Abstract
Chromosomal aberrations are useful in assessing treatment options and clinical outcomes of acute myeloid leukemia (AML) patients. However, 40∼50% of the AML patients showed no chromosomal abnormalities, i.e., with normal cytogenetics aka the CN-AML patients. Testing of molecular aberrations such as FLT3 or NPM1 can help to define clinical outcomes in the CN-AML patients but with various successes. Goal of this study was to test the possibility of Wilms’ tumor 1 (WT1) gene overexpression as an additional molecular biomarker. A total of 103 CN-AML patients, among which 28% had overexpressed WT1, were studied over a period of 38 months. Patient’s response to induction chemotherapy as measured by the complete remission (CR) rate, disease-free survival (DFS) and overall survival (OS) were measured. Our data suggested that WT1 overexpression correlated negatively with the CR rate, DFS and OS. Consistent with previous reports, CN-AML patients can be divided into three different risk subgroups based on the status of known molecular abnormalities, i.e., the favorable (NPM1mt/no FLT3ITD), the unfavorable (FLT3ITD) and the intermediate risk subgroups. The WT1 overexpression significantly reduced the CR, DFS and OS in both the favorable and unfavorable groups. As the results, patients with normal WT1 gene expression in the favorable risk group showed the best clinical outcomes and all survived with complete remission and disease-free survival over the 37 month study period; in contrast, patients with WT1 overexpression in the unfavorable risk group displayed the worst clinical outcomes. WT1 overexpression by itself is an independent and negative indicator for predicting CR rate, DFS and OS of the CN-AML patients; moreover, it increases the statistical power of predicting the same clinical outcomes when it is combined with the NPM1 mt or the FLT3 ITD genotypes that are the good or poor prognostic markers of CN-AML.
Introduction
Acute myeloid leukemia (AML) is defined as hematopoietic stem cell malignancy characterized by clonal expansion of myeloid blasts. It is typically divided into three different risk groups, i.e, the favorable, the intermediate and the unfavorable group based on the types of chromosomal aberrations. About half (40∼50%) of the AML patients have normal karyotype or normal cytogenetics that typically belong to the intermediate risk group in terms of patient’s survival [1], [2]. However, inconsistencies were found among this group of patients in their responses to chemotherapy and prognosis that sometimes makes it difficult to make the right decision for therapeutic treatment and/or assessment of the possible treatment outcome of the patients.
Adding examination of molecular aberrations is thought to be helpful in addressing the differences as described above. Few molecular markers have been used to predict treatment response and prognosis in cytogenetically normal acute myeloid leukemia (CN-AML), such as the nucleoplasmin (NPM1) gene and the fms-like tyrosine kinase 3 (FLT3) gene. A typical NPM1 gene mutation includes small insertions (4∼11 bp) in the coding region of exon 12. The FLT3 gene mutations usually include a D835 point mutation in the tyrosine kinase domain (TKD) of the exon 20 or internal tandem duplications (ITD) in the exon of 14 or 15. Detection of these NPM1 and FLT3 gene mutations has been used to evaluate clinically biological behavior of leukemia cell in the CN-AML patients [2], [3].
The Wilms’ tumor 1 (WT1) gene, which is located on the chromosome 11p13, encodes a zinc-finger transcriptional factor that has emerged as an important regulator of normal and malignant hematopoiesis. WT1 is also one of the molecules that are known to control cellular apoptosis [4]. Resistance of leukemia blasts to apoptosis may cause poor clinical outcomes. Therefore, regulation of apoptotic or anti-apoptotic pathways has high clinical relevance with regard to the remission therapy and the overall survival of the AML patients. Interestingly, high WT1 gene expression was consistently found in peripheral blood (PB) or bone marrow (BM) in the AML patients in comparison with normal controls [5]. However, the significance of WT1 overexpression in therapeutic response and prognosis are still elusive in CN-AML [6]–[8]. Goal of this study was to examine possible correlation of WT1 gene expression with therapeutic response and prognosis in the CN-AML patients. In addition, we also examined the possible interactions of WT1 gene overexpression with the NPM1 or FLT3 gene status, which are known molecular markers associated with the survival and treatment outcomes of AML patients.
Materials and Methods
Patient Population
A total of 103 CN-AML patients consisting of 58 males and 45 females with a median age of 42 years (range, 17–82 years) were recruited for the WT1 overexpression study. All patients were newly diagnosed patients with CN-AML at the Henan Cancer Hospital from the time period of September of 2009 to October, 2012, i.e., a total of 38 months. The diagnosis of AML was made according to the FAB classification. The M3 patients were not included in this study because of the success in chemotherapy based on all-trans-retinoic acid and arsenic trioxide. The standard RHG banding techniques were employed in the karyotyping of leukemia. One milliliter of bone marrow was collected in the EDTA vacutainer from all 103 patients before treatment. This clinical study was approved by the Committee of International and Scientific Research at the Affiliated Tumor Hospital of Zhengzhou University. Written informed consents were obtained from all patients for this study. If a minor was enrolled in this study, a letter of authorization will be first obtained from minor’s guardian along with a signed informed consent by the guardian.
The WT1 overexpression was defined in this study as ≥250 copies/104 ABL as previously recommended [9]. When the WT1 gene was not detected by real-time PCR or the copy number was under the lower limit of detection (3×102 copies/mL), we denoted this WT1 gene copy number as “non-detectable” or “ND”. Based on this criterion, the WT1 gene overexpression was detected in 29 of 103 patients (28%) with a median value of 720 copies/104 ABL (ranged from ND to 8.2×106 copies/mL).
Among these patients, 29% (30/103) of them carried 3 different NPM1 mt mutant genotypes. The most common NPM1 mt mutation was the type A (80%, 24/30), which had a “TCTG” insertion in exon 12. In addition, 4 of 30 patients (13%) had the type B mutation (“CATG” insertion), and 2 of 30 patients (7%) had the type 13 mutation (“TAAG” insertion) [10]. Seven of the 30 NPM1 mt carrying patients (23%) also showed high WT1 gene expression.
The FLT3 mutation was detected in 27% of the total patient cohort (28/103), which included the FLT3 TKD and FLT3 ITD mutations, respectively. In which 10 out of the 28 patients (36%) carried the FLT3 TKD mutation including the D835H (80%, 8/10), D835V (10%, 1/10) and D835Y (10%, 1/10) point mutations, respectively. The FLT3 ITD, which is generally associated with unfavorable outcome [2], [3], was identified in 18 of the 28 patients (64%) that carried the internal tandem duplication in exon 14 (61%, 11/18), intron 14 (6%, 1/18) and exon 15 (33%, 6/18). As shown in Table 1 , 30% (3/10) of the FLT3 TKD–carrying patients and 44% (8/18) of the FLT3 ITD–carrying patients also showed WT1 gene overexpression.
Table 1. Correlation of WT1 overexpression with clinical data, FAB subtypes, and molecular abnormalities in CN-AML patients.
| Variant | Total (N = 103) | WT1op (N = 29, 28%) | P |
| Median age, years (range) | 42 (17∼82) | 44 (29∼74) | .809 |
| Age in groups, years | .549 | ||
| ≤60 | 91 | 27 (30) | |
| >60 | 12 | 2 (17) | |
| Sex | .238 | ||
| Male | 58 | 19 (33) | |
| Female | 45 | 10 (22) | |
| WBC count, 109/L | .906 | ||
| 20 or below | 40 | 11 (28) | |
| Above 20 | 63 | 18 (29) | |
| FAB subtype | |||
| M0 | 4 | 2 (50) | .672 |
| M1 | 16 | 3 (19) | .543 |
| M2 | 41 | 13 (32) | .515 |
| M4 | 16 | 4 (25) | .998 |
| M5 | 26 | 7 (27) | .872 |
| Molecular abnormalities | |||
| NPM1mt | 30 | 7 (23) | .485 |
| FLT3TKD | 10 | 3 (30) | 1 |
| FLT3ITD | 18 | 8 (44) | .091 |
| Risk molecular subgroups * | |||
| Favorable | |||
| NPM1mt/no FLT3ITD | 23 | 4 (17) | .193 |
| Unfavorable | |||
| FLT3ITD | 18 | 8 (44) | .091 |
| Intermediate | |||
| Others excluding NPM1mt/no FLT3ITD and FLT3ITD | 62 | 17 (27) | .838 |
WT1op, WT1 overexpression; FAB, French-American-British; CN-AML, cytogenetically normal acute myeloid leukemia; WBC, white blood cell.
*stratification based on molecular abnormalities [2].
Treatment
All patients received one or two courses of induction chemotherapy with DA (daunorubicin 45 mg/m2×3 days; cytarabine 100–200 mg/m2 every 12 hours×7 days) or HA (harringtonine 4–6 mg/m2×7 days; cytarabine 100–200 mg/m2 every 12 hours ×7 days). If patients get complete remission, they would receive consolidation chemotherapy with high-dose cytarabine (3 g/m2 every 12 hours on days 1, 3, 5 and 7 for a total of 24 g/m2). Otherwise they would continue to receive other induction chemotherapies. Whether patient will receive hematopoietic stem cell transplantation (HSCT) will depend on comprehensive clinic situation.
DNA and RNA Extraction
Genomic DNA was extracted by using the Genomic DNA Extraction Kit (Tiangen, Beijing, China). Total RNA was isolated using the Trizol reagent (Invitrogen, Carlsbad, USA). All protocols were conducted according to manufacturer’s instructions. The quality and concentration of DNA and RNA were analyzed with a Biophotometer (Eppendorf AG, Hamburg, Germany).
Quantification of WT1 Gene Expression
Real-time reverse-transcriptase polymerase chain reactions (RT-PCR) using patient-derived RNA were carried out on the 7300 real-time PCR system (Applied Biosystems, Foster City, CA, USA). Commercially available WT1 mRNA quantification kit (Yuanqi, Shanghai, China) was used to detect WT1 gene expression. A housekeeping gene ABL was used as an internal control for calibration of possible variations caused by the variable efficiencies of RNA extraction, RT-PCR and operation. The relative levels of the WT1 expression to the ABL control of the clinical samples were calculated by simultaneous reaction with series standards of known concentrations (3×103–6 copies/ml). To calculate copy number of a specific sample, we first established a standard curve by using a series of known commercial standards from 3×103 to 3×106 copies/mL. The linear dynamic range of this method was from 3×102 copies/mL to 3×107 copies/mL. If a samples copy number was above the top limit, the sample will be re-measured with dilutions.
The data were analyzed using the Sequence Detection Software Version 1.2 (Applied Biosystems). For analysis of samples, detectable WT1 copy numbers were expressed as copies per 104 ABL copies according to manufacturer’s instruction. The cutoff for normal WT1 expression was defined as 250 copies/104 ABL as previously used in BM [10].
Detection of NPM1 and FLT3 Gene Mutations
For detection of the NPM1 and FLT3 mutations, we carried out PCR gene amplification by using the 9700 PCR amplification system (Applied Biosystems). Exon 12 of the NPM1 gene and exon 14, 15 and 20 of the FLT3 gene were amplified using respective primer pairs, NPM1 ex12-F (5′-TTAACTCTCTGGTGGTAGAATGAA-3′), NPM1 ex12-R (5′- TGTT ACAGAAATGAAATAAGACGG-3′), FLT3 ex14-F (5′-TTCCCTTT CATCCAAGAC-3′), FLT3 ex14-R (5′-AAACATTTGGCACATTCC-3′), FLT3 ex15-F (5′-GCAATTTAGGTATGAAAGCCAGC-3′), FLT3 ex15-R (5′-CTTTCAGCATTTTG ACGGCAACC-3′), FLT3 ex20-F (5′-CCAGGAACGTGCTTGTCA-3′) and FLT3 ex20-R (5′-TCAAAAA TGCACCACAGTGAG-3′). PCR reaction was performed in a total volume of 25 μL containing 100 ng of DNA, 10 μM of each primer and 12.5 μL 2×PCR buffer (containing MgCl2, dNTP mix and Taq polymerase) (Tiangen). PCR reactions were carried out as follows: denaturation at 95°C for 5 minutes, annealing at 55°C for 1 min. and extension at 72°C for 1 min. for 40 cycles. Amplification products were detected by 1% agarose gel electrophoresis. If the PCR product size was correct, amplification products were subsequently confirmed by nucleotide sequencing using the ABI PRISM 3100 genetic analyzer (Applied Biosystems) after purification. The sequencing results were then compared with the reference and wild type sequences of (NPM1: GenBank, NG-016018.1) or FLT3 (GenBank, NG-007066.1). Gene mutations were confirmed by nucleotide sequencing with coverage of double-strand DNA by using the forward and reverse primers.
Definitions and Statistical Analysis
Complete remission was characterized by morphologically normal marrow with <5% blasts, neutrophil count >1×109/L, platelet count >100×109/L and normal physical for more than 1 month. The CR rate was evaluated after received one or two courses of induction chemotherapy. Relapse was defined as the reappearance of blasts in the blood or the finding of more than 5% blasts in the bone marrow or any other evidence of leukemia recurrence. The DFS in patients who achieved CR was estimated from the date of CR to relapse or death. The OS was defined as the time from diagnosis to death by any causes [11].
For descriptive statistics, we calculated median, range and percentage of the cases. Proportion was compared using the Chi-Square test. Survival probability was estimated using the Kaplan–Meier curves and the difference between groups was analyzed by the log-rank test. Multivariate analysis was performed applying the COX regression model. For all statistical analyses, the p value that was 2-tailed with less than 0.05 was considered to be statistically significant. Statistical analyses were performed by using the statistical software package SPSS Version 19.0 (SPSS Science).
Results
Correlation of WT1 Overexpression with Clinical Parameters
A total of 103 CN-AML patients, consisting of 58 males and 45 females with a median age of 42 years (range, 17–82 years) were recruited to this study. The WT1 overexpression was defined as ≥250 copies/104 ABL. Objective of this part of the study was to examine the possible correlation of the WT1 gene overexpression with various common clinical features or molecular abnormalities.
Specifically, the WT1 gene overexpression was detected in 29 of 103 patients (28%) with a median value of 720 copies/104 ABL (range: ND to 8.2×106). Comparisons of WT1 gene overexpression with various clinical parameters and their possible statistical significance are summarized in Table 1 . Initial comparisons of the WT1 gene overexpression with common clinical parameters such as age, sex, WBC counts and the FAB subtypes showed no significant differences between the patient groups with or without WT1 gene overexpression. In order to examine the possible role of WT1 gene overexpression in assessing treatment response, prognosis and survival of AML patients with normal cytogenetics, All CN-AML patients were divided into three different risk subgroups based on their known molecular abnormalities such as the FLT3 and NPM1 gene mutation statuses. As recommended previously [2], [3], the favorable outcome subgroup (n = 23) are those CN-AML patients who have the NPM1 gene mutation (NPM1 mt) but do not have the internal tandem duplication in the FLT3 gene (FLT3 ITD). The unfavorable outcome subgroup (n = 18) are those patients with the FLT3ITD gene mutations. The remaining patients (n = 62), who do not have the NPM1 mt/no FLT3 ITD nor the FLT3 ITD genotypes were defined as the intermediate risk subgroup [2], [3]. No statistical significant differences were found in all of these risk groups. Together, these data suggest that the WT1 overexpression is not associated with any particular risk group or a clinical parameter.
Role of WT1 Overexpression in Response to Induction Chemotherapy
To examine the potential role of WT1 overexpression in predicting treatment outcome of the CN-AML patients, we first evaluated the impact of the WT1 overexpression on patient’s response to induction chemotherapy. Complete remission (CR) rates were used and calculated for this evaluation. As shown in Figure 1A , the CR rate was significantly influenced by the level of WT1 gene expression [p = .003]. While about 76% (56/74) of the CN-AML patients in the normal WT1 gene control group had complete remission after treatment, close to half of those patients 45% (13/29) with WT1 overexpression had complete remission.
Figure 1. Complete remission (CR) rate analysis based on the WT1 expression status and other molecular abnormalities in the CN-AML patients.
(A) Comparison of the CR rates between the CN-AML patients with normal (WT1 ctr) or high (WT1 op) WT1 gene expression. (B) Comparison of the CR rates among three different risk subgroups that were stratified based on molecular abnormalities, i.e., the favorable risk group included CN-AML patients that are carrying the NPM1 mt/no FLT3 ITD genotypes; the unfavorable risk group with the FLT3 ITD genotypes; and the intermediate group are those patients other than the two other risk groups, i.e., lack of the NPM1 mt/no FLT3 ITD and FLT3 ITD genotypes [8]. (C) Possible role of WT1 overexpression in determining the CR rates among the three risk subgroups. Abbreviations: WT1 ctr, normal WT1 expression; WT1 op, WT1 overexpression.
Since the WT1 overexpression clearly showed its impact on patient’s response to induction chemotherapy, we next tested the possible role of WT1 overexpression in measuring CR rates among the 3 different risk groups as defined by the FLT3 and NPM1 mutation status. Consistent with the previous classification [2], [3], the CR rates in our patient cohort were positively correlated with the three defined risk molecular subgroups ( Figure 1B ), i.e., the CR rates were found to be 91% (21/23), 69% (43/62) and 28% (5/18) in the favorable (NPM1 mt/no FLT3 ITD), intermediate and unfavorable (FLT3 ITD) groups, respectively.
The WT1 gene expression status was then added to the data analyses and compared for their CR rates among the 3 different risk groups. As shown in Figure 1C , it is evident that WT1 gene expression status had no effect on CR rate in the intermediate risk group. However, statistically significant differences were revealed between patient groups with or without high WT1 gene expression in the favorable (NPM1 mt/no FLT3 ITD) and the unfavorable (FLT3 ITD) risk groups. For example, in the favorable (NPM1 mt/no FLT3 ITD) patient group, complete remission was seen in all 19 patients when the WT1 gene was expressed at the normal level. However, only half of those patients (2 out of 4) showed complete remission when WT1 gene was overexpressed (p = .024). Remarkably, in the unfavorable (FLT3 ITD) risk group, none of the patient with WT1 overexpression showed complete remission whereas about half of this group of patient (5 out of 10) with normal WT1 expression had complete remission (p = .036).
Altogether, the CR rate analyses suggested that WT1 overexpression alone has no clear role in predicting CR in the intermediate risk group. It however could potentially play a functional role in predicting CR of the CN-AML patients either in the favorable (NPM1 mt/no FLT3 ITD) or the unfavorable (FLT3 ITD) groups. Specifically, when WT1 overexpression is added to the predefined risk groups, it may increase the risk or abridge the favorable outcome of the CN-AML patients by interactions with the favorable group or with the unfavorable group, respectively.
Role of WT1 Overexpression in Predicting Disease-free and Overall Survivals
Since WT1 overexpression appeared to play a functional role in predicting complete remission of the CN-AML patients especially when it is combined with the favorable NPM1 mt/no FLT3 ITD or the unfavorable FLT3 ITD genotypes, we next examined the potential impact of WT1 overexpression on the disease-free survival (DFS) and the overall survival (OS) of the studied CN-AML patient cohort. The CN-AML patients were followed in a time period of 1–38 months with a median value of 18 months for DFS or 13 months for OS, respectively. The Kaplan-Meier overall survival analysis was used to calculate the DFS and OS. The potential difference between each testing groups was analyzed by the log-rank test. The final results are summarized in Figure 2 .
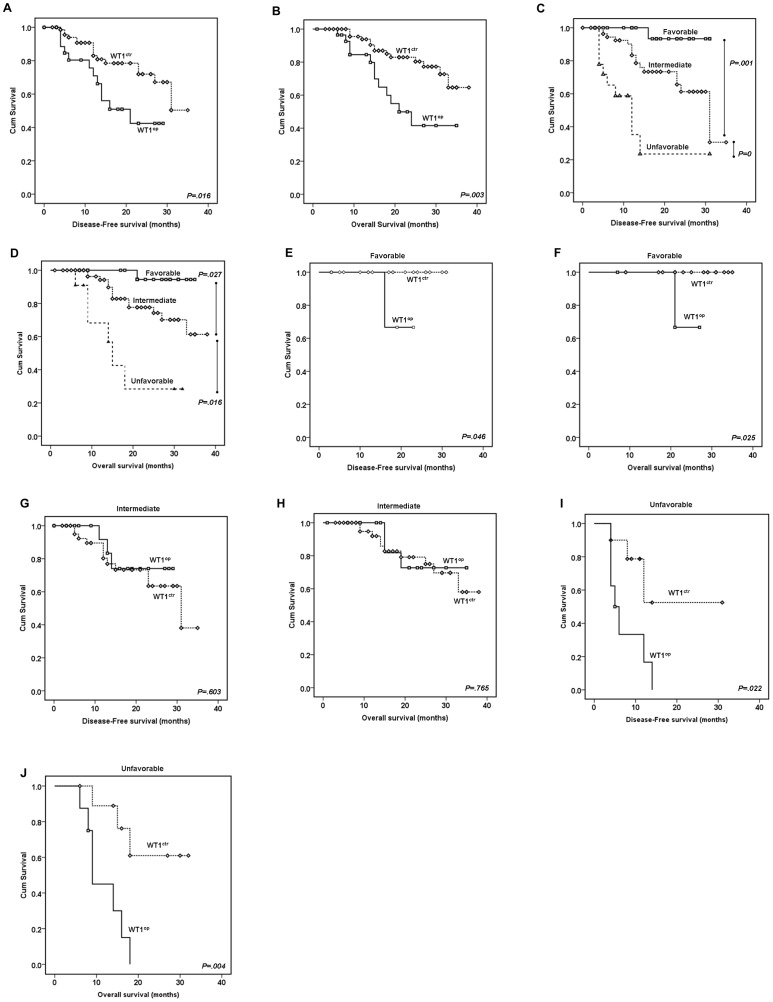
Figure 2. Determination of the diease-free survival (DFS) and overall survival (OS) in the CN-AML patients based on the WT1 expression status and other molecular abnormalities.
(A–B) Comparison of the DFS (A) and OS (B) between the CN-AML patients with normal (WT1 ctr) or high (WT1 op) WT1 gene expression. The mean DFS and OS of patients with WT1 op (n = 29) or WT1 ctr (n = 74) were 18.9±2.1 vs. 27.8±1.4 mo (p = .016); and 23.6±2.3 vs. 32.5±1.3 mo (p = .003), respectively. (C–D) Comparison of the DFS (C) and OS (D) among three different risk subgroups that were stratified based on molecular abnormalities, i.e., the favorable risk group included CN-AML patients that are carrying the NPM1 mt/no FLT3 ITD genotypes; the unfavorable risk group with the FLT3 ITD genotypes; and the intermediate group are those patients other than the two other groups, i.e., lack of the NPM1 mt/no FLT3 ITD and FLT3 ITD genotypes [8]. The average DFS of the patients with favorable (n = 23), intermediate (n = 62) or unfavorable genotype (n = 18) were 30.0±1.0, 26.6±1.6 and 13.9±3.0 mo, respectively. The average OS of the patients with favorable (n = 23), intermediate (n = 62) or unfavorable genotype (n = 18) were 34.2±0.85, 31.4±1.5 and 19.0±2.5 mo, respectively. (E–J) Possible role of WT1 overexpression in determining the DFS (E, G and I) and OS (F, H, and J) among the favorable (E–F; NPM1 mt/no FLT3 ITD), the intermediate (G–H) and the unfavorable (I–J; FLT3 ITD) molecular and risk subgroups. Note that patients with WT1 op in the favorable (n = 4) or unfavorable (n = 8) had inferior DFS and OS than their control WT1 ctr groups (favorable, n = 19; unfavorable, n = 10). No significant differences of DFS and OS were observed between normal and high WT1 gene expression in the intermediate group. Abbreviations: WT1 ctr, normal WT1 expression; WT1 op, WT1 overexpression.
Comparison of the DFS and OS between the WT1 overexpression group with the WT1 control group showed that WT1 overexpression has significantly reduced patient’s DFS ( Figure 2A ; log rank = 5.847, p = .016) and the OS ( Figure 2B ; log rank = 8.616, p = .003). As comparison references for the effect of WT1 overexpression, the impact of other single gene mutational effect such as the FLT3 ITD, NPM1 mt or FLT3 TKD mutations on DFS and OS was also evaluated. Consistent with the prior report [2], the FLT3 ITD genotype played a strong role in predicting DFS and OS (log rank = 20.641, p = 0 for DFS and log rank = 19.157, p = 0 for OS). In contrast, little or no significant influence was detected in patients with the NPM1 mt genotype (log rank = 2.146, p = .143 for DFS and log rank = 2.325, p = .127 for OS); or with the FLT3 TKD genotype (log rank = 0.712, p = .399 for DFS and log rank = 0.176, p = .675 for OS).
Disease-free survival and the overall survival were also calculated against the three different risk subgroups. Consistent with previous classifications, significant differences in DFS and OS were indeed found among the three risk subgroups. Specifically, patients in the favorable risk group (NPM1 mt/no FLT3 ITD) showed excellent DFS and OS; while the high risk or the unfavorable risk (FLT3 ITD) group displayed very poor outcomes of the DFS and OS ( Figure 2C and 2D ) with the intermediate groups lied in between.
When combining the WT1 overexpression with the three different groups, a very similar contributing patterns of the WT1 overexpression, as we saw in calculation of the CR rates, were observed with regard to its interaction with the three different risk subgroups. Specifically, the WT1 overexpression did not seem to affect the DFS and OS in the intermediate subgroup ( Figure 2G and 2H ; log rank = 0.270, p = .603 for DFS, log rank = 0.089, p = .765 for OS). The average survivals of this group of patients with normal WT1 (WT1 ctr; n = 45) or high WT1 (WT1 op; n = 17) expression were 25.4±1.9 and 24.8±2.1 mo (p = .603) for DFS, and 30.6±1.8 and 29.9±2.5 mo (p = .765) for OS, respectively. However, the WT1 overexpression significantly reduced the DFS and OS in the favorable risk (NPM1 mt/no FLT3 ITD) patient group ( Figure 2E and 2F ; log rank = 4, p = .046 for DFS, log rank = 5; p = .025 for OS). The average survivals of this group of patients with normal WT1 (WT1 ctr; n = 19) lived well beyond the study period, however, 7 out the 8 (88%) patients in the high WT1 (WT1 op; n = 8) group had DFS of less than 16.0 mo (p = .046) or OS of about 21.0 mo (p = .025), respectively. The other patient was only followed up to 10 months; thus no specific DFS or OS could be assigned at the completion of this study. Most significantly, the WT1 overexpression appeared to abruptly shorten patent’s survival of the unfavorable risk group patients in both categories ( Figure 2I and 2J ; log rank = 5.246; p = .022 for DFS, log rank = 8.481; p = .004 for OS).
Based on these results, we suggest that WT1 overexpression by itself could play an important but negative role in predicting DFS and OS of the CN-AML patients; moreover, the WT1 overexpression, when combined with the NPM1 mt or the FLT3 ITD genotypes, will serve as a poor prognostic marker in reducing DFS and OS in the favorable risk (NPM1 mt/no FLT3 ITD) patient group or worsening the patient outcomes in the unfavorable risk (FLT3 ITD) group.
Multivariate Analysis of the WT1 Overexpression and its Role in Prognosis of CN-AML
Here we were interested in further testing whether WT1 overexpression is an independent prognostic factor in predicting DFS or OS the CN-AML patients. Multivariate analysis was carried out by using the cause-specific Cox regression model to analyze the relationship among age (age ≤60 years vs. >60 years), the WT1 overexpression, other single gene mutations and the three molecular risk subgroups. As shown in Table 2 that age, FLT3 ITD and NPM1 mt/no FLT3 ITD are all independent prognostic markers as previously reported [2], [3]. Significantly and indeed, the WT1 overexpression also appeared to be an independent prognostic marker for DFS and OS (p = .028).
Table 2. Multivariate analysis (Cox regression) for clinical and molecular variables of DFS and OS in CN-AML patients.
| Variant | DFS | OS | ||
| Hazard ratio (95% CI) | P | Hazard ratio (95% CI) | P | |
| Age# | 0.32 (0.12∼0.87) | .025 * | 0.37 (0.13∼1.07) | .037 * |
| WT1op | 2.17 (0.96∼4.92) | .034 * | 2.50 (1.10∼5.68) | .028 * |
| NPM1mt | 1.68 (0.41∼6.82) | .470 | 0.92 (0.25∼3.40) | 0.9 |
| FLT3TKD | 2.71 (0.89∼8.33) | .082 | 2.01 (0.55∼7.28) | .289 |
| FLT3ITD | 3.35 (1.26∼8.92) | .016 * | 3.91 (1.42∼10.72) | .008 * |
| Risk molecular subgroups | ||||
| Favorable | ||||
| NPM1mt/no FLT3ITD | 0.07 (0.01∼0.88) | .039 * | 0.16 (0.07∼1.01) | .041 * |
| Unfavorable | ||||
| FLT3ITD | 3.35 (1.26∼8.92) | .016 * | 3.91 (1.42∼10.72) | .008 * |
| Intermediate | ||||
| Others excluding NPM1mt/no FLT3ITD and FLT3ITD | 1.15 (0.25∼5.39) | .86 | 1.27 (0.34∼6.16) | .81 |
CN-AML, cytogenetically normal acute myeloid leukemia; DFS, disease-free survival; OS, overall survival; CI, confidence interval; WT1op, WT1 overexpression.
age ≤60 years vs. >60 years.
*P values <.05.
Discussion
Chromosomal aberrations have traditionally been used to assess treatment response, prognosis and survival of the AML patients. Subsequent studies showed, however, that about 85% of the CN-AML patients carried one or more molecular mutations [12]. Indeed, characterization of gene mutations such as FLT3 or NPM1 further helped to define the clinical outcomes of AML patients especially when these patients present with normal cytogenetics. Therefore, identification of new molecular biomarker and testing its association with the existing biomarkers are of particular importance for better characterization and risk stratification of CN-AML patients and thus for better patient care.
WT1 overexpression has been shown to play a role in hematologic malignancy [5]. However, molecular mechanism of WT1 overexpression in CN-AML remains to be elusive. There are a number of downstream effectors of the WT1 genes [13]. For example, the heparin-binding growth factor midkine (MK) gene is a prognostic biomarker for various cancers [14], [15]. The insulin-like growth factor I receptor (IGF-I-R) is another known downstream effector [16]. More importantly, many of those downstream effectors are indeed involved in cellular growth or survival. Early study by using antisense oligonucleotides has showed that WT1 is required not only for proliferation but also for inhibiting apoptosis in tumor cell cultures [17]. Therefore, WT1 overexpression could potentially be used as a tumor-specific target for cancer treatment. Intriguing, an early trial by using peptide vaccines against WT1 in leukemia patients did show promising results [18].
In this study, we examined the possible role of WT1 gene overexpression in CN-AML patient’s responses to induction chemotherapy and in predicting the treatment outcome such as the disease-free survival or overall survival. We further evaluated the possible contribution of WT1 overexpression to the identification of risk groups that are typically stratified by genotypes such as the NPM1 or FLT3 mutation status, which are known molecular markers associated with the survival and treatment outcomes of the AML patients.
From this study, we found that WT1 overexpression by itself conversely correlated with the CR rate, DFS and OS ( Figure 1 and 2 ). Furthermore, WT1 overexpression also contributes, as a negative prognostic marker, to the prognosis and therapeutic response of the CN-AML patients in the favorable (NPM1 mt/no FLT3 ITD) and the unfavorable (FLT3 ITD) molecular subgroups ( Figure 1 and 2 ). Specifically, based on the observations of this CN-AML patient cohort, patients with the WT1 overexpression/FLT3 ITD genotypes showed the worst CR rate, OS and DFS (WT1 op in Figure 2I and 2J ). Conversely, patients with normal WT1 expression and the NPM1 mt/no FLT3 ITD genotypes displayed the best outcome (WT1 ctr in Figure 2E and 2F ). Altogether, our data suggest that the WT1 gene overexpression is an independent and negative prognostic factor in predicting patient’s response to induction chemotherapy and treatment outcomes. In addition, when combined with the NPM1 mt or the FLT3 ITD genotypes, the WT1 overexpression also contribute negatively to patient’s response to induction chemotherapy and treatment outcomes. Please note that any clear therapeutic effect of complete remission described here is almost certainly contributed by multifactorial factors such as age, patient’s physical status, peripheral white blood counts among other molecular factors, e.g., NPM1 mt, FLT3 ITD, etc. What we have showed here suggesting WT1 gene overexpression is at least one of the molecular factors that could play a negative prognostic role in predicting complete remission.
WT1 plays an important role in pathogenesis of AML, but its specific function remains elusive or controversial [5]. In some of the earlier studies, the WT1 overexpression at diagnosis was shown to be as an adverse predictor for CR rate, DFS and OS in AML patient. In contrast, other studies suggested that WT1 overexpression was not associated with disease outcome [19], [20]. Moreover, previous CN-AML studies on the WT1 overexpression was mainly used as a molecular marker to detect minimal residual disease. Little was known about the prognostic significance of the WT1 overexpression in CN-AML. Intriguingly, Frederik Damm and co-workers did suggest earlier that WT1 overexpression could potentially be used as one of the several biomarkers to formulate some kind of integrative prognostic risk score for the stratification of CN-AML [21]. Indeed, our data showed here that the WT1 overexpression can not only be used as a negative prognostic marker for CN-AML but also, for the first time to the best of our knowledge, contributes to the identification of the CN-AML risk subgroups that are normally stratified by the NPM1 and FLT3 mutation status.
It was noticed that the percentage of the WT1 overexpression showed in our study cohort was somewhat lower than the previous reports (28% vs. 48%–73%) [7], [8]. One obvious difference between our study and the other reports was the use of the reference gene in determining the level of WT1 gene expression. For example, the housekeeping gene ABL was used in our study as an internal control for the calculation of the WT1 gene expression. The cutoff for normal WT1 expression was defined as 250 copies/104 ABL as previously recommended [9]. In contrast, different cutoff value of the ABL gene or other housekeep gene such as GAPDH gene was used in the other studies [7], [20]. It is also possible that other molecular aberrations, e.g., MiR-15a/16-1, which is as a tumor suppressor that down-regulates WT1 expression in the process of leukemia cell proliferation [22], could be another contributing factor to the observed differences in the WT1 overexpression. In spite of the observed differences in the percentages of the WT1 gene expression, this difference should not affect the fact that WT1, when it is overexpressed, contributes negatively to the pathogenesis of CN-AML.
Combining with all of our data, our results strongly suggest that WT1 overexpression is an independent and negative prognostic biomarker that could potentially be used to evaluate response to induction chemotherapy and prognosis of AML patients with normal cytogenetics. In addition, the use of WT1 overexpression as an additional biomarker seems to enhance the statistical power in the identification of risk subgroups that are normally stratified solely based on the NPM1 or the FLT3 mutational status. Therefore, adding profiling of WT1 gene expression level in the future decision-making of patient’s response to induction chemotherapy or prognosis of the CN-AML patients could potentially provide better or more effective care of this group of patients.
Acknowledgments
The authors would like to thank Ge Li for helpful discussions and revision.
Funding Statement
This work was supported by the Social Public Service Foundation of Henan Technology Department [072103810503] to Xiaodong Lyu who was a visiting physician scientist in the RZ’s laboratory at the University of Maryland School of Medicine when this manuscript was written. The authors would like to thank Ge Li for helpful discussions and revision. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mrozek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD (2007) Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood 109: 431–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lyu XD, Zhao RY, Chen Q (2013) Personalized Approach to Diagnosis and Treatment of Acute Myeloid Leukemia. J Clin Exp Patho 3: 143–145. [Google Scholar]
- 3. Dohner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, et al. (2010) Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 115: 453–474. [DOI] [PubMed] [Google Scholar]
- 4. Yang L, Han Y, Suarez Saiz F, Minden MD (2007) A tumor suppressor and oncogene: the WT1 story. Leukemia 21: 868–876. [DOI] [PubMed] [Google Scholar]
- 5. Bergmann L, Maurer U, Weidmann E (1997) Wilms tumor gene expression in acute myeloid leukemias. Leuk Lymphoma 25: 435–443. [DOI] [PubMed] [Google Scholar]
- 6. Garg M, Moore H, Tobal K, Liu Yin JA (2003) Prognostic significance of quantitative analysis of WT1 gene transcripts by competitive reverse transcription polymerase chain reaction in acute leukaemia. Br J Haematol 123: 49–59. [DOI] [PubMed] [Google Scholar]
- 7. Miglino M, Colombo N, Pica G, Grasso R, Clavio M, et al. (2011) WT1 overexpression at diagnosis may predict favorable outcome in patients with de novo non-M3 acute myeloid leukemia. Leuk Lymphoma 52: 1961–1969. [DOI] [PubMed] [Google Scholar]
- 8. Barragán E, Cervera J, Bolufer P, Ballester S, Martín G, et al. (2004) Prognostic implications of Wilms’ tumor gene (WT1) expression in patients with de novo acute myeloid leukemia. Haematologica 89: 926–933. [PubMed] [Google Scholar]
- 9. Cilloni D, Renneville A, Hermitte F, Hills RK, Daly S, et al. (2009) Real-Time Quantitative Polymerase Chain Reaction Detection of Minimal Residual Disease by Standardized WT1 Assay to Enhance Risk Stratification in Acute Myeloid Leukemia: A European LeukemiaNet Study. J Clin Oncol 27: 5195–5201. [DOI] [PubMed] [Google Scholar]
- 10. Rau R, Brown P (2009) Nucleophosmin (NPM1) mutations in adult and childhood acute myeloid leukaemia: towards definition of a new leukaemia entity. Hematological Oncology 27: 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheson BD, Bennett JM, Kopecky KJ, Büchner T, Willman CL, et al. (2003) Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol 21: 4642–4649. [DOI] [PubMed] [Google Scholar]
- 12. Marcucci G, Haferlach T, Dohner H (2011) Molecular genetics of adult acute myeloid leukemia: Prognostic and therapeutic implications. J Clin Oncol 29: 475–486. [DOI] [PubMed] [Google Scholar]
- 13. Adachi Y, Matsubara S, Pedraza C, Ozawa M, Tsutsui J, et al. (1996) Midkine as a novel target gene for the Wilms’ tumor suppressor gene (WT1). Oncogene 13: 2197–2203. [PubMed] [Google Scholar]
- 14. Jono H, Ando Y (2010) Midkine: a novel prognostic biomarker for cancer. Cancers (Basel) 2: 624–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma Z, Li H, Wang B, Shen Q, Cui E, et al. (2013) Midkine mRNA level in peripheral blood mononuclear cells is a novel biomarker for primary non-small cell lung cancer: a prospective study. J Cancer Res Clin Oncol 139: 557–562. [DOI] [PubMed] [Google Scholar]
- 16. Werner H, Roberts CT Jr, Rauscher FJ 3rd, LeRoith D (1996) Regulation of insulin-like growth factor I receptor gene expression by the Wilms’ tumor suppressor WT1. J Mol Neurosci 7: 111–123. [DOI] [PubMed] [Google Scholar]
- 17. Tuna M, Chavez-Reyes A, Tari AM (2005) HER2/neu increases the expression of Wilms’ tumor 1 (WT1) protein to stimulate S-phase proliferation and inhibit apoptosis in breast cancer cells. Oncogene 24: 1648–1652. [DOI] [PubMed] [Google Scholar]
- 18. Oka Y, Tsuboi A, Taguchi T, Osaki T, Kyo T, et al. (2004) Induction of WT1 (Wilms’ tumor gene)-specific cytotoxic T lymphocytes by WT1 peptide vaccine and the resultant cancer regression. Proc Natl Acad Sci USA 101: 13885–13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spassov BV, Stoimenov AS, Balatzenko GN, Genova ML, Peichev DB, et al. (2011) Wilms’ tumor protein and FLT3-internal tandem duplication expression in patients with de novo acute myeloid leukemia. Hematology 16: 37–42. [DOI] [PubMed] [Google Scholar]
- 20. Miyawaki S, Hatsumi N, Tamaki T, Naoe T, Ozawa K, et al. (2010) Prognostic potential of detection of WT1 mRNA level in peripheral blood in adult acute myeloid leukemia. Leuk Lymphoma 51: 1855–1861. [DOI] [PubMed] [Google Scholar]
- 21. Damm F, Heuser M, Morgan M, Wagner K, Görlich K, et al. (2011) Integrative prognostic risk score in acute myeloid leukemia with normal karyotype. Blood 117: 4561–4568. [DOI] [PubMed] [Google Scholar]
- 22. Gao SM, Xing CY, Chen CQ, Lin SS, Dong PH, et al. (2011) MiR-15a and miR-16-1 inhibit the proliferation of leukemic cells by down-regulating WT1 protein level. J Exp Clin Cancer Res 30: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]