain, Desulfovibrio africanus, was inhibited over a shorter incubation (5 h) with chloroform. Longer incubation (3 days) eventually affected culture growth; however, no significant difference in the specific Hg methylation rates was observed between shorter- and longer-incubated cultures.36 The result that the addition of chloroform only impacted Hg methylation in biofilm cultures in this study indicated a role for the acetyl-CoA pathway for methylation in biofilm but not planktonic cultures of the organisms tested. Further, while Hg methylation in chloroform-amended biofilm cultures was inhibited, it was not inhibited completely, suggesting that multiple pathways for Hg methylation may take place in biofilm cells. Parks et al. proposed a mechanism by which the proteins encoded by the genes they identified as necessary for methylation (hgcA and hgcB) might methylate Hg, which may involve the acetyl-CoA pathway as well as other pathways to provide a C1 source. Because it is unknown whether the proteins encoded by hgcA and hgcB are involved with additional metabolic pathways in SRB, our findings that an unknown pathway may also be involved are not inconsistent with these results.
The importance of the acetyl-CoA pathway in biofilm cultures relative to that in planktonic cultures was also supported by the result of the cooS expression tests because biofilm cultures showed up to 4 times higher expression of cooS than planktonic cultures on a per cell basis. While similar results were observed when copy numbers were normalized to the geometric mean of expression of 16S rRNA using two primer sets (Figure 3B), it should be kept in mind that standardization of mRNA in gene expression studies has been a complex issue that has not been fully resolved. Ideally, expression would be normalized to the cell count but the accuracy of the cell count can be a concern.50 Although the use of reference genes is preferable, expression of reference genes can vary significantly. It has been suggested that reference genes can adapt to growth conditions, which is especially important for biofilms because their metabolic processes may differ significantly from planktonic cultures.47,48 Desirable reference genes would express equally despite experimental treatments; however, in cases when they do not, geometric means of multiple reference genes are used.49–51 The use of reference, or housekeeping, genes for biofilms, in particular, is complex and has not yet been thoroughly investigated.
It is known that, in response to environmental signals, biofilm bacteria can express new phenotypes that distinguish themselves from their planktonic counterparts.9,58–60 This is illustrated by data that have shown higher resistance to toxic compounds, including antimicrobials, metals, and metalloids, in biofilm cultures than in planktonic cultures.61–63 In particular, work on the susceptibility of planktonic and biofilm cells of the same microorganism to antibiotics has shown that the structures of biofilms do not simply provide a diffusion barrier to these compounds; instead, the biofilms employ distinct resistance mechanisms and exhibit differential gene expression compared to planktonic modes of growth.64–66 In biofilms, significant upregulation of multiple genes related to cellular function and metabolic pathways may occur;67,68 thus, the formation of biofilms may favor one pathway over another. Presumably, it is possible that such alternation of gene and protein expressions may result in different metabolic pathways between biofilm and planktonic cultures, thus causing differences in Hg methylation. Indeed, the results of our work showing that differences in expression of a gene are likely important in methylation between biofilm and planktonic cultures suggest a metabolic basis for the high methylation rates observed in environmental biofilms.
A likely connection between Hg methylation and the acetyl-CoA pathway in biofilm cultures can also be inferred from the significantly higher methylation rates observed in biofilm versus planktonic cultures for both M8 and ND132 (Figures 1 and 2). King et al. have suggested that Hg methylation potential may be related to genetic composition and/or carbon metabolism in SRB.69 Results from their pure culture experiments showed that Hg methylation rates on a per cell basis were up to 3 orders of magnitude higher in the family Desulfobacteriaceae, which is the family of SRB that uses the acetyl-CoA pathway for complete oxidation of carbon substrates compared to the other family of SRB, Desulfovibrionaceae. The same pattern was also observed by Ekstrom and Morel from their cobalt limitation assays, which showed that D. multivorans, a complete oxidizer that uses the acetyl-CoA pathway for major carbon metabolism, made 10–50 times more MeHg per cell than a Desulfovibrio strain, D. africanus (DSMZ 2603).37
It is noted that the process of intracellular MeHg synthesis has been postulated to be most likely a metabolic mistake rather than a mechanism that confers Hg resistance in SRB;30,70 however, a tight coupling between Hg methylation and MeHg export from the cell of Hg-methylating strains observed in a recent study has brought up the question of whether methylation may be a strategy for bacteria to avoid buildup and subsequent toxicity of cellular Hg, or possibly a part of the Hg detoxification process.29 In the present study, it was observed that (1) chloroform only inhibited Hg methylation and not growth in biofilms and (2) the acetyl-CoA pathway seems to influence the rate of formation of MeHg in biofilm cultures only, possibly by providing C1 for Hg methylation.38 These results, combined with the observation that, in general, biofilm cells typically show higher resistance to toxic metals than planktonic cells, suggest that whether methylation may serve as a mechanism for resistance to or reduction of HgII toxicity for D. desulfuricans biofilms warrants future study.
One of the major goals of Hg methylation research is to determine factors influencing Hg transformation potentials in order to develop quantitative, reliable models to predict Hg methylation rates in different environments. Theoretically, such a model should rely on parameters that adequately describe (i) the microbial community composition, (ii) the bioavailability of inorganic HgII, and (iii) the principal biochemical pathways responsible for Hg methylation.71,72 Recent research has attempted to predict in situ methylation rates using information on SRB activity and methylation.69,73 Our study contributes to a deeper understanding of the actual methylation rates in sediments and in attached communities. Extending this work to field samples would indicate the environmental importance of a metabolic basis for increased methylation in attached communities. Whether detached biofilm cells would retain increased methylation capability is an open question. If so, events such as rain could disturb attached communities and transport cells with higher Hg methylation rates. In addition, having an understanding of the mechanisms by which SRB methylate Hg holds promise for informing remediation strategies in engineered treatment systems.
ACKNOWLEDGMENTS
This work was supported through National Science Foundation CAREER Grant BES-0348783. We thank all of our laboratory colleagues for their constant support, and we are appreciative of laboratory assistance from Saeedreza Hafenezami, Myfanway Rowlands, Jonathan Lucio, and Ahana Mukherjee.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Fitzgerald WF, Clarkson TW. Mercury and monomethylmercury: present and future concerns. Environ. Health Perspect. 1991;96:159–166. doi: 10.1289/ehp.9196159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolfe MF, Schwarzbach S, Sulaiman RA. Effects of mercury on wildlife: a comprehensive review. Environ. Toxicol. Chem. 1998;17(2):146–160. [Google Scholar]
- 3.Compeau GC, Bartha R. Sulfate-reducing bacteria: principal methylators of mercury in anoxic esturaine sediment. Appl. Environ. Microbiol. 1985;50(2):448–502. doi: 10.1128/aem.50.2.498-502.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Compeau GC, Bartha R. Effect of salinity on mercury-methylating activity of sulfate-reducing bacteria in estuarine sediments. Appl. Environ. Microbiol. 1987;53(2):261–265. doi: 10.1128/aem.53.2.261-265.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilmour CC, Henry EA. Mercury methylation in aquatic systems affected by acid deposition. Environ. Pollut. 1991;71(2–4):131–169. doi: 10.1016/0269-7491(91)90031-q. [DOI] [PubMed] [Google Scholar]
- 6.Gilmour CC, Henry EA, Mitchell R. Sulfate stimulation of mercury methylation in fresh-water sediments. Environ. Sci. Technol. 1992;26(11):2281–2287. [Google Scholar]
- 7.Fleming EJ, Mack EE, Green PG, Nelson DC. Mercury methylation from unexpected sources: molybdate-inhibited freshwater sediments and an iron-reducing bacterium. Appl. Environ. Microbiol. 2006;72(1):457–464. doi: 10.1128/AEM.72.1.457-464.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerin EJ, Gilmour CC, Roden E, Suzuki MT, Coates JD, Mason RP. Mercury methylation by dissimilatory iron-reducing bacteria. Appl. Environ. Microbiol. 2006;72(12):7919–7921. doi: 10.1128/AEM.01602-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu. Rev. Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 10.Potera C. Biofilms invade microbiology. Science. 1996;273(5283):1795–1797. doi: 10.1126/science.273.5283.1795. [DOI] [PubMed] [Google Scholar]
- 11.Cleckner LB, Gilmour CC, Hurley JP, Krabbenhoft DP. Mercury methylation in periphyton of the Florida Everglades. Limnol. Oceanogr. 1999;44(7):1815–1825. [Google Scholar]
- 12.Achá D, Iniguez V, Roulet M, Guimarães J, Luna R, Alanoca L, Sanchez S. Sulfate-reducing bacteria in floating macrophyte rhizospheres from an Amazonian floodplain lake in Bolivia and their association with Hg methylation. Appl. Environ. Microbiol. 2005;71(11):7531–7535. doi: 10.1128/AEM.71.11.7531-7535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desrosiers M, Planas D, Mucci A. Mercury methylation in the epilithon of boreral shield aquatic ecosystems. Environ. Sci. Technol. 2006;40(5):1540–1546. doi: 10.1021/es0508828. [DOI] [PubMed] [Google Scholar]
- 14.Hamelin S, Amyot M, Brkay T, Wang Y, Planas D. Methanogens: principal methylators of mercury in lake periphyton. Environ. Sci. Technol. 2011;45(18):7693–7700. doi: 10.1021/es2010072. [DOI] [PubMed] [Google Scholar]
- 15.Okabe S, Ito T, Satoh H, Watanabe Y. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ. Microbiol. 1999;65(11):5107–5116. doi: 10.1128/aem.65.11.5107-5116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schramm A, Santegoeds CM, Nielsen HK, Ploug H, Wagner M, Pribyl M, Wanner J, Amann R, de Beer D. On the occurrence of anoxic microniches, denitrification, and sulfate reduction in aerated activated sludge. Appl. Environ. Microbiol. 1999;65(9):4189–4196. doi: 10.1128/aem.65.9.4189-4196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito T, Nielsen JL, Okabe S, Watanabe Y, Nielsen PH. Phylogenetic identification and substrate uptake patterns of sulfate-reducing bacteria inhabiting an oxic–anoxic sewer biofilm determined by combining microautoradiography and fluorescent in situ hybridization. Appl. Environ. Microbiol. 2002;68(1):356–364. doi: 10.1128/AEM.68.1.356-364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart P. Diffusion in biofilms. J. Bacteriol. 2003;185(5):1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braissant O, Decho AW, Dupraz C, Glunk C, Przekop KM, Visscher PT. Exopolymeric substances of sulfate-reducing bacteria: interactions with calcium at alkaline pH and implication for formation of carbonate minerals. Geobiology. 2007;5(4):401–411. [Google Scholar]
- 20.Braissant O, Decho AW, Przekop KM, Gallagher KL, Glunk C, Dupraz C, Visscher PT. Characteristics and turnover of exopolymeric substances in a hypersaline microbial mat. FEMS Microbiol. Ecol. 2009;67(2):293–307. doi: 10.1111/j.1574-6941.2008.00614.x. [DOI] [PubMed] [Google Scholar]
- 21.Dupraz C, Reid RP, Braissant O, Decho AW, Norman RS, Visscher PT. Processes of carbonate precipitation in modern microbial mats. Earth-Sci. Rev. 2009;96(3):141–162. [Google Scholar]
- 22.Compeau GC, Bartha R. Effects of sea salt anions on the formation and stability of methylmercury. Bull. Environ. Contam. Toxicol. 1983;31(4):486–493. doi: 10.1007/BF01622282. [DOI] [PubMed] [Google Scholar]
- 23.Craig PJ, Moreton PA. Total mercury, methyl mercury and sulfide in River Carron sediments. Mar. Pollut. Bull. 1983;14(11):408–411. [Google Scholar]
- 24.Winfrey MR, Rudd JWM. Environmental factors affecting the formation of methylmercury in low pH lakes. Environ. Toxicol. Chem. 1990;9(7):853–869. [Google Scholar]
- 25.Benoit JM, Gilmour CC, Mason RP, Riedel GS, Riedel GF. Behavior of mercury in the Patuxent River estuary. Biogeochemistry. 1998;40(2–3):249–265. [Google Scholar]
- 26.Kampalath RA, Lin C-C, Jay JA. Influences of zero-valent sulfur on mercury methylation in bacterial cocultures. Water Air Soil Pollut. 2013;224(2) [Google Scholar]
- 27.Lin C-C, Jay JA. Mercury methylation by planktonic and biofilm cultures of Desulfovibrio desulfuricans. Environ. Sci. Technol. 2007;41(19):6691–6697. doi: 10.1021/es062304c. [DOI] [PubMed] [Google Scholar]
- 28.Schaefer JK, Morel FMM. High methylation rates of mercury bound to cysteine by Geobacter sulfurreducens. Nat. Geosci. 2009;2:123–126. [Google Scholar]
- 29.Schaefer JK, Rocks S, Zheng W, Liang L, Gu B, Morel FMM. Active transport, substrate specificity, and methylation of Hg(II) in anaerobic bacteria. Proc. Natl. Acad. Sci. U.S.A. 2011;108(21):8714–8719. doi: 10.1073/pnas.1105781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilmour CC, Elias DA, Kucken AM, Brown SD, Palumbo AV, Schadt CW, Wall JD. Sulfate-reducing bacterium Desulfovibrio desulfuricans ND132 as a model for understanding bacterial mercury methylation. Appl. Environ. Microbiol. 2011;77(12):3938–3951. doi: 10.1128/AEM.02993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman M, Chase T, Jr, Bartha R. Carbon flow in mercury biomethylation by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 1990;56(1):298–300. doi: 10.1128/aem.56.1.298-300.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi SC, Bartha R. Cobalamin-mediated mercury methylation by Desulfovibrio desulfuricans LS. Appl. Environ. Microbiol. 1993;59(1):290–295. doi: 10.1128/aem.59.1.290-295.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi SC, Chase T, Jr, Bartha R. Enzymatic catalysis of mercury methylation by Desulfovibrio desulfuricans LS. Appl. Environ. Microbiol. 1994;60(4):1342–1346. doi: 10.1128/aem.60.4.1342-1346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi SC, Chase T, Jr, Bartha R. Metabolic pathways leading to mercury methylation in Desulfovibrio desulfuricans Ls. Appl. Environ. Microbiol. 1994;60(11):4072–4077. doi: 10.1128/aem.60.11.4072-4077.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scholten J, Conrad R, Stams A. Effect of 2-bromoethane sulfonate, molybdate and chloroform on acetate consumption by methanogenic and sulfate-reducing populations in freshwater sediment. FEMS Microbiol. Ecol. 2000;32(1):35–42. doi: 10.1111/j.1574-6941.2000.tb00696.x. [DOI] [PubMed] [Google Scholar]
- 36.Ekstrom EB, Morel FMM, Benoit JM. Mercury methylation independent of the acetyl-coenzyme A pathway in sulfate-reducing bacteria. Appl. Environ. Microbiol. 2003;69(9):5414–5422. doi: 10.1128/AEM.69.9.5414-5422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ekstrom EB, Morel FMM. Cobalt limitation of growth and mercury methylation in sulfate-reducing bacteria. Environ. Sci. Technol. 2008;42(1):93–99. doi: 10.1021/es0705644. [DOI] [PubMed] [Google Scholar]
- 38.Parks JM, Johs A, Podar M, Bridou R, Hurt RA, Jr, Smith SD, Tomanicek SJ, Qian Y, Brown SD, Brandt CC, Palumbo AV, Smith JC, Wall JD, Elias DA, Liang L. The genetic basis for bacterial mercury methylation. Science. 2013;339(6125):1332–1335. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 39.Pak K-R, Bartha R. Mercury methylation by interspecies hydrogen and acetate transfer between sulfidogens and methanogens. Appl. Environ. Microbiol. 1998;64(6):1987–1990. doi: 10.1128/aem.64.6.1987-1990.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jay JA, Murray KJ, Mason RP, Gilmour CC, Morel FMM, Roberts AL, Hemond HF. Mercury methylation by Desulfovibrio desulfuricans ND 132 in the presence of polysulfides. Appl. Environ. Microbiol. 2002;68(11):5741–5745. doi: 10.1128/AEM.68.11.5741-5745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown SD, Gilmour CC, Kucken AW, Wall JD, Elias DA, Brandt CC, Podar M, Chertkov O, Held B, Bruce DC, Detter JC, Tapia R, Han CS, Goodwin LA, Cheng JF, Pitluck S, Woyke T, Mikhailova N, Ivanova NN, Han J, Lucas S, Lapidus AL, Land ML, Hauser LJ, Palumbo AV. Genome sequence of the mercury-methylating strain Desulfovibrio desulfuricans ND132. J. Bacteriol. 2011;193(8):2078–2079. doi: 10.1128/JB.00170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Truper HG, Dowrkin M, Harder W, Schleifer KH, editors. The Prokaryotes: A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications. New York: Springer-Verlag; 1992. pp. 3352–3378. [Google Scholar]
- 43.Moench TT, Zeikus JG. An improved preparation method for a titanium(III) media reductant. J. Microbiol. Methods. 1983;1:199–202. [Google Scholar]
- 44.Henry E. Ph.D. Dissertation. Cambridge, MA: Harvard University; 1992. The role of sulfate-reducing bacteria in environmental mercury methylation. [Google Scholar]
- 45.Oremland RS, Capone DG. Use of “specific” inhibitors in biogeochemistry and microbial ecology. Adv. Microb. Ecol. 1988;10:287–383. [Google Scholar]
- 46.Chen X, Stewart PS. Biofilm removal caused by chemical treatment. Water Res. 2000;34(17):4229–4233. [Google Scholar]
- 47.Vandecasteele SJ, Peetersmans WE, Merckx R, Eldere JV. Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J. Bacteriol. 2001;183(24):7094–7101. doi: 10.1128/JB.183.24.7094-7101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413(6858):860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 49.Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 2008;9 doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vandesompele J, DePreter K, Pattyn F, Poppe B, VanRoy N, DePaepe A. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000;25(2):169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 54.Method 1630- Methyl mercury in water by disfillation, aqueous ethylation, purge and trap, and CVAFS. Washington, DC: U.S. Environmental Protection Agency Office of Water, Office of Science and Technology, Engineering and Analysis Division (4303); 2001. [Google Scholar]
- 55.Wilson LG, Bandurski RS. Enzymatic reactions involving sulfate, sulfite, selenate, and molybdate. J. Biol. Chem. 1958;233(4):975–981. [PubMed] [Google Scholar]
- 56.Lin C-C, Yee N, Barkay T. Microbial transformations in the mercury cycle. In: Liu G, Cai Y, O’Driscoll N, editors. Environmental Chemistry and Toxicology of Mercury. Hoboken, NJ: John Wiley & Sons, Inc.; 2012. pp. 155–192. [Google Scholar]
- 57.Benoit JM, Gilmour CC, Mason RP. Aspects of bioavailability of mercury for methylation in pure cultures of Desulfobulbus propionicus (1pr3) Appl. Environ. Microbiol. 2001;67(1):51–58. doi: 10.1128/AEM.67.1.51-58.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davies DG, Chakrabarty AM, Geesey GG. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 1993;59(4):1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies DG, Geesey GG. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl. Environ. Microbiol. 1995;61(3):860–867. doi: 10.1128/aem.61.3.860-867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schembri MA, Kjærgaard K, Klemm P. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 2003;48(1):253–267. doi: 10.1046/j.1365-2958.2003.03432.x. [DOI] [PubMed] [Google Scholar]
- 61.Costerton JW, Steward PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 62.Teitzel GM, Parsek MR. Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2003;69(4):2313–2320. doi: 10.1128/AEM.69.4.2313-2320.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harrison JJ, Rabiei M, Turner RJ, Badry EA, Sproule KM, Ceri H. Metal resistance in Candida biofilms. FEMS Microbiol. Ecol. 2006;55(3):479–491. doi: 10.1111/j.1574-6941.2005.00045.x. [DOI] [PubMed] [Google Scholar]
- 64.Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GAA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426(6964):306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 65.Harrison JJ, Ceri H, Stremick CA, Turner RJ. Biofilm susceptibility to metal toxicity. Environ. Microbiol. 2004;6(12):1220–1227. doi: 10.1111/j.1462-2920.2004.00656.x. [DOI] [PubMed] [Google Scholar]
- 66.Harrison JJ, Ceri H, Stremick C, Turner RJ. Differences in biofilm and planktonic cell mediated reduction of metalloid oxyanions. FEMS Microbiol. Lett. 2004;235(2):357–362. doi: 10.1016/j.femsle.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 67.Resch A, Rosenstein R, Nerz C, Götz F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 2005;71(5):2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lattif AA, Chandra J, Chang J, Liu S, Zhou G, Chance MR, Ghannoum MA, Mukherjee PK. Proteomics and pathway mapping analyses reveal phase-dependent over-expression of proteins associated with carbohydrate metabolic pathways in Candida albicans biofilms. Open Proteomics J. 2008;1:5–26. [Google Scholar]
- 69.King JK, Kostka JE, Frischer ME, Saunders FM. Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments. Appl. Environ. Microbiol. 2000;66(6):2430–2437. doi: 10.1128/aem.66.6.2430-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henry E. Ph.D. Dissertation. Cambridge, MA: Harvard University; 1992. The role of sulfate-reducing bacteria in environmental mercury methylation. [Google Scholar]
- 71.Regnell O, Tunlid A, Ewald G, Sangfors O. Methyl mercury production in freshwater microcosms affected by dissolved oxygen levels: role of cobalamin and microbial community composition. Can. J. Fish. Aquat. Sci. 1996;53(7):1535–1545. [Google Scholar]
- 72.Siciliano SD, Lean DRS. Methyltransferase: an enzyme assay for microbial methylmercury formation in acidic soils and sediments. Environ. Toxicol. Chem. 2002;21(6):1184–1190. [PubMed] [Google Scholar]
- 73.King JK, Kostka JE, Frischer ME, Saunders FM, Jahnke RA. A quantitative relationship that demonstrates mercury methylation rates in marine sediments are based on the community composition and activity of sulfate-reducing bacteria. Environ. Sci. Technol. 2001;35(12):2491–2496. doi: 10.1021/es001813q. [DOI] [PubMed] [Google Scholar]