Abstract
The microRNA319 (miR319) family is conserved among diverse plant species. In rice (Oryza sativa L.), the miR319 gene family is comprised of two members, Osa-miR319a and Osa-miR319b. We found that overexpressing Osa-miR319b in rice resulted in wider leaf blades and delayed development. Here, we focused on the biological function and potential molecular mechanism of the Osa-miR319b gene in response to cold stress in rice. The expression of Osa-miR319b was down-regulated by cold stress, and the overexpression of Osa-miR319b led to an enhanced tolerance to cold stress, as evidenced by higher survival rates and proline content. Also, the expression of a handful of cold stress responsive genes, such as DREB1A/B/C, DREB2A, TPP1/2, was increased in Osa-miR319b transgenic lines. Furthermore, we demonstrated the nuclear localization of the transcription factors, OsPCF6 and OsTCP21, which may be Osa-miR319b-targeted genes. We also showed that OsPCF6 and OsTCP21 expression was largely induced by cold stress, and the degree of induction was obviously repressed in plants overexpressing Osa-miR319b. As expected, the down-regulation of OsPCF6 and OsTCP21 resulted in enhanced tolerance to cold stress, partially by modifying active oxygen scavenging. Taken together, our findings suggest that Osa-miR319b plays an important role in plant response to cold stress, maybe by targeting OsPCF6 and OsTCP21.
Introduction
Rice is a kind of important food and economic crop, and its production is frequently affected by cold stress. Cold stress is one of the most important limiting factors affecting rice production in temperate and high elevation areas [1]. During the early growth stages, cold stress severely inhibits rice seedling establishment and eventually leads to non-uniform maturation [2]. Cold stress always imposes a series of negative effects on photosynthesis, respiration, and the production and accumulation of reactive oxygen species (ROS) [3], [4]. However, rice has evolved efficient mechanisms to rapidly sense and respond to cold stress. To understand the underlying molecular mechanisms of the cold stress response, many intensive studies have been performed. MicroRNAs (miRNAs) are small single-stranded non-coding RNAs, usually ∼21 bp in length, which regulate biological processes through mediating translational inhibition or cleavage of target transcripts [5], [6], [7], [8]. Several studies have reported the involvement of miRNAs and their target genes in plant responses to stress conditions [9], [10], [11]. Under cold stress, the up- or down-regulation of miRNAs modifies the transcript abundance of their target genes [12], [13].
The miR319 family is one of the most ancient and conserved miRNA families, and has been found in a large number of plant species, from Physcomitrella to flowering plants [14], [15], [16], [17], [18], [19]. MiR319, previously known as ‘miR-JAW’, was firstly described in Arabidopsis because of its role in controlling leaf morphogenesis [20], [21]. It has been reported that miR319 targeted TEOSINTE BRANCHED/CYCLOIDEA/PCF (TCP) genes, the plant-specific transcription factors containing bHLH motifs that allow DNA binding and protein-protein interaction [22], [23], [24], [25], [26], [27]. Microarray data of the shoot apical meristem of miR319 transgenic plants, compared to wild-type (WT), showed that expression of all miR319-targeted TCP genes decreased up to 30-fold, which strongly indicated that TCP genes were degraded by miR319 activity [20], [28], [24]. Expression of TCPs, lacking the miR319 recognition site were not affected. Previous studies showed that miR319-TCPs regulated leaf developmental processes, including leaf senescence, cell proliferation, and cell differentiation [20], [29], [30].
In addition, several researchers have suggested the involvement of miR319 in the cold stress response. In sugarcane, changes in miR319 expression under cold stress were observed in both roots and shoots [31]. In rice, expression of Osa-miR319 was down-regulated by cold stress [13]. Yang et al. (2013) observed that overexpression of Osa-miR319b and repression of its targets, OsPCF5 and OsPCF8 led to enhanced cold tolerance (4°C) after chilling acclimation (12°C). MiR319 was supposed to participate in plant cold tolerance by regulating its target genes or functioning together with other miRNAs [27]. However, little is known about the regulatory relationship between miR319 and its targets, OsPCF6 and OsTCP21, under cold stress.
In previous studies, we constructed a rice miRNA expression profile under cold stress based on the microarray data, and identified a total of 18 cold stress responsive miRNAs, including Osa-miR319. In this study, we further characterized the biological function of Osa-miR319 in response to cold stress by using overexpression transgenic lines. To elucidate the regulatory relationship between OsmiR319 and OsPCF6/OsTCP21, we evaluated the effect of Osa-miR319 overexpression on OsPCF6 and OsTCP21 expression under cold stress. We also investigated the roles of OsPCF6 and OsTCP21 in plant cold response and ROS accumulation under cold stress by using overexpression and RNAi transgenic rice. Collectively, the results suggested that, as Osa-miR319 targets, OsPCF6 and OsTCP21 negatively regulated plant cold tolerance.
Materials and Methods
Plant materials and stress treatments
De-hulled seeds of Oryza sativa L. (Kong Yu 131, from the College of Agriculture, Northeast Agricultural University, Harbin, Heilongjiang Province, China) were sterilized with 5% sodium hypochlorite (NaClO) for 15 min and rinsed with sterilized water for three times. The sterilized seeds were germinated on 1/2 Murashige and Skoog (MS) medium with 1% (W/V) agar for 4 d, and then transferred to Yoshida nutrient solution under normal culture conditions (25–28°C, 600 µmol photons m−2 s−1, 14 h light/10 h dark cycles, 75% relative humidity) [52]. For gene expression analysis, the 2-week-old seedlings were exposed to 4°C, and equal amounts of leaves were harvested at certain time points. After being frozen in liquid nitrogen, the samples were stored at −80°C before RNA extraction.
RNA isolation, cDNA synthesis and real-time RT-PCR analysis
Total RNA was extracted by using a plant RNA isolation reagent (Cat. No. 12322-012, Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. Reverse transcription reactions were performed by using M-MLV reverse transcriptase (Cat. No. M0253V, NEB, Ipswich, MA, USA). OsEf1-α (elongation factor 1-α) was used as an internal control. The 2−ΔΔCt method was used for real-time PCR analysis. To enable statistical analysis, three fully independent biological replicates were obtained and subjected to real-time PCR in triplicate. Primers for quantitative real-time PCR are listed in Table 1.
Table 1. Primers of genes used for quantitative real-time PCR validation.
| Gene | Prime sequence |
| Pre-OsR319b | Forward: 5′- GTCGAATTAGCTGCCGACTC -3′ |
| Reverse: 5′-TCGAAGAGATCGAGGAGGAG-3′ | |
| OE-OsPCF6 | Forward: 5′-GTGCCAATAGGGGGACCCT-3′ |
| Reverse: 5′-CCAAGCAGGAAGGAAATGGT-3′ | |
| OE-OsTCP21 | Forward: 5′-CACTTCATGCCCGTCCACG-3′ |
| Reverse: 5′-TGGCTGGAGAGGTGAGACTGC-3′ | |
| Ri-OsPCF6 | Forward: 5′-ATCGGTTGCCTGTCAATTTT-3′ |
| Reverse: 5′-GATTTATCTCAACTTTATAG-3′ | |
| Ri-OsTCP21 | Forward: 5′-TTTTTGGCCATCATGAACAC-3′ |
| Reverse: 5′-GATTTATCTCAACTTTATAG-3′ | |
| J033099M14 | Forward:5′-CTCAAATCAAGGCGTCAACTAAGA-3′ |
| Forward:5′-TTTGTCAATATATACGTGGCATATACCA-3′ | |
| OsSIK1 | Forward:5′-TCTGGTAGTCTGCCCGAGGAA-3′ |
| Forward:5′-TATGTACTGGTTGCAATCAG-3′ | |
| Os01g22249 | Forward:5′-AACGGAGTGGAAGCAGCGT-3′ |
| Forward:5′-CAGCACCTCTATGTTGCCCA-3′ | |
| DREB1A | Forward:5′-GGAGCAAGCAGAAACACACA-3′ |
| Forward:5′-TCGTCTCCCTGAACTTGGTC-3′ | |
| DREB2A | Forward:5′-AAGCACGGCATAATTTTTGG-3′ |
| Forward:5′-CGTTCTTTCCAGCTTTCCTG-3′ | |
| DREB1B | Forward:5′-CTCGCACTGAAAAGTGTGGA-3′ |
| Forward:5′-CAAAAGGAGGGAGAAATCTGG-3′ | |
| DREB1C | Forward:5′-CTACGCGTACTACGGCAACA-3′ |
| Forward:5′-GAGGAGCAAAGCTGGTTGAG-3′ | |
| Tpp1 | Forward:5′-CCTTCAGCAAATCATGAGCA-3′ |
| Forward:5′-AGCCTCCAGCACTTCGTTTA-3′ | |
| Tpp2 | Forward:5′-AGGATGCATTCAAGGTTCTGA-3′ |
| Forward:5′ -CAAGATGCCAGTTTCTTCAGG-3′ | |
| OsCPT1 | Forward:5′-AGTCGCGTGTTCTCCTTTGT-3′ |
| Forward:5′-CATGACAGCAGCTTGCAAAT-3′ |
Vector construction and generation of transgenic rice
Osa-miR319b pre-sequences were downloaded from miRBase (http://www.mirbase.org) and PCR amplified using gene specific primers (5′-GGCTTAAUGCTCCAAAAGTTTCGTGGTTGTT-3′ and 5′-GGTTTAAUACTGGTGCTATCATTTCATGCCC-3′). The PCR products were then cloned into the binary vector pCAMBIA330035sU to create the Osa-miR319b overexpression construct driven by the CaMV35s promoter [60]. The full-length coding region of OsPCF6 was amplified with gene specific primers 5′-GAGCCTCTCTCGCGGAACAAGGCAT-3′ and 5′-GAGGATGCGTTGCTGGGATCGATCA-3′; the OsTCP21 gene was amplified using the specific primers 5′-TCCTAGGGCATATGACCGGTACTACG-3′ and 5′-GCTGCTGATCTGATCAGTGCTTGCC-3′. The PCR products were ligated into the pBluescript SK vector (TransGen Biotech Beijing China) for sequence, and then, digested by BamHI and XbaI, and cloned into pCAMBIA3300 to obtain the pCAMBIA3300-OsPCF6 and pCAMBIA3300-OsTCP21 constructs. To generate the RNA interference (RNAi) constructs, OsPCF6 and OsTCP21 were amplified using specific primers, OsPCF6: 5′-ATCGGTTGCCTGTCAATTTT-3′ and 5′-GATTTATCTCAACTTTATAG-3′; and OsTCP21: 5′-TTTTTGGCCATCATGAACAC-3′ and 5′-GATTTATCTCAACTTTATAG-3′, respectively, and cloned into pCAMBIA3300 by forward and reverse insertions. The recombinant constructs were introduced into Agrobacterium tumefaciens EHA105 and then transformed into rice embryonic calli. The re-generated seedlings were selected on 1/2 MS medium containing 15 mg L−1 glufosinate (Roche, Germany).
The presence of the Osa-miR319b, OsPCF6 or OsTCP21 genes in the glufosinate-positive plants were confirmed by PCR analysis using CaMV35S promoter-specific forward primer and the Bar gene-specific reverse primer. The PCR-positive seedlings were further confirmed by southern blot analysis. Briefly, genomic DNA extracted from 14-day-old seedlings was digested with HindIII, electrophoresed on a 0.8% (W/V) agar gel, and blotted onto a nylon membrane (Roche, Germany) under alkaline conditions. Digoxingenin-dUTP amplified Bar from pCAMBIA330035sU was used as a probe for hybridization. The membrane was exposed to X-ray film (Eastern Kodak). Transcript levels of Osa-miR319b, OsPCF6 and OsTCP21 genes in transgenic rice were analyzed by using semi-quantitative RT-PCR or quantitative real-time PCR analysis using gene specific primers. The OsEf1-α (elongation factor 1-α) was used as an internal control.
Phenotypic analysis of transgenic rice under cold stress
For the cold tolerance test, three-leaf seedlings of both WT and transgenic lines were exposed to 4°C for 7 d, and then recovered under normal conditions for 10 d. Survival rate of WT and transgenic plants lines after 10 days of recovery. The proline content in rice leaves was determined by the method described previously [53], [54].
DAB is oxidized by hydrogen peroxide in the presence of some heme-containing proteins, such as peroxidases, to generate a dark brown precipitate [55]. DAB staining of the rice aerial parts were determined according to the method described previously [56], [57], [58].
Subcellular localization of the OsPCF6 and OsTCP21-GFP fusion protein
To determine the subcellular localization, the full-length OsPCF6 and OsTCP21 coding regions were inserted into the pBSK:eGFP vector to generate pBSK-OsPCF6-GFP and pBSK-OsTCP21-GFP. The constructs were confirmed by sequencing and used for transient transformation of onion (Allium cepa) epidermal cells by biolistic bombardment. After 24 h of incubation, GFP fluorescence in transformed onion cells was observed at 488 nm with a confocal microscope (Zeiss, Germany). GFP fluorescence was detected between 505 nm and 550 nm with an excitation wavelength of 488 nm. GFP fluorescence and light field visions were recorded in separate channels and then merged into an overlay image.
Statistics
All numerical data was analyzed using the Student's t-test. Statistical differences were referred as significant when *P<0.05 and **P<0.01.
Results
Production and molecular characterization of Osa-miR319b transgenic Oryza sativa plants
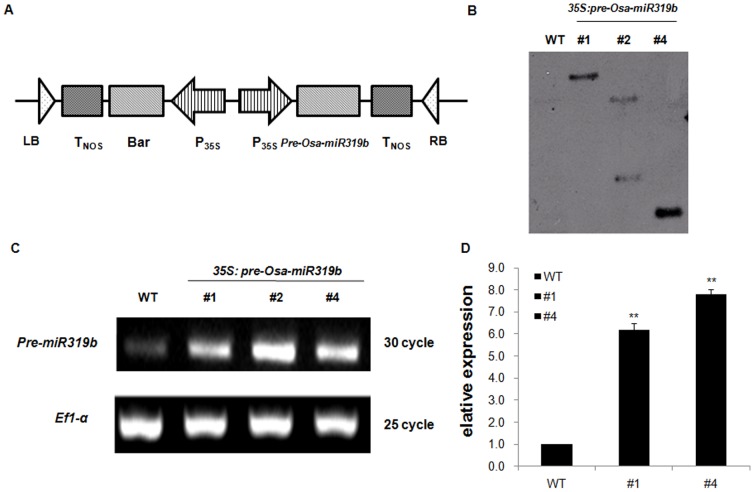
In previous studies, we identified a total of 18 cold stress responsive miRNAs using microarray data, including miR-156k, miR-166k, miR-166m, miR-167a/b/c, miR-168b, miR-169e, miR-169f, miR-169h, miR-171a, miR-535, miR-319a/b, miR-1884b, miR-444a.1, miR-1850, miR-1868, miR-1320, miR-1435, and miR-1876 [13]. In the current study, we focused on the Osa-miR319b gene, which was obviously down-regulated under cold stress. To explore the potentiality of manipulating Osa-miR319b expression in annual species for enhancing plant resistance to cold stress, we prepared a chimeric construct, p35S-Osa-miR319b/p35S-Bar, under the control of the CaMV 35S promoter (Fig. 1A). The transgenic lines were identified by glufosinate selection, PCR and southern blot analysis. Southern blotting results showed that Osa-miR319b was integrated into the rice genome of three independent transgenic lines (Fig. 1B). Semi-quantitative RT-PCR analysis showed relatively higher expression levels of Osa-miR319b in all three lines (Fig. 1C). Expression levels of the mature-Osa-miR319b in transgenic lines were further confirmed by quantitative real-time PCR analysis (Fig. 1D). The homozygous T2 generation of two single-copy transgenic lines (#1 and #4) was used in the following experiments.
Figure 1. Generation and molecular analysis of Osa-miR319b transgenic lines.
(A) Schematic diagram of the Osa-miR319b overexpression gene construct, p35S-Osa-miR319b/p35S-Bar, in which the Osa-miR319b gene is under the control of the CaMV 35S promoter. The CaMV 35S promoter-driven Bar is included for glufosinate resistance. LB, Left border; RB, right border. (B) Southern blot analysis of transgenic rice. Rice DNA and vector DNA were digested by HindIII, run on a 0.8% agarose gel, transferred to a nylon membrane, and probed with digoxingenin-dUTP against the amplified bar gene sequence. The wild-type rice DNA was used as negative control. (C) Expression pattern of Osa-miR319b in wild-type and transgenic lines by semi-quantitaive RT-PCR. cDNAs were normalized using the Ef1-α gene. (D) Verification of the mature-Osa-miR319b overexpression in Osa-miR319b-overexpressing transgenic rice lines by real-time RT-PCR. RNA was extracted from the flag leaves of the second generation of the transgenic rice plants. snRNA U6 served as a reference gene for the detection of miRNAs.
Overexpression of Osa-miR319b in rice results in increased leaf width
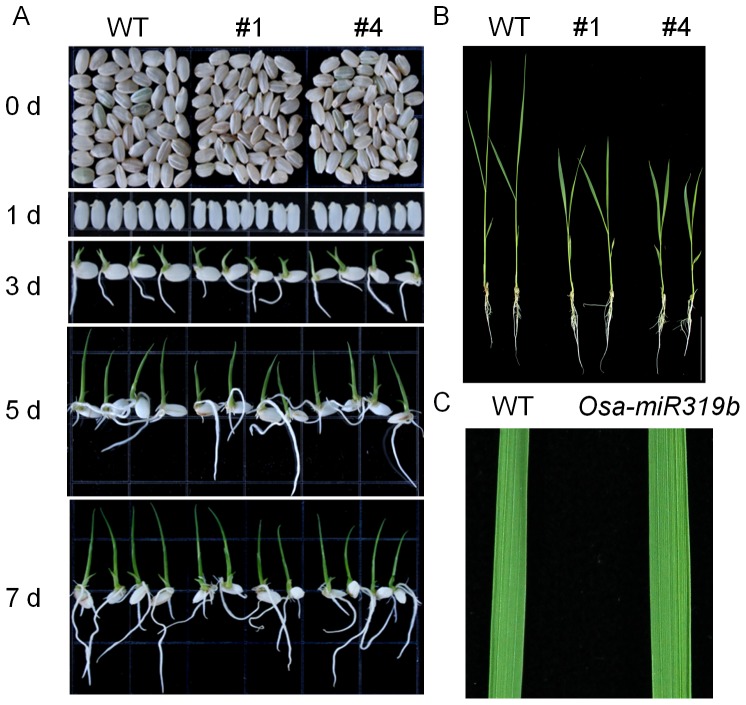
The most conspicuous feature of Osa-miR319b-overexpressing transgenic plants (OE) is delayed development and wider leaves. It is noteworthy that the size of OE seed is smaller than wild-type seed. Gross morphology at 1–5 day old seeding phase of wild-type and transgenic plants have no remarkable different. However, gross morphology of transgenic plants are smaller than wild-type plants at two-week-old (Fig. 2B), but the leaf blade of transgenic plant is larger compared with wild-type blade (Fig. 2C). There is no obviously difference in plant height and the number of productive tillers per plant at final period (data not shown). These results indicated that overexpression of Osa-miR319b possibly conferred plants developments.
Figure 2. Osa-miR319b overexpressing plants showed delayed development and widen leaf.
(A) Gross morphology at 0 to 7-day-old seedlings phase of WT and transgenic plants; (B) Transgenic plants showed 14 d delaying of heading time comparing with WT. (C) The Osa-miR319b-overexpressing transgenic plants exhibited wider leaves than WT controls.
Overexpression of Osa-miR319b in O.sativa enhance plant tolerance to cold stress
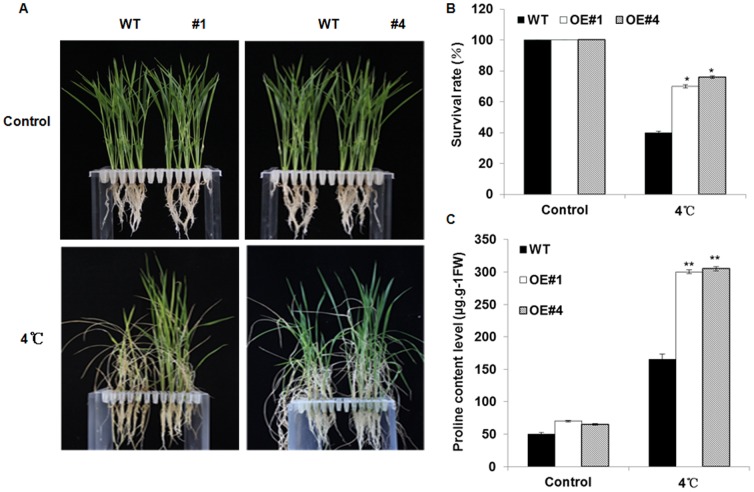
To test whether Osa-miR319b overexpression affects plant tolerance to cold stress, the WT and overexpression (OE) seedlings were hydroponically cultured in Yoshida solution until three leaves appeared. The seedlings were directly exposed to 4°C for 7 d, and then recovered for 10 d. As shown in Figure 3A, the OE lines showed significantly better growth performances than WT. The statistical analysis revealed that the survival rate of WT seedlings under cold stress was approximately 30%, while OE lines #1 and #4 displayed approximately 80% and 90% survival rates respectively (Fig. 3B) (P<0.05, by Student's t-test). These results demonstrated that Osa-miR319b overexpression enhanced plant cold tolerance at the seedling stage.
Figure 3. Stress tolerance of Osa-miR319b transgenic rice and wild-type rice subjected to cold stress treatment.
(A) Growth performance of WT and transgenic rice (#1 and #4) under normal condition and cold stress. The WT and transgenic rice seedlings were exposed to 4°C for 7 d followed by 10 d of recovery. (B) The survival rates of WT and transgenic seedlings under normal condition and cold stress. The numbers of survived seedlings/total seedlings used to calculate the survival rate. (C) The free proline content of WT and transgenic seedlings under normal condition and cold stress. Wild-type and transgenic rice seedlings of 2-week-old were exposed to 4°C for 3 days and then collected for determination of proline. Data are the mean ± SE of three biological replicates. Asterisks indicate statistically significant differences (*P<0.05, **P<0.01) between wild-type and transgenic lines.
The accumulation of proline to facilitate osmoregulation is a common adaptive mechanism for plant tolerance to cold stress. To test whether the enhanced cold tolerance of Osa-miR319b OE plants is related to the capacity of proline accumulation, the free proline contents of WT and OE plants were investigated. As shown in Figure 3C, no significant difference was observed between WT and OE plants under normal conditions. Cold stress obviously promoted the free proline accumulation in both WT and OE plants. However, the proline content of OE plants was significantly higher than that of WT (Fig. 3C) (P<0.01, by Student's t-test). These results indicated that overexpression of Osa-miR319b conferred increased cold tolerance, perhaps through promoting the free proline accumulation under cold stress.
Overexpression of Osa-miR319b increases the transcript levels of cold stress responsive genes
To explore the role of Osa-miR319b in the cold stress response, the expression of the well-known cold stress responsive marker genes was analyzed in both WT and OE plants. Marker genes linked to cold stress, such as DREB1/CBF, DREB2A, OsCPT1, OsTPP1/2, could contribute to plant cold enhanced tolerance. To test whether these genes were responsible for enhanced the cold tolerance of Osa-miR319b OE plants, quantitative real-time PCR was performed to evaluate the expression levels. As shown in Figure 4, cold stress obviously induced expression of all the above genes, and Osa-miR319b OE lines exhibited significantly higher expression levels than WT, except OsCPT (P<0.05, by Student's t-test). OsCPT is a putative target member of the dehydration-responsive element-binding factor 1 DREB1/CBF pathway, and showed opposite expression to DREB1/CBF under cold stress. Thus, Osa-miR319b may confer increased cold tolerance through up-regulating cold stress responsive genes, including DREB1/CBF, DREB2A, and TPP1/2.
Figure 4. Expression levels of some cold-responsive genes in wild-type plants and overexpressing Osa-miR319b plants.
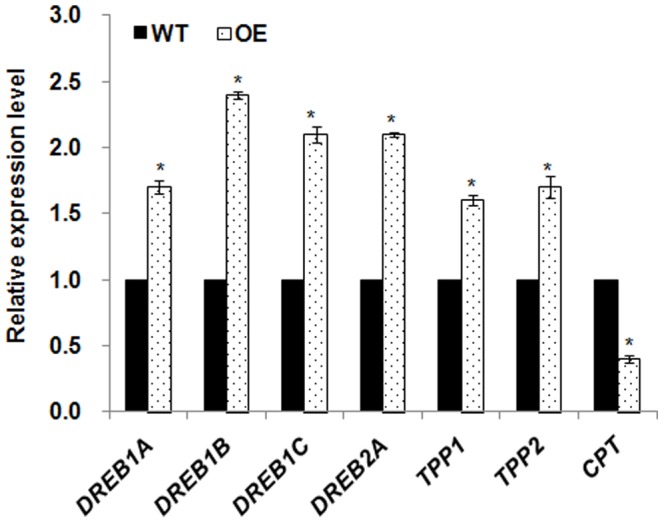
Marker gene expression levels were set equal to 1 in the WT plants in cold stress with 4°C for 8 h. The expression level of marker genes are represented as a ratio in OE plants compared with WT plants. Data are the mean ± SE of three biological replicates. Asterisks indicate statistically significant differences (*P<0.05) between WT and transgenic lines.
The cold induction of OsPCF6 and OsTCP21 is obviously repressed in Osa-miR319b OE lines
In previous studies, PCF6 gene was identified as one of the miR319b targets by using 5′RACE analysis in sugarcane [32]. Through Blastp search, we identified two homologous genes OsPCF6 and OsTCP21. Thus, we analyzed the expression profiles of OsPCF6 and OsTCP21 in WT and Osa-miR319b OE lines in response to cold stress, to verify whether their expression was affected by Osa-miR319b overexpression. The quantitative real-time PCR results showed that expression of OsPCF6 and OsTCP21 was significantly induced by cold stress in WT plants (Fig. 5). However, in the Osa-miR319b OE lines, the cold induction of OsPCF6 and OsTCP21 was obviously inhibited (P<0.05, by Student's t-test), which indicated that Osa-miR319b overexpression repressed the induced expression of OsPCF6 and OsTCP21 under cold stress. These results suggested that OsPCF6 and OsTCP21 were indeed target genes of Osa-miR319b, and were possibly involved in plant response to cold stress.
Figure 5. qRT-PCR analysis of OsPCF6 and OsTCP21 expression patterns under 4°C in wild-type and Osa-miR319b-overexpressing plants.
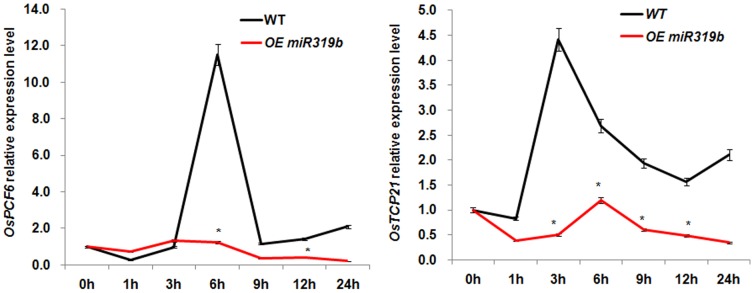
Expression patterns of the OsPCF6 and OsTCP21 genes under 4°C in rice seedlings at 0, 1, 3, 6, 9, 12, 24 h after cold stress onset in WT and Osa-miR319b-overexpressing lines. The expression level at 0 h was set at 1.0. Data are the mean ± SE of three biological replicates. Asterisks indicate statistically significant differences (*P<0.05) between wild-type and transgenic lines.
OsPCF6 and OsTCP21 negatively regulates plant cold tolerance
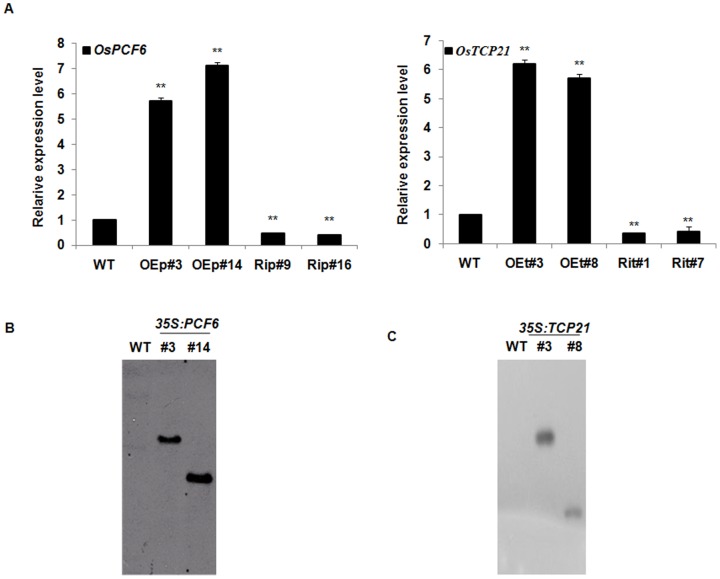
To test whether OsPCF6 and OsTCP21 were directly involved in plant response to cold stress, we generated OE and RNAi (Ri) transgenic lines, which were confirmed by glufosinate selection, quantitative real-time PCR (Fig. 6A) and southern blot analysis (Fig. 6B). Compared with WT, the transcript abundance of OsPCF6 and OsTCP21 was significantly higher in the OsPCF6 and OsTCP21 OE lines (OEp#3 and #14; OEt#3 and #8) but lower in the OsPCF6 and OsTCP21 Ri lines (Rip#9 and #18; Rit#1 and #7) (P<0.01, by Student's t-test). Seedlings of WT and homozygous T2 progeny were grown in Yoshida solution and used in the following phenotypic experiments. No differences were observed among WT, OE and Ri lines when grown under normal, non-stressed conditions.
Figure 6. Molecular confirmation of OsPCF6 and OsTCP21 transgenic rice.
(A) OsPCF6 and OsTCP21 expression in wild-type and transgenic rice. Total RNAs from 2-week-old wild-type and transgenic rice plants were isolated, reverse-transcribed, and analysed by real-time reverse-transcription PCR. cDNAs were normalized using the Ef1-α gene. Data are the means ± SE of three biological replicates. Asterisks indicate statistically significant differences (**P<0.01) between WT and transgenic lines (OE and Ri). (B) Southern blot analysis of OsPCF6. (C) Southern blot analysis of OsTCP21. The WT rice genomic DNA was used as negative control.
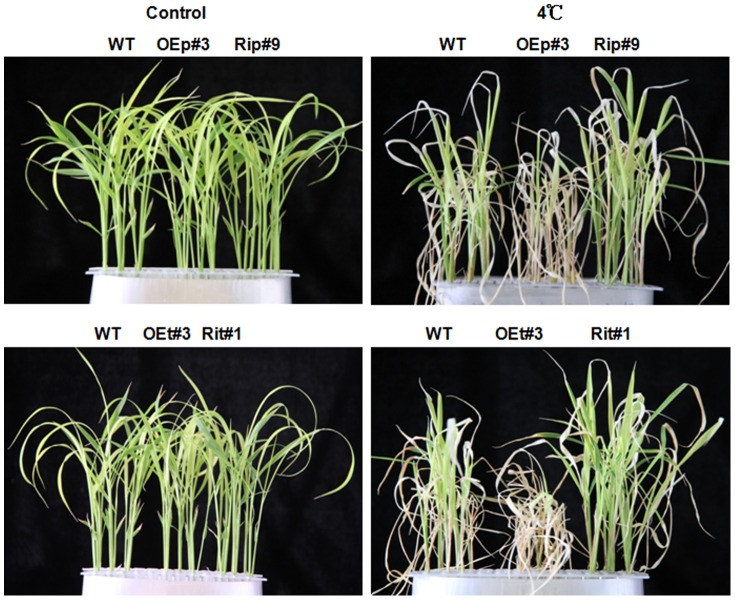
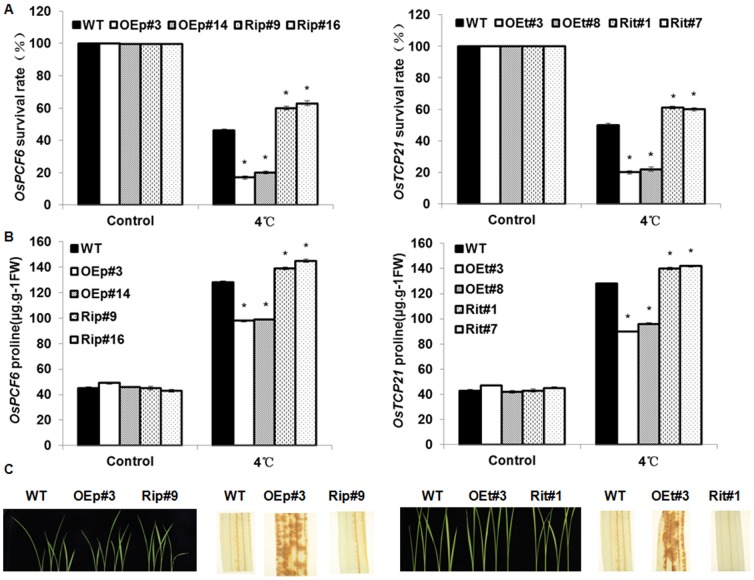
To investigate the involvement of OsPCF6 and OsTCP21 in cold stress, the WT, OE and Ri seedlings were exposed to 4°C for 7 d, after which they were recovered for 10 d. Phenotypically, most OsPCF6 and OsTCP21 Ri seedlings remained green and showed continuous growth, whereas both WT and OE seedlings showed severe leaf rolling and wilting after cold stress (Fig. 7). As shown in Figure 8A, the survival rates of the OE and/or Ri lines were significantly lower and/or higher, respectively, than that of WT when exposed to cold stress, indicating that the alteration of OsPCF6 and OsTCP21 expression levels altered plant cold tolerance.
Figure 7. Effect of cold stress on OsPCF6 and OsTCP21 in wild-type and transgenic plants.
Left: wild-type, OsPCF6 and OsTCP21 transgenic plants (OE and Ri) without stress treatment; right: wild-type and transgenic seedlings treated with 4°C for 7 d followed by 10 d of recovery.
Figure 8. Transgenic plants (OE and Ri) of OsPCF6 and OsTCP21 confer cold tolerance in rice.
(A) The survival rates of WT, OsPCF6 and OsTCP21 transgenic seedlings under normal condition and cold stress. (B) The proline contents of WT, OsPCF6 and OsTCP21 transgenic seedlings under normal condition and cold stress. Wild-type and transgenic rice seedlings of 2-week-old were exposed to 4°C for 3 days and then collected for determination of proline. (C) H2O2 accumulation in WT, OsPCF6 and OsTCP21 transgenic plants under cold stress. The 2-week-old seedling were exposed to 4°C for 3 d, and then seedling roots were cut off and directly incubated in the DAB dye solution for 8 h for H2O2 analysis. Left: WT, OsPCF6 and OsTCP21 transgenic plants without stress treatment; right: wild-type and transgenic plants, after treatment and staining. OE is dark brown; WT and Ri lines have the lightest color.
In addition, we also evaluated the free proline accumulation in OsPCF6 and OsTCP21 OE and Ri transgenic lines under cold stress. As shown in Fig. 8B, there was no significant difference in the proline contents among WT, OE and Ri transgenic plants under control conditions. As expected, an obvious increase in the free proline content was observed under cold stress in both WT and transgenic plants. However, the OsPCF6 and OsTCP21 RNAi lines accumulated more free proline than WT under cold stress (Fig. 8B). These results indicate that OsPCF6 and OsTCP21 negatively regulated plant cold tolerance, perhaps through modulating the accumulation of free proline under cold stress.
Reactive oxygen species (ROS) generation is usually observed in damaged cells during cold stress. Considering the damage caused by ROS, we evaluated the ROS generation in leaves of WT, OE and Ri seedlings which were exposed to cold stress. H2O2 formation was visualized by polymerization with 3, 3′ -diaminobenzidine (DAB). As shown in Fig. 8C, in the OE seedling leaves, H2O2 accumulation was observed as dark brown deposits, indicating that OE plants exhibited the weakest ability to eliminate ROS. The OsPCF6 and OsTCP21 Ri plants had the strongest ability to eliminate ROS, and thus, displayed the lightest color. These results suggested that OsPCF6 and OsTCP21 regulated plant cold tolerance by mediating ROS generation or scavenging.
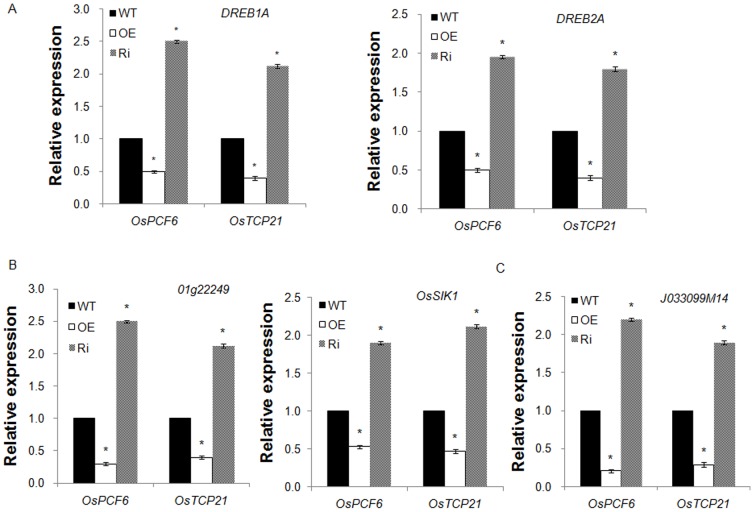
To further elucidate the mechanism by which OsPCF6- and OsTCP21-RNAi plants enhanced cold tolerance the effects of cold stress on expression of genes responsible for proline biosynthesis and H2O2 scavenging were investigated. As shown in Figure 9, treatment with cold stress led to a great increase in transcripts of DREB1A and DREB2A and H2O2 scavenging (Os01g22249 and OsSIK1) and proline biosynthesis genes (D-1-pyrroline-5-carboxylate synthase genes, J033099M14) in the OsPCF6- and OsTCP21-RNAi lines than wild-type and overexpression lines was observed in response to cold stress.
Figure 9. Expression levels of some cold-responsive genes in wild-type plants and OsPCF6 and OsTCP21 transgenic plants.
(A) Expression levels of DREB1A and DREB2A gene in transgenic lines and wild-type plants. (B) Expression levels of ROS scavenging genes (Os01g22249 and OsSIK1) in transgenic lines and wild-type plants. (C) Expression levels of putative proline synthase gene (J033099M14) in transgenic lines and wild-type plants. Data are the mean ± SE of three biological replicates. Asterisks indicate statistically significant differences (*P<0.05) between WT and transgenic lines.
OsPCF6 and OsTCP21 proteins are located in the nuclei and the spatial-temporal expression profiles of Osa-miR319b and OsPCF6 and OsTCP21
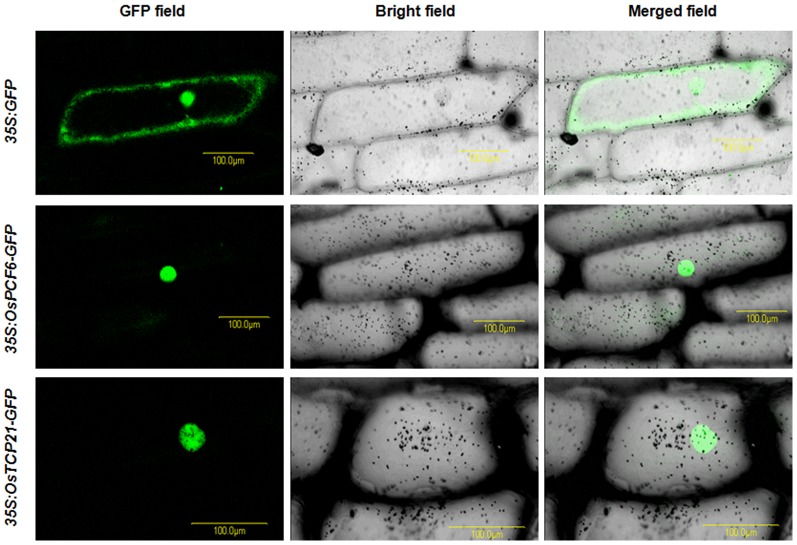
As transcription factors, OsPCF6 and OsTCP21 are supposed to be located in the nuclei. To examine the subcellular localization of OsPCF6 and OsTCP21 proteins, the full-length coding regions of OsPCF6 and OsTCP21 were fused in-frame to the 5′-terminus of the GFP reporter under the control of the CaMV35S promoter. The OsPCF6-GFP and OsTCP21-GFP recombinant constructs and GFP alone were introduced into onion (Allium cepa) epidermal cells by particle bombardment. An examination of protein fluorescence by confocal laser-scanning microscopy revealed that the OsPCF6-GFP and OsTCP21-GFP fusion proteins were specifically localized in the nuclei, whereas GFP alone showed a ubiquitous distribution in the cell (Fig. 10).
Figure 10. OsPCF6-GFP and OsTCP21-GFP proteins were located in the nuclei.
Confocal images of onion epidermis cells under the GFP channel showing the constitutive localization of GFP and the nuclear localization of OsPCF6- and OsTCP21-GFP fusions driven by the cauliflower mosaic virus 35S promoter. Onion epidermal peels were bombarded with DNA-coated gold particles and GFP expression was visualized 24 h later.
Previous studies showed higher expression of Osa-miR319b in the young panicle and culm, and lower expression in root, mature panicle, and leaf. Also, OsTCP21 was mainly detected in later panicle stages, ovaries and stigma, whereas, OsPCF6 showed higher accumulation in shoot apical meristem, embryo and endosperm [19], [59]. Osa-miR319b was not inversely correlated with the OsPCF6 and OsTCP21 in spatio-temporal expression specificity, suggesting that there exists a complex relationship between Osa-miR319b and its above target genes in rice.
Discussion
It has been suggested that miRNA play essential roles in regulation of gene expression for normal growth and development and adaptation to environmental stresses [33]. In previous study, ectopic expression of miR319 induced larger leaf blades and continuous growth of leaf margins in snapdragon, Arabidopsis, tomato and other species though each with its distinct leaf form [20], [22]. In our study, we observed that overexpressing Osa-miR319b could result in wider leaf and delayed development in seeding stage of rice (Fig. 2). This result is generally similar to the larger leaves phenotype observed in transgenic dicotyledonous plants with miR319 overexpression [18], [20], [22]. Yang (2013) showed that wider leaves of overexpressing Osa-miR319b plants are not due to change in their cell size but to their increase cell number reflected mainly by increased leaf small veins. However, how affected the leaf width and delayed development of Osa-miR319b in our further study.
Numerous cold-regulated miRNAs have been identified in Arabidopsis [34], [35] and rice [13]. Previous studies suggested that miR319b responded to cold stress and that TCP transcription factors were the predicted targets of miR319b [13], [25], [19]. Transcription factors up-regulated by cold stress contained the TCP-domain families [36], leading us to conduct further experiments to characterize the function of miR319b and TCP in plant cold stress response. Both our and Yang study demonstrated that overexpressing Osa-miR319b led to enhanced cold tolerance (4°C) in transgenic rice seeding. Furthermore, cold stress osa-miR319b expression was down-regulated while the expression of miR319-targeted genes was up-regulated. The study by Yang observed that down-regulating the expression of either of the two miR319-targeted genes, OsPCF5 and OsPCF6, in RNAi plants also resulted in enhanced cold tolerance. In this study, we focused on another two targets of Osa-miR319b, OsPCF6 and OsTCP21. We found that OsPCF6 and OsTCP21 expression was largely greatly by cold stress, and the degree of induction was obviously repressed in Osa-miR319b transgenic lines. Consistent with previous study we also demonstrated that overexpression or repression OsPCF6 and OsTCP21 significantly decreased or increased plant cold torlerance. Moreover, we also reported that OsPCF6 and OsTCP21 negatively regulated ROS generation or scavenging under cold stress.
To evaluate the role of Osa-miR319b in plant tolerance to cold stress, in this study, we generated transgenic plants over-expressing Osa-miR319b by Agrobacterium-mediated transformation (Fig. 1B, C). One important finding was that Osa-miR319b-overexpressing plants exhibited higher survival rate than WT plants when exposed to cold stress (Fig. 3B). Therefore, Osa-miR319b possibly participates in and responds to cold stress, and over-expression of Osa-miR319b enhances plant tolerance to cold stress.
To elucidate the potential mechanisms for the enhanced cold tolerance of Osa-miR319b OE plants, an analysis of the proline content was conducted to monitor changes in physiological processes associated with plant cold stress tolerance. The accumulation of free proline [37], [38] is a common phenomenon in response to cold stress. The accumulated free proline to facilitates osmoregulation and protect plants from the dehydration that results from cold stress by reducing the water potential of plant cells. This study found a greater accumulation of free proline in the Osa-miR319b OE plants, which may partially account for the higher tolerance of Osa-miR319b OE plants to cold stress (Fig. 3C).
In addition to the Osa-miR319b OE plants can accumulation of free proline content, we also found that overexpression of Osa-miR319b led to higher expression levels of cold stress related genes, such as Hsieh showed that transgenic expression of the transcriptional activator, CRT/DRE-binding factor 1 (CBF1), enhanced the cold tolerance of tomato plants(Fig. 4). DREB1/2 have been shown to play irreplaceable roles in cold stress [39], [40]. In rice, OsDREB1A, OsDREB1B, OsDREB1C and OsDREB2A have been studied in depth [41]. We demonstrated up-regulated expression of DREB1 genes in OE plants under cold stress compared with wild-type plants. OsCPT is the deduced target gene of the DREB pathway via the DRE/CRT cis-element [42]. In our study, OsCPT displayed down-regulated expression in Osa-miR319b OE plants under cold stress. These results demonstrated that Osa-miR319b played an important role in cold tolerance by regulating a number of cold stress related genes.
In rice, miR319 was predicted to target five TCP genes, OsPCF5, OsPCF6, OsPCF7, OsPCF8, and OsTCP21. The abundance of the five TCP genes in different tissues of rice. As Yang showed that the highest expression of OsPCF5 and OsPCF8 was detected in the leaf and root, and that of OsPCF7 and OsTCP21 in the leaf and young panicle, OsPCF6 was highly expressed in the young and mature panicle. Moreover, the miR319-targeted genes were up-regulated by low temperature treatments, the inverse-correlated with Osa-miR319b. For example, down-regulation of either OsPCF5 or OsPCF8 significantly improved the cold tolerance of transgenic seedlings cold tolerance [19], in sugarcane PCF6, which was identified as miR319b target gene by 5′RACE was induced by cold tolerance [32]. The expression patterns of OsPCF6 and OsTCP21 suggested potential roles in cold stress response (Fig. 5). The OsPCF6/OsTCP21 Ri and OE plants showed enhanced and/or decreased tolerance to cold stress than WT, respectively (Fig. 7). One important finding was that RNAi plants exhibited a higher survival rate than WT plants (Fig. 8A). The cold stress phenotype of OsPCF6 and OsTCP21 OE plants counters the phenotype of the Osa-miR319b-overexpressing plants, while the RNAi plant's phenotypes are in keeping with those of the Osa-miR319b-overexpressing plants (compare Fig. 3A and 7). The phenotypic analysis indicated that both OsPCF6 and OsTCP21 were involved in cold responses, and that they may be regulated by Osa-miR319b.
To elucidate the mechanisms responsible for the enhanced tolerance of Ri lines, we analyzed the levels of free proline in OsPCF6/OsTCP21 Ri and OE seedlings. The levels of free proline has been increased in Ri lines under cold stress (Fig. 8B), which may conferring the cold tolerance. Cold stress also causes increased levels of ROS, the elevated concentrations of ROS can damage cellular structures and macromolecules, leading to cell death [43], [44]. An increased ROS-scavenging ability might be beneficial to plant cold stress tolerance. As Huang reported, cold tolerance was enhanced when OsZFP245 up-regulated the activities of ROS-scavenging enzymes [45]. The less accumulation of H2O2 under cold stress may result from an enhanced capacity for scavenging ROS, and the finding that the Ri lines had the lightest colors than in the wild-type and OE plants (Fig. 8C). The greater tolerance of RNAi plants to cold stress found in this study may be accounted for, at least in part, by mitigating oxidative damage due to suppression of ROS production.
To further elucidate the mechanism underlying the greater accumulation of ROS and proline in the OsPCF6/OsTCP21-over-expressing plants under cold stress, this study examined the changes in proline synthase (D-1-pyrroline-5-carboxylate synthase genes, J033099M14) and ROS scavenging genes (Os01g22249 and OsSIK1) in OsPCF6- and OsTCP21-overexpressing, Ri, and wild-type plants in response to cold stress at the transcriptional level. The present results demonst rate that express ion of proline synthase and ROS scavenging genes was higher in OsPCF6- and OsTCP21-Ri plants than in wild-type and OsPCF6- and OsTCP21-OE plants under cold stress, suggesting that the higher proline content s and lower ROS contents in OsPCF6- and OsTCP21-Ri plants are likely to result from the greater up-regulation of these genes under cold stress (Fig. 9B,C). These results indicate that OsPCF6 and OsTCP21play an important role in cold tolerance in rice by regulating proline contents and ROS scavenging.
Based on this study, a hypothetical mechanistic model of the regulation by Osa-miR319b and its targets in rice has been proposed (Fig. 11), but how Osa-miR319b and its targets participate in cold stress remains to be determined. Exposure of rice to cold induces decreased Osa-miR319b expression, which regulates the expression of the proposed target genes OsPCF6 and OsTCP21. Hsieh showed that transgenic expression of the transcriptional activator, CRT/DRE-binding factor 1 (CBF1), enhanced the cold tolerance of tomato plants [44], [46]. Zat12, an ROS-response zinc finger protein was shown to regulate cold-induced genes. Microarray analysis demonstrated that cold-responsive genes were upregulated by overexpression of Zat12 [47], and Zat12 downregulated CBF transcript expression suggesting a role for Zat12 in suppressing the CBF cold-response pathway. These studies demonstrate a close link between CBF, ROS signaling, and the cold stress response [4]. Up-regulated Osa-miR319b and down-regulated OsPCF6 and OsTCP21 can increase the account of CBF (Fig. 4, 9A), but Zat12 down-regulated CBF transcript expression, ROS up-regulated Zat12 transcript expression suggesting that decrease amount of ROS improve cold resistance. This result suggested that up-regulated Osa-miR319b and down-regulated OsPCF6 and OsTCP21 enhanced cold tolerance possibly through decreased levels of ROS.
Figure 11. Possible molecular mechanism of cold stress in rice.
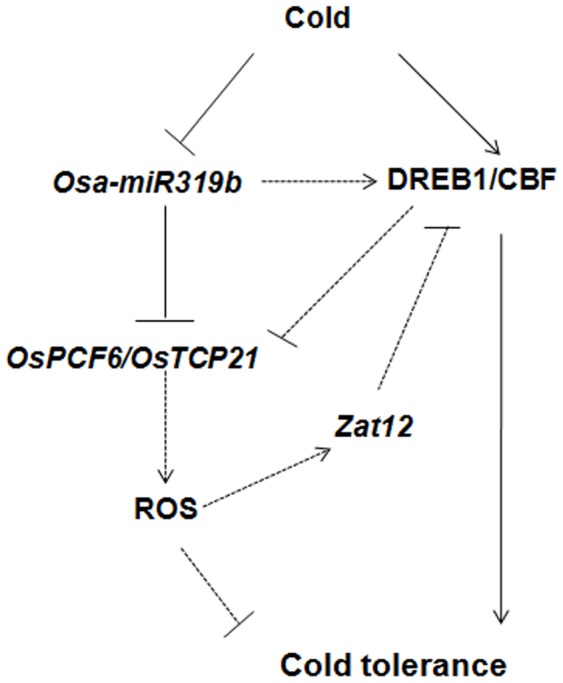
Under cold stress Osa-miR319b is down-regulated, leading to the repression of its targets and possibly the up-regulation of ROS (arrow with dashed line), which in turn inhibits cold tolerance. Under cold stress DREB1/CBF is up-regulated, and Zat12 down-regulated CBF transcript expression, and account of ROS up-regulated Zat12 transcript expression which in turn increases cold tolerance. However, it is possible that Osa-miR319b induces the expression of DREB1/CBF (arrow with dashed line and question mark).
Cold temperatures negatively affect plant growth and development by causing tissue injury and delaying growth [48], and thereby significantly restrict the spatial distribution of plants and the productivity of forests. A plant's ability to withstand cold temperatures depends on its ability to regulate gene expression that modifies their physiology, metabolism, and growth [49]. Diverse plant species endure cold stress to varying degrees, and miR319 was up-regulated in two varieties of sugarcane [32] and down-regulated in the two major subspecies of rice [13] exposed to cold stress. The ability to tolerate low temperatures is a major distinguishing factor in classifying the two major rice subspecies, japonica and indica [50]. Andaya reported that the japonicas are more cold tolerant than the indicas [51]. The expression of Osa-miR319b in japonica rice was reduced beginning at 3 h after exposed to cold, and there was almost no expression at 24 h [13]. However, in indica rice, the expression of Osa-miR319b increased between 12 and 24 h after exposed to cold [19]. The result show differences in Osa-miR319b regulation among different rice genotypes contrasting in tolerance to cold. These results suggest that differences in the intensity of the regulation of Osa-miR319b could be tested as a marker for selecting cold-tolerant rice cultivars.
In summary, this study indicates that Osa-miR319b functions as a positive regulator to mediate the tolerance of rice seedlings to cold stress. The overexpression of Osa-miR319b and RNAi of OsPCF6 and OsTCP21 led to a greater accumulation of osmolytes, such as free proline, and DREB1/CBF proteins in rice, and suppressed the accumulation of ROS under conditions of cold stress through down-regulating OsPCF6 and OsTCP21, the targets of Osa-miR319b. The up-regulation of Osa-miR319b or RNAi of OsPCF6 and OsTCP21 may allow rice plants to effectively osmoregulate by accumulating free proline and to minimize oxidative damage in plants under abiotic stress. OsPCF6 and OsTCP21 are a kind of transcription factors [25], located in the nuclei (Fig. 10) and the function under cold stress have been characterized by generating OE and RNAi transgenic rice plants. More importantly, overexpression of Osa-miR319b and the OE and RNAi of OsPCF6 and OsTCP21 in rice seedlings did not affect their phenotypes under control conditions. Therefore, Osa-miR319b, OsPCF6 and OsTCP21 provide a promising tool for improving the tolerance of rice to abiotic stress in general and to cold stress in particular.
Acknowledgments
We thank Dr. Yucheng Wang of Northeast Forestry University (Heilongjiang, China) for kindly providing the confocal microscope (Zeiss, Germany).
Funding Statement
This work was supported by the National Natural Science Foundation of China (31171578), the “863” project (2008AA10Z153), Heilongjiang Provincial Higher School Science and Technology Innovation Team Building Program (2011TD005), National basic scientific talent training fund projects (J1210069), Graduate student innovation research project in Heilongjiang province (YJSCX2012-047HLJ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lou QJ, Chen L, Sun ZX, Xing YZ, Li J, et al. (2007) A major QTL associated with cold tolerance at seedling stage in rice (Oryza sativa L.). Euphytica 158: 87–94. [Google Scholar]
- 2. Peyman S (2010) Evalution on sixty-eight rice germplasms in cold tolerance at germination stage. Rice Sci 1: 77–81. [Google Scholar]
- 3. Xin Z, J Browse (2000) Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ 23: 893–902. [Google Scholar]
- 4. Suzuki N, Mittler R (2006) Reactive oxygen species and temperature stresses: a delicate balance between signaling and destruction. Physiol Plantarum 126: 45–51. [Google Scholar]
- 5. Bartel PD (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 6.Kim VN (2005) MicroRNA biogenesis: coordinated cropping and dicing. Mol Cell Biol pp. 376–385. [DOI] [PubMed]
- 7. Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Ann Rev Plant Biol 57: 19–53. [DOI] [PubMed] [Google Scholar]
- 8. Kim VN, Nam JW (2006) Genomics of microRNA. Trends Genet 22: 165–173. [DOI] [PubMed] [Google Scholar]
- 9. Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799. [DOI] [PubMed] [Google Scholar]
- 10. Phillips JR, Dalmay T, Bartel D (2007) The role of small RNAs in abiotic stress. FEBS Letters 581: 3592–3597. [DOI] [PubMed] [Google Scholar]
- 11.Eldem V, Okay S, Unver T (2013) Plant microRNAs: new players in functional genomics. Turk J of Agr and Forestry doi:10.3906/tar-1206-50.
- 12. Jagadeeswaran G, Saini A, Sunkar R (2009) Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229: 1009–1014. [DOI] [PubMed] [Google Scholar]
- 13. Lv DK, Bai X, Li Y, Ding XD, Ge Y, et al. (2010) Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 459: 39–47. [DOI] [PubMed] [Google Scholar]
- 14. Talmor-Neiman M, Stav R, Frank W, Voss B, Arazi T (2006) Novel micro-RNAs and intermediates of micro-RNA biogenesis from moss. Plant J 47: 25–37. [DOI] [PubMed] [Google Scholar]
- 15. Fattash I, Voss B, Reski R, Hess WR, Frank W (2007) Evidence for the rapid expansion of microRNA-mediated regulation in early land plant evolution. BMC Plant Biol 7: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Axtell MJ, Bowman JL (2008) Evolution of plant microRNAs and their targets. Trends in Plant Sci 13: 343–349. [DOI] [PubMed] [Google Scholar]
- 17. Warthmann N, Chen H, Ossowski S, Weigel D, Herve P (2008a) Highly specific gene silencing by artificial miRNAs in rice. PLoS One 3: e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang CH, Li DY, Mao DH, Liu X, Ji CJ, et al. (2013) Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ 36: 2207–2218. [DOI] [PubMed] [Google Scholar]
- 20. Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, et al. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- 21. Ozhuner E, Eldem V, Ipek A, Okay S, Sakcali S, et al. (2013) Boron stress responsive microRNAs and their targets in barley. PLoS One 8: e59543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, et al. (2007) Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39: 787–791. [DOI] [PubMed] [Google Scholar]
- 23. Palatnik JF, Wollmann H, Schommer C, Schwab R, Boishouvier J, et al. (2007) Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319 . Dev Cel1 3: 115–125. [DOI] [PubMed] [Google Scholar]
- 24. Schommer C, Palatnik JF, Aggarwal P, Chetelat A, Cubas P, et al. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martin-Trillo M, Cubas P (2009) TCP genes: a family snapshot ten years later. Cell 11: 1360–1385. [DOI] [PubMed] [Google Scholar]
- 26. Nag A, King S, Jack T (2009) miR319a targeting of TCP4 is critical for petal growth and development in arabidopsis. P Natl Acad Sci USA 106: 22534–22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schommer C, Bresso EG, Spinelli SV, Palatnik JF (2012) Role of microRNA miR319 in plant development. MicroRNAs in Plant Development and Stress Responses Signaling and Communication in Plants 15: 29–47. [Google Scholar]
- 28. Efroni I, Blum E, Goldshmidt A, Eshed Y (2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yanai O, Shani E, Russ D, Ori N (2011) Gibberellin partly mediates LANCEOLATE activity in tomato. Plant J 68: 571–582. [DOI] [PubMed] [Google Scholar]
- 30. Ben-Gera H, Ori N (2012) Auxin and LANCEOLATE affect leaf shape in tomato via different developmental processes. Plant Signal Behav 10: 1255–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saini A, Li YF, Jagadeeswaran G, Sunkar R (2012) Role of microRNAs in plant adaptation to environmental stresses. MicroRNAs in Plant Development and Stress Responses Signaling and Communication in Plants 15: 219–232. [Google Scholar]
- 32. Thiebaut F, Rojas CA, Almeida KL, Grativol C, Domiciano CG, et al. (2012) Regulation of miR319 during cold stress in sugarcane. Plant Cell Environ 35: 502–512. [DOI] [PubMed] [Google Scholar]
- 33. Nonogaki H (2010) MicroRNA gene regulation cascades during early stages of plant development. Plant Cell Physiol 51: 1840–1856. [DOI] [PubMed] [Google Scholar]
- 34. Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis . Plant Cell 16: 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Q, Zhang YC, Wang CY, Luo YC, Huang QJ, et al. (2009) Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett 583: 723–728. [DOI] [PubMed] [Google Scholar]
- 36. Hannah MA, Heyer AG, Hincha DK (2005) A global survey of gene regulation during cold acclimation in Arabidopsis thaliana . PLoS Genet 1: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu JP, Zhu JK (1997) Proline accumulation and salt-stress-induced gene expression in a salt-hypersensitive mutant of Arabidopsis. Plant Physiol 114: 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Armengaud P, Thiery L, Buhot N, Grenier-de March G, Savouré A (2004) Transcriptional regulation of proline biosynthesis in Medicago truncatula reveals developmental and environmental specific features. Physiologia Plantarum 120: 442–450. [DOI] [PubMed] [Google Scholar]
- 39. Mao D, Chen C (2012) Colinearity and similar expression pattern of rice DREB1s reveal their functional Conservation in the cold-responsive pathway. PLoS One 7: e47275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao K, Shan XJ, Yuan HZ, Liu Y, Liao X, et al. (2013) Isolation and characterization of dehydration-responsive element-binding factor 2C (MsDREB2C) from Malus sieversii Roem. Plant Cell Physiol 54: 1415–1430. [DOI] [PubMed] [Google Scholar]
- 41. Agarwal PK, Agarwal P, Reddy MK, Sopory SK (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25: 1263–1274. [DOI] [PubMed] [Google Scholar]
- 42. Ma Q, Dai X, Xu Y, Guo J, Liu Y, et al. (2009) Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 150: 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410. [DOI] [PubMed] [Google Scholar]
- 44. Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Plant Boil 55: 373–399. [DOI] [PubMed] [Google Scholar]
- 45. Huang J, Sun SJ, Xu DQ, Yang X, Bao YM, et al. (2009) Increased tolerance of rice to cold, drought and oxidative stresses mediated by the overexpression of a gene that encodes the zinc finger protein ZFP245. Biochem Biophys Res Commun 389: 556–561. [DOI] [PubMed] [Google Scholar]
- 46. Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hsieh TH, Lee JT, Yang PT, Chiu LH, Charng YY, et al. (2002) Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol 129: 1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis . Plant J 41: 195–211. [DOI] [PubMed] [Google Scholar]
- 49. Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444: 139–158. [DOI] [PubMed] [Google Scholar]
- 50. Chinnusamy V, Zhu JK, Sunkar R (2010) Gene regulation during cold stress acclimation in plants. Methods Mol Biol 639: 39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Glaszmann JC, Kaw RN, Khush GS (1990) Genetic divergence among cold tolerant rices (Oryza sativa). Euphytica 45: 95–104. [Google Scholar]
- 52. Andaya VC, Mackill DJ (2003) QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica×indica cross. Springer 106: 1084–1090. [DOI] [PubMed] [Google Scholar]
- 53.Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice. IRRI Los Banos.
- 54. Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39: 205–207. [Google Scholar]
- 55. Song SY, Chen Y, Chen J, Dai XY, Zhang WH (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234: 331–345. [DOI] [PubMed] [Google Scholar]
- 56. Ueno H, Shibata H, Kihara J, Honda Y, Arase S (2003) Increased tryptophan decarboxylase and monoamine oxidase activities induce sekiguchi lesion formation in rice infected with Magnaporthe grisea . Plant J 36: 215–228. [DOI] [PubMed] [Google Scholar]
- 57. Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellar localization of H2O2 in plants. H2O2 accumulation in papillae hypersensitive response during the barley-podery mildew interaction. Plant J 11: 1187–1194. [Google Scholar]
- 58. Bindschedler LV, Dewdney J, Blee KA, Stone JM, Asai T, et al. (2006) Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J 47: 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Daudi A, Cheng Z, O'Brien JA, Mammarella N, Khan S, et al. (2012) The apoplastic oxidative burst peroxidase in Arabidosis is a major component of pattern-triggered immunity. Plant Cell 24: 275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sharma R, Kapoor M, Tyagi A K, Kapoor S (2010) Comparative transcript profiling of TCP family genes provide insight into gene functions and diversification in rice and Arabidopsis[J]. Journal of Plant Molecular Biology and Biotechnology 1: 24–38. [Google Scholar]
- 61. Geu-Flores F, Nour-Eldin HH, Nielsen MT, Halkier BA (2007) USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res 35: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]